Abstract
In this study, the tet(W) gene region of a human clinical isolate of Clostridium difficile resistant to tetracycline was characterized. This gene was a new allele showing 99% sequence identity to the gene found in the human strain Bifidobacterium longum F8, and it is not transferable by “in vitro” mating experiments.
The majority of the tetracycline resistance genes coding for ribosomal protection proteins reside on mobile or conjugative elements, and this is one of the reasons for their wide distribution among bacterial genera (8, 12). tet(M) is the most widespread gene class, and it is usually found on conjugative elements of the Tn916 family (7), whereas tet(W) has the second-largest host range and has been detected in both gram-positive and gram-negative bacteria, especially in those isolated from environmental samples (2, 3, 9-11). tet(W) is associated with conjugative or nonconjugative elements which may vary among different bacteria (4, 6, 16, 17). In Clostridium difficile, tetracycline resistance is usually mediated by a tet(M) gene (1, 14). Although, it has been demonstrated that C. difficile is able to exchange antibiotic resistance genes with other bacteria of the gastrointestinal tract and that tetracycline resistance genes can be transferred between intestinal bacteria (1, 11, 13), tet(W) has not been detected in C. difficile, so far. This paper is the first report of the presence of a tet(W) gene in a human clinical strain of C. difficile isolated from a child with C. difficile-associated disease.
A sample of 35 toxigenic C. difficile strains, isolated from symptomatic patients, was selected from our national collection as representative of the tetracycline-resistant strains isolated from 1987 to 2005. All of these strains were intermediate or resistant to tetracycline (MICs of ≥8 μg/ml), according to the Clinical and Laboratory Standards Institute (5), when analyzed by Etest (AB Biodisk, Solna, Sweden) following the manufacturer's instructions. The presence of a tet(M) gene was assessed by the primer couple TETMd and TETMr (12), whereas the amplification of a 457-bp internal fragment of tet(W) was performed using the primer couple WRC1 and WRC2, designed on the conserved region of tet(W) genes (Table 1). All C. difficile isolates were positive for tet(M), but one strain, C. difficile CD5, showing a MIC of 8 μg/ml, was also positive for tet(W).
TABLE 1.
Primers used in this study
| Primer | Sequence (5′→3′) | Location in C. difficile CD5 tet(W) regiona |
|---|---|---|
| Detection and sequencing of tet(W) gene | ||
| WRC1 | CATCTCTGTGATTTTCAGCTTTTCTCTCCC | 1871-1897 |
| WRC2 | AGTCTGTTCGGGATAAGCTCTCCGCCG | 2328-2299 |
| W9 | ATGAAAATAATCAATATTGGAATTCT | 1447-1472 |
| W11 | TTACATTACCTTCTGAAACATATG | 3366-3343 |
| Sequencing of tet(W) flanking regions | ||
| TSP1 | GAGAGCTTATCCCGAACAGA | 1892-1873 |
| TSP2 | CATCTGTGCCACTGGAAGGA | 1649-1630 |
| TSP3 | GCTCTCCGTCAAGGTCGTCT | 1512-1493 |
| TER1 | GTGTTGTCTGCATTTTACT | 979-961 |
| AUN1 | GGTGAGCCGGATTGGG | NR |
| TSP4 | TGGAACGTAACGGACTGTAA | 2920-2939 |
| TSP5 | CTCTATGCGCCCCAGGAATA | 3076-3095 |
| TSP6 | GCGGAGCGTATGCCTTACAG | 3243-3262 |
| TSP6A | TTAAGGGCTTGTCCTCTCTGCC | 3642-3663 |
| Wrev6 | GCTCTGCGTCTATGCGTCT | 4046-4064 |
| Wrev4 | AGATGTTCCTCGCGCAATTTT | 4497-4517 |
| Wrev2 | ATGAAACGTTTACCTAAATATAC | 5059-5037 |
| UNI3A | TCACAGAAGTATGCCAAGCGA | NR |
NR, the positions of AUN1 and UNI3A are not reported since these primers were used to sequence the 5′ and the 3′ ends of the C. difficile CD5 tet(W) region, respectively.
To characterize the tet(W) region, the genomic DNA of this strain was extracted using the NucleoBond buffer set III and the NucleoBond AXG 20 (Macherey-Nagel, Düren, Germany), following the manufacturer's instructions, except that bacterial lysis was carried out for 45 min at 37°C, using double the recommended quantity of lysozyme and proteinase K.
C. difficile CD5 DNA was then digested with HindIII and analyzed by specific hybridization assays using the tet(W) and tet(M) PCR products as probes. The PCR products were purified with the NucleoSpin extract kit (Macherey-Nagel, Düren, Germany) and labeled with the ECL enhanced chemiluminescence direct nucleic acid labeling and detection system (Amersham Biosciences, Buckinghamshire, United Kingdom). Hybridization analysis showed that strain CD5 had one copy of both tet(M) and tet(W) genes. In fact, one band of about 5.0 kb was observed with the tet(M) probe and one of about 2.5 kb with the tet(W) probe (data not shown).
The tet(W) gene was completely amplified using primers W9 and W11, designed on the 5′- and 3′-end conserved sequences, respectively, of many tet(W) genes already known (Table 1). The PCR fragment of about 1.9 kb was cloned into Escherichia coli using the Qiagen PCR cloning kit (Qiagen, Milden, Germany). Nucleotide and amino acid comparisons were accomplished using the European Bioinformatics Institute ClustalW server (http://www.ebi.ac.uk/Tools/clustalw/index.html), and the output was used for the construction of the phylogenetic tree by TreeView 1.4. Sequence analysis revealed that strain CD5 tet(W) was a new allele, showing a range of identity with the other tet(W) genes comprising between 85 and 99%. In particular, the sequence of C. difficile CD5 Tet(W) showed an identity of 99% to Tet(W) of Bifidobacterium longum F8. The phylogenetic tree obtained by nucleotide sequence comparison indicated that this gene belonged to the same cluster of two Bifidobacterium strains isolated from humans (Fig. 1). Since B. longum is abundant in the gastrointestinal tract of humans, a genetic transfer of tet(W) between these two anaerobic bacteria “in vivo” may be possible, even if further evidence and studies will be necessary to confirm this hypothesis.
FIG. 1.
Unrooted neighbor-joining phylogenetic tree obtained from the nucleotide multiple alignment of the C. difficile tet(W) gene identified in this study and other 13 tet(W) gene sequences available in GenBank. Among the species included are Megasphaera elsdenii, Selenomonas ruminantium, Arcanobacterium pyogenes, and Mitsuokella multiacida. The branch lengths are scaled in proportion to the extent of the change per position, as indicated by the scale bar. GenBank accession numbers are in parentheses.
The entire sequence of strain CD5 tet(M) was also obtained, and the sequence was 100% identical to that of the Enterococcus faecalis DS16 tet(M) gene, as already observed in other C. difficile strains (15).
The analysis of tet(W) and tet(M) transcription was performed with the Qiagen OneStep reverse transcription-PCR kit, using the same primers designed for tet(W) and tet(M) detection. C. difficile CD5 RNA was extracted using the NucleoSpin RNA II (Macherey-Nagel, Düren, Germany) from colonies grown for 24 h either in the presence or in the absence of tetracycline (10 μg/ml) and treated with recombinant DNase (DNA-free kit; Ambion, Austin, TX). The results confirmed that C. difficile CD5 tet genes were actively transcribed in both the presence and absence of tetracycline (data not shown).
Filter matings between C. difficile CD5 and the recipient strains C. difficile CD37R and Butyrivibrio fibrisolvens 2221R were performed as already described (13). B. fibrisolvens 2221R is a rumen strain which, in the past, proved to be a good recipient for a C. difficile erythromycin resistance determinant, erm(B) (13). The transfer of tet(W) and the simultaneous transfer of both tet(W) and tet(M) were unsuccessful. On the contrary, the tet(M) gene was transferred to the recipient strain C. difficile CD37R with average frequencies of 6.3 × 10−5 per donor and 3.1 × 10−4 per recipient. All transconjugants analyzed showed a MIC for tetracycline of 8 μg/ml. The conjugative element carrying the tet(M) gene was characterized as an element with a genetic organization similar to that of Tn916 from E. faecalis DS16 (data not shown), as already described in other C. difficile strains (14). Any attempt to transfer tet(M) from C. difficile CD5 to B. fibrisolvens 2221R was unsuccessful.
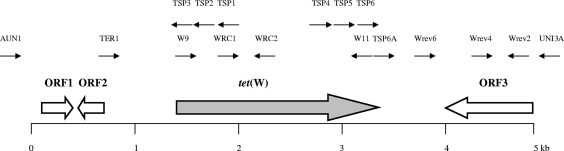
Genome walking of the regions flanking tet(W) was carried out using the DNA walking SpeedUp premix kit (Seegene, Seoul, South Korea), and the primers used for sequencing are reported in Table 1. Sequence analysis was performed using BLAST software (http://www.ncbi.nlm.nih.gov/BLAST/) and ORF Finder (http://www.ncbi.nlm.nih.gov/gorf/gorf.html). The C. difficile CD5 tet(W) region of 5,059 bp is shown in Fig. 2. The sequences surrounding tet(W) showed three potential open reading frames (ORFs). The hypothetical protein of 115 amino acids encoded by ORF1, located upstream of tet(W), produced a significant alignment with the putative conserved domain DUF955. A conserved H-E-X-X-H motif of this domain is suggestive of a catalytic active site and shows similarity to that of the metallopeptidase family M48 and the cluster of orthologous groups COG2856, described as zinc metallopeptidases. This C. difficile CD5 protein showed high identity (99%) to that found in an uncultured bacterium from a human stool sample (GenBank protein accession no. CAM12483.1; from amino acids 63 to 177). ORF2 is oppositely oriented compared to both tet(W) and ORF1 and codes for a hypothetical protein of 50 amino acids that produced a significant alignment with the putative conserved domain DUF24, characteristic of transcriptional regulators related to the pfam01047 family, as observed for similar bacterial proteins identified during a sequence-based survey of members of the normal human gut microbiota (http://genome.wustl.edu/sub_genome_group.cgi?GROUP=3&SUB_GROUP=4) currently in progress. In particular, the first 47 amino acids of C. difficile CD5 ORF2 protein showed identities of 82% to that of Coprococcus eutactus ATCC 27759 (GenBank protein accession no. EDP 25685; from amino acids 54 to 100), 80% to that of Clostridium leptum DSM 753 (GenBank protein accession no. ED059633; from amino acids 63 to 107), 72% to that of B. longum DJO10A (GenBank protein accession no. ZP_00120895; from amino acids 60 to 106), and 70% to that of B. longum NCC2705 (GenBank protein accession no. NP_695657; from amino acids 70 to 116), respectively.
FIG. 2.
Genetic organization of the C. difficile CD5 tet(W) DNA region. The approximate extent and organization of the region are not necessarily to scale. ORFs and their orientation are represented by arrows: the tet(W) gene is designated by a gray arrow, whereas white arrows represent the three ORFs found in the DNA regions surrounding tet(W). Individual primers and their orientations are indicated by small black arrows.
The region downstream of tet(W), from nucleotides 3550 to 5059, was 100% identical to the chromosomal region from nucleotides 5008 to 3499 of C. difficile QCD-32g58 (GenBank nucleotide accession no. NZ_AAML04000015). This strain was recently isolated from a patient with severe C. difficile-associated disease in Quebec, and the sequencing of its genome is currently in progress. In this region, strain CD5 showed an ORF (ORF3; located from nucleotides 5059 to 4076) that codes for a hypothetical protein identical to that encoded by the locus CdifQ_04003905 of C. difficile QCD-32g58 (GenBank protein accession no. ZP_01801544). This protein shows a putative conserved domain that characterizes the radical SAM, a protein superfamily catalyzing diverse reactions, including unusual methylations, isomerization, sulfur insertion, ring formation, anaerobic oxidation, and protein radical formation.
This paper documents for the first time the presence of tet(W) in C. difficile and, due to the high identity of C. difficile tet(W) with those of other human gastrointestinal bacteria, supports the hypothesis of horizontal gene transfer events in the rapid diffusion of this tetracycline resistance determinant.
Nucleotide sequence accession number.
The nucleotide sequence described in this paper has been deposited in the EMBL database (http://www.ebi.ac.uk/embl/) under accession no. AM749838.
Acknowledgments
This work was partially supported by the FIRB Project “Costruzione di un laboratorio nazionale per lo studio delle resistenze batteriche agli antibiotici” of the Italian Ministry of Research and University.
We are indebted to Karen Scott (Rowett Research Institute, Bucksburn, Aberdeen, United Kingdom) for supplying B. fibrisolvens strain. We are also grateful to Tonino Sofia for editing the manuscript.
Footnotes
Published ahead of print on 10 December 2007.
REFERENCES
- 1.Adams, V., D. Lyras, K. A. Farrow, and J. I. Rood. 2002. The clostridial mobilisable transposons. Cell. Mol. Life Sci. 12:2033-2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aires, J., F. Doucet-Populaire, and M. J. Butel. 2007. Tetracycline resistance mediated by tet(W), tet(M), and tet(O) genes of Bifidobacterium isolates from humans. Appl. Environ. Microbiol. 73:2751-2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Billington, S. J., J. G. Songer, and B. H. Jost. 2002. Widespread distribution of a Tet W determinant among tetracycline-resistant isolates of the animal pathogen Arcanobacterium pyogenes. Antimicrob. Agents Chemother. 46:1281-1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Billington, S. J., and B. H. Jost. 2006. Multiple genetic elements carry the tetracycline resistance gene tet(W) in the animal pathogen Arcanobacterium pyogenes. Antimicrob. Agents Chemother. 50:3580-3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clinical and Laboratory Standards Institute. 2007. Methods for antimicrobial susceptibility testing of anaerobic bacteria: approved standard, 7th ed. CLSI document M11-A7. Clinical and Laboratory Standards Institute, Wayne, PA.
- 6.Kazimierczak, K. A., H. J. Flint, and K. P. Scott. 2006. Comparative analysis of sequences flanking tet(W) resistance genes in multiple species of gut bacteria. Antimicrob. Agents Chemother. 50:2632-2639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rice, L. B. 1998. Tn916 family conjugative transposons and dissemination of antimicrobial resistance determinants. Antimicrob. Agents Chemother. 42:1871-1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Roberts, M. C. 1996. Tetracycline resistance determinants: mechanisms of actions, regulation of expression, genetic mobility, and distribution. FEMS Microbiol. Rev. 19:1-24. [DOI] [PubMed] [Google Scholar]
- 9.Scott, K. P., T. M. Barbosa, K. J. Forbes, and H. J. Flint. 1997. High-frequency transfer of a naturally occurring chromosomal tetracycline resistance element in the ruminal anaerobe Butyrivibrio fibrisolvens. Appl. Environ. Microbiol. 63:3405-3411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Scott, K. P., C. M. Melville, T. M. Barbosa, and H. J. Flint. 2000. Occurrence of the new tetracycline resistance gene tet(W) in bacteria from the human gut. Antimicrob. Agents Chemother. 44:775-777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Scott, K. P. 2002. The role of conjugative transposons in spreading antibiotic resistance between bacteria that inhabit the gastrointestinal tract. Cell Mol. Life Sci. 59:2071-2082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Speer, B. S., N. B. Shoemaker, and A. A. Salyers. 1992. Bacterial resistance to tetracycline: mechanisms, transfer, and clinical significance. Clin. Microbiol. Rev. 5:387-399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Spigaglia, P., F. Barbanti, and P. Mastrantonio. 2005. Horizontal transfer of erythromycin resistance from Clostridium difficile to Butyrivibrio fibrisolvens. Antimicrob. Agents. Chemother. 49:5142-5145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Spigaglia, P., V. Carucci, F. Barbanti, and P. Mastrantonio. 2005. ErmB determinants and Tn916-like elements from clinical isolates of Clostridium difficile. Antimicrob. Agents Chemother. 49:2550-2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Spigaglia, P., F. Barbanti, and P. Mastrantonio. 2006. New variants of the tet(M) gene in Clostridium difficile clinical isolates harbouring Tn916-like elements. J. Antimicrob. Chemother. 57:1205-1209. [DOI] [PubMed] [Google Scholar]
- 16.Villedieu, A., M. L. Diaz-Torres, N. Hunt, R. McNab, D. A. Spratt, M. Wilson, and P. Mullany. 2003. Prevalence of tetracycline resistance genes in oral bacteria. Antimicrob. Agents Chemother. 47:878-882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Villedieu, A., A. P. Roberts, E. Allan, H. Hussain, R. McNab, D. A. Spratt, M. Wilson, and P. Mullany. 2007. Determination of the genetic support for tet(W) in oral bacteria. Antimicrob. Agents Chemother. 51:2195-2197. [DOI] [PMC free article] [PubMed] [Google Scholar]