Abstract
Peptides that mediate dimerization of attached zinc finger DNA-binding domains have been evolved in vitro starting from random sequences. We first used phage display to select dimerization elements from libraries of random 15-residue polypeptides that were fused to the N terminus of the zinc finger domains. We then reoptimized these peptides by sequentially randomizing five-residue blocks (proceeding across the peptide in three steps) and selecting variant peptides that further stabilized the protein–DNA complex. Biochemical experiments confirmed that the selected peptides promote dimerization of the zinc fingers on an appropriate DNA target site. These results demonstrate that dimerization units can be obtained readily from random polypeptide libraries of moderate complexity. Our success reemphasizes the utility of searching random peptide libraries in protein design projects, and the sequences presented here may be useful when designing novel transcription factors.
The affinity and specificity of DNA-binding proteins depend not only on interactions with the DNA but also on interactions with proteins that bind at neighboring sites. Such protein–protein interactions may involve homo- or heterodimerization or the assembly of multiprotein complexes. Dimerization strategies already have been tested, in structure-based design efforts, to create DNA-binding proteins with enhanced affinity and specificity. In the first such study (1), computer modeling was used to design a fusion between zinc finger subdomains from Zif268 and the dimerization element from Gal4. Recent design efforts with zinc fingers also have used leucine zipper dimerization motifs in an analogous manner (S. Wolfe, E. Ramm, and C.O.P., unpublished data).
The selection of dimerization elements from libraries of random peptides represents an intriguing alternative to structure-based design and raises many interesting questions. How common are functional dimerization units? Do the selected structures always resemble known motifs? Can we obtain new dimerization units that would be useful when designing transcription factors for potential applications in gene therapy?
Phage display (reviewed in refs. 2 and 3) provides a powerful method for selecting functional peptides from large populations of random polypeptides. Peptide libraries displayed on phage often have been screened for peptide–protein interactions in studies that focus on epitope mapping, analysis of substrate specificity, and the development of leads for drug design. Peptides that can substitute for larger protein domains also have been generated, either through stepwise minimization and reoptimization of a naturally occurring domain or by selection from random sequence libraries (reviewed in ref. 4). In one study, a peptide selected to bind the erythropoietin receptor (5) was found to induce dimerization of the receptor–peptide complex (6), demonstrating that self-associating peptides can—at least under some circumstances—be isolated from random polypeptide sequences.
In this study, we used phage display to select and optimize peptides that mediate dimerization of DNA-binding modules. Our work may have practical implications in the design of DNA-binding proteins and, more generally, demonstrates how random peptide extensions provide a basis for selecting proteins with desired functions. These results also may have implications regarding the role of protein–protein interactions in the evolution of transcription factors.
MATERIALS AND METHODS
Phage Display Libraries.
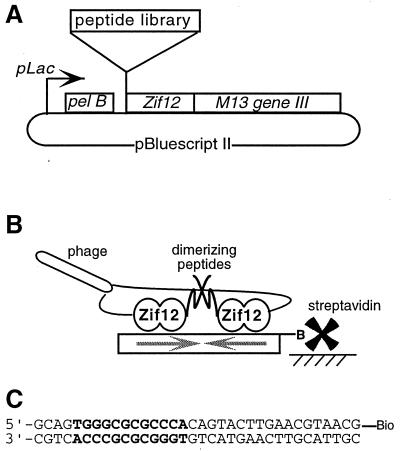
Phagemid vectors used in the selections were created from pZif12 (7) by restoring the reading frame between the Zif12-coding region and gene III and by introducing convenient restriction sites at the start of Zif12. Libraries containing randomized peptides were constructed by cassette mutagenesis, using NN(G/C/T) randomized codons for the initial libraries and NN(G/T) for the reoptimization libraries. The complete fusion protein used for phage display (Fig. 1A) contained a PelB signal sequence; a short leader peptide (NH2-EPRAQNS in initial selections and NH2-EP in reoptimizations); the random peptide; residues 4–60 of Zif268 (numbering as in ref. 8); a linker that includes an amber codon; and residues 23–424 of M13 gene III product. The ligated phagemid libraries were electroporated into XL-1 Blue E. coli cells, yielding ≈108 transformants for the initial selection libraries and ≈109 transformants for each of the reoptimization libraries.
Figure 1.
(A) Sketch showing key segments of the phagemid. (B) Expected arrangement of fusion proteins at the target DNA. Phage displaying two copies of a dimerizing peptide–Zif12 fusion can form stable complexes with the biotinylated target DNA site, which contains an inverted repeat of the Zif12-binding site. The phage–DNA complexes are captured by streptavidin coupled to a solid support, and phage that bind less tightly are washed away. (C) The DNA site used for affinity selection of phage, with the two juxtaposed Zif12-binding sites in bold.
Phage Selections.
For the initial selections, phage were grown, harvested, and processed essentially as described previously for zinc finger phagemid selections (7). Selections during the block-reoptimization steps were conducted similarly, but with the following set of changes. Binding reactions included 2 mM DTT to minimize the risk of selecting disulfide-bonded dimer interfaces. Phage–DNA complexes were captured by streptavidin-coated paramagnetic beads (Dynal, Great Neck, NY) that had been equilibrated in pZif12 wash buffer (7). Five microliters of a 10-mg/ml suspension of beads was used to capture up to 10 pmol of DNA site (with bound phage). The beads were washed (five times using 0.5 ml for each 8-min wash) and treated with high-salt elution buffer (7). To increase stringency, (i) binding, capture, and elution were performed at 37°C in the second and third reoptimization steps, and (ii) in the third step, the target DNA site contained a mutation in one of the Zif12-binding sites (the half-site distal to the biotin was TGAGCG). The target DNA concentration also was lowered through the course of the reoptimization to help force competition among members of the phage pool and to further increase stringency. In the first block-reoptimization step, the target DNA concentration was reduced from 40 nM (cycles 1–3) to 8 nM (cycles 4–6) and then to 2 nM (cycles 7–9), with the salmon sperm competitor DNA concentration lowered proportionately at each stage. In the second step, concentrations were 20 nM (cycle 1) or 2 nM. In the third step, the mutant DNA was present at 1 nM throughout. In the later stages of each block-reoptimization step, we estimate that the phage concentration was 10- to 20-fold higher than the target DNA.
Protein Production and Purification.
DNA fragments encoding peptide-Zif12 fusions (with Met-Ala at their N terminus) were cloned into pET-21d (Novagen) and expressed in BL21 (DE3) or BL21(DE3) pLysS cells. Cultures were induced, lysed, and sonicated as recommended (Novagen). Peptides were present in insoluble inclusion bodies and were purified by reversed-phase batch extraction (using Waters Sep-pak C18 cartridges) and reversed-phase HPLC as described (8). The Zif12 peptide (amino acids 2–59 of Zif268) and peptide 2 were expressed as glutathione S-transferase (GST) fusion proteins from pGEX-2T and pGEX-6P-3 (Pharmacia), respectively. These peptides were purified by affinity chromatography and cleaved from the GST as directed (Pharmacia), leaving a Gly-Ser dipeptide at the N terminus of Zif12 and a heptapeptide (GPLGSDP) at the N terminus of peptide 2. The cleaved peptides were purified further by reversed-phase HPLC. All peptides were reconstituted from lyophilized HPLC fractions and refolded as described (9), and their concentrations were quantified by comparison with BSA standards in SDS/PAGE using Coomassie staining. For peptides that gave a stable gel shift (peptides 1*, 3, and 5*), the active concentration of peptide was determined as described (9), and we found that each of these samples was fully active for DNA binding.
DNA-Binding Assays.
Gel mobility-shift assays (10) and DNase I footprinting experiments (11) were used to assess the DNA-binding activity of various peptides. Only peptides 1*, 3, and 5* produced complexes that were sufficiently stable for quantitative gel-shift assays, and footprinting was used to measure the affinity of the other peptides.
Labeled DNA probes were generated as follows. For the gel-shift studies shown in Fig. 2B, oligos corresponding to the phage-selection target site (5′-GGTTGCAGTGGGCGCGCCCACAGTACTTGAACGTAACG-3′ and 5′-CGTTACGTTCAAGTACTGTGGGCGCGCCCACTGC-3′, Zif12 sites in bold) or a single-site mutant (bold regions above replaced with the sequences 5′-TGGGCGTATGCT-3′ and 5′-AGCATACGCCCA-3′) were annealed and end-labeled with Klenow. A labeled restriction fragment was used for quantitative studies. The oligos 5′-GGAATTCCTGATCAAGATCTGGTCACGTCCATAGGCTAGGCATGTCAAGGCTGTATG-3′ and 5′-GGGATCCACTCGCGAACGCGTCCTTGTAGTGGGCGCGCCCACATACAGCCTTGACAT-3′ (Zif12 sites in bold) were annealed, extended by mutually primed extension, and cloned into the EcoRI and BamHI sites of pBluescript II SK(+). The probe was prepared by digesting the plasmid with EcoRI and NotI; labeling the DNA with Klenow, [α-32P]dCTP, and [α-32P]dGTP; and purifying the small fragment by native PAGE.
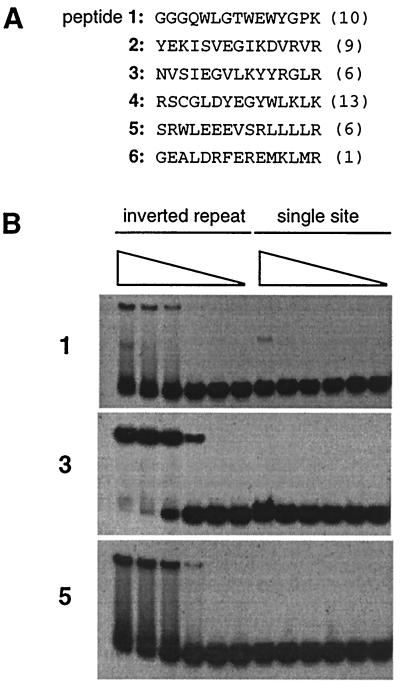
Figure 2.
(A) Sequences of peptide extensions isolated from the initial selection. Numbers in parentheses give the frequency of occurrences among the 45 clones sequenced. The clones for peptide 2 included a Glu-21-to-Asp mutation in the zinc finger region that may have been partially responsible for the affinity of this peptide. (B) Gel mobility-shift assays using purified fusion peptides 1, 3, and 5. Protein (2.5 μM, 250 nM, 25 nM, 2.5 nM, 250 pM, and no protein) was incubated with DNA containing either an inverted repeat of Zif12 sites or a single Zif12 site and then electrophoresed through native polyacrylamide gels. The reduced mobility of the inverted repeat probe in the presence of protein indicates the formation of protein–DNA complexes. Similar results were obtained with fusion peptides 2 and 6, but data are not shown because these peptides were not studied further. Binding of peptide 4, also not shown, appeared to depend on disulfide bond formation, and this peptide was not pursued further.
Binding reactions (typically 10 μl) contained the labeled DNA site (at >100-fold below the protein concentration at half-maximal binding), protein (for quantitative assays, we used 1.3- to 2.0-fold dilution steps over a range of four orders of magnitude), and a buffer containing 15 mM Hepes, pH 7.8, 60 mM potassium acetate, 60 mM potassium glutamate, 5 mM MgCl2, 20 μM ZnSO4, 5% glycerol, 0.1% Nonidet P-40, 1 mM DTT, and 0.1 mg/ml acetylated BSA. After equilibrating the binding reactions at 4°C (1.5–16 hr, depending on the peptide), the reactions either were resolved by native PAGE (7.5% 37.5:1 acrylamide/bisacrylamide with 2.5% glycerol, run at 4°C in electrophoresis buffer containing 25 mM Tris, 190 mM glycine, and 1 mM EDTA) or treated with DNase I. DNase I reactions (4 min, 4°C using 2.5 μl of 30 μg/ml DNase I for 10-μl reaction) were terminated, prepared, and electrophoresed as described (12). Data were collected by using a PhosphorImager (Molecular Dynamics).
To determine dissociation constants (Kd) for the fusion peptides, binding data for the selected peptides were fit by nonlinear regression to the equation θ = 1/(1 + Kd/[P]2), where θ is the fraction of DNA bound and [P] is the concentration of free protein, which approximately equals the total protein concentration in our experiments. This equation describes the binding of two protein molecules to a DNA molecule with strict cooperativity. Data for Zif12, which bound essentially noncooperatively, were fit to the equation θ = 1/(1 + Kd′/[P]), where Kd′ is the dissociation constant for a Zif12 monomer; the corresponding Kd for the overall reaction of two Zif12 monomer with the DNA was calculated as (Kd′)2.
Sedimentation Equilibrium.
Peptide samples at three concentrations, ranging from ≈10 μM to ≈100 μM, were centrifuged to equilibrium in a Beckman Optima XL-A at 20,000 and 30,000 rpm at 4°C in a buffer containing 15 mM Tris, pH 7.8, 150 mM KCl, 5 mM MgCl2, 20 μM ZnSO4, and 0.2 mM DTT. Solvent density and partial specific volumes of peptides were calculated as described (13). The sedimentation data were analyzed by methods described in refs. 14 and 15, using the program nonlin (16).
RESULTS
To select dimerization motifs, we attached random peptides to a DNA-binding domain and selected those fusion proteins that could bind more stably to a symmetric DNA site (Fig. 1). Random 15- and 30-residue peptides were expressed at the amino terminus of the first two zinc fingers of Zif268 (8, 17) (we refer to this two-finger peptide as Zif12), and these peptide-Zif12 fusions were displayed on filamentous bacteriophage. Phage from the 15- and 30-mer libraries, representing 108 different sequences from each library, were pooled, and our affinity-selection protocol was used with a target DNA duplex containing an inverted repeat of the Zif12-binding site. The original Zif12 peptide, which lacks any N-terminal extension, binds specifically, but weakly, to the “half-site” sequence TGGGCG, and Zif12 phage are not retained by the target DNA. Therefore, our protocol enriches for phage that display peptides that augment the DNA-binding activity of the zinc fingers.
After seven cycles of selection and amplification, the phage pool bound more than 100-fold more efficiently than the initial random libraries, indicating successful enrichment of higher-affinity phage. We sequenced 45 clones from this final phage pool and found 6 different 15-mer peptide sequences (Fig. 2A). These peptides did not share any obvious homology, aside from a basic residue present at the final position of each sequence. blast searches (18) showed no significant similarity between the selected peptides and known natural proteins.
To assess their DNA-binding activity, peptides obtained in these initial selections were expressed in Escherichia coli (as fusions with Zif12), purified, and tested in gel mobility-shift assays (Fig. 2B). These fusion peptides bound specifically to duplex DNA containing an inverted repeat of the Zif12 sites, showing little or no activity with DNA containing a single site. The complexes with the inverted-repeat site all migrated similarly in the gel, with a mobility for the complexes that was consistent with the formation of dimers. The isolated Zif12 zinc finger domains did not shift either DNA site under these conditions.
Quantitative DNA-binding assays were used to investigate further the affinity and cooperativity for binding of several of these fusion proteins (Table 1). When the DNA contained an inverted repeat of the Zif12-binding site, fusion peptides 1, 3, and 5 bound substantially tighter than did Zif12 alone. Scatchard analysis demonstrated that binding of these peptides to the inverted repeat is second order with respect to protein, as expected for a species that exists as a monomer in solution, but that binds the DNA site as a dimer. (This analysis also showed that Zif12 binds the inverted repeat with slight cooperativity, but the data for Zif12 were more consistent with a first-order than a second-order reaction.)
Table 1.
Affinities for DNA duplexes containing the inverted repeat site 5′-TGGGCGCGCCCA-3′
| Peptide | Half-maximal binding, nM | Kd, M2 |
|---|---|---|
| Zif12 | 9,600 | 9.2 (±3.3) × 10−11 |
| 1 | 410 | 1.7 (±0.44) × 10−13 |
| 3 | 37 | 1.4 (±0.04) × 10−15 |
| 5 | 440 | 1.9 (±0.13) × 10−13 |
| 1* | 15 | 2.3 (±0.35) × 10−16 |
| 5* | 12 | 1.4 (±0.27) × 10−16 |
Half-maximal binding is given in concentration of monomer. Kd represents that for binding of a dimer. Data shown are mean (±SD) for three independent experiments.
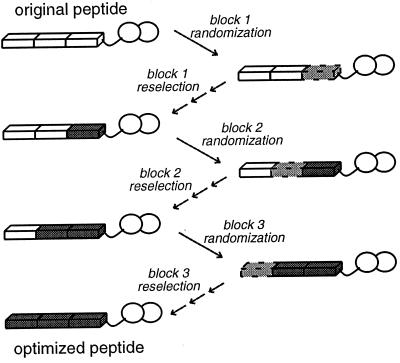
Because our initial search could test only a tiny fraction of all possible 15-mer sequences, we expected that we could use our initial peptides as a starting point for the “evolution” of even more efficient dimerization motifs. We developed a sequential reoptimization strategy (Fig. 3) to try to improve the dimerization properties of fusion peptides obtained in our initial selections. Our strategy conceptually divided each 15-mer peptide into three five-residue blocks. In the first reoptimization step, we completely randomized the block closest to the fingers (with the other residues held constant) and selected for sequences with even higher affinity for the symmetric DNA site. The second and third blocks were randomized and reselected in subsequent reoptimization steps. During this procedure, we took several measures to increase the stringency of the selection conditions. The concentration of DNA target was lowered 40-fold over the course of the reoptimization steps, and the temperature of the binding reaction was raised from 23 to 37°C. In the third reoptimization step, a mutation was introduced into one of the Zif12 sites to weaken the protein–DNA interface (and thus create greater selective pressure for effective dimerization). In the later cycles of each reoptimization step, phage were present in excess of the DNA target, forcing direct competition among the remaining phage for the limited number of binding sites and, thus, favoring selection of the tightest-binding sequences from each pool.
Figure 3.
Overall scheme for sequential reoptimization of peptides. A 15-residue peptide obtained from the initial selection was divided conceptually into three blocks of 5 aa each and reoptimized in three steps. In the first step, the five-residue block closest to the fingers was completely randomized, with the other 10 aa held constant. Phage display of the new fusion proteins, with six to nine selection and amplification cycles, was used to obtain the best sequences from this pool. In the second reoptimization step, the central five-residue block was completely randomized, with the finger-proximal region held constant as the newly optimized sequence and the finger-distal region corresponding to the initially selected sequence. The best sequences from this pool were obtained again via phage display with a series of selection and amplification cycles. In the final reoptimization step, the finger-distal five-residue block was completely randomized and then reselected in the context of the two other reoptimized blocks.
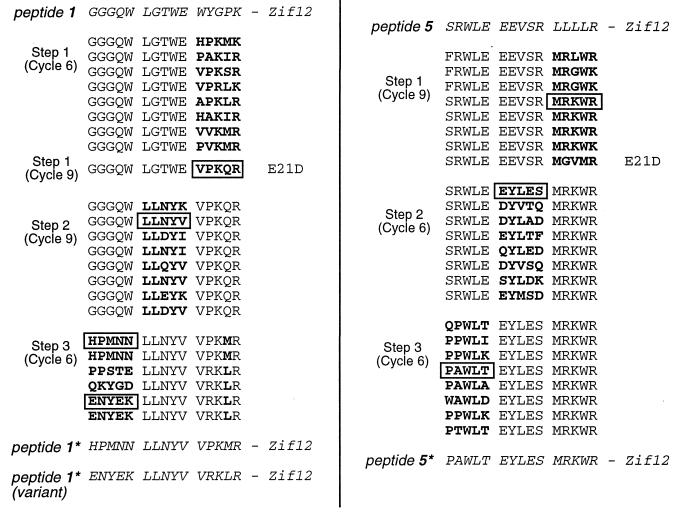
Progress of the sequential reoptimization protocol was monitored by sequencing phage pools at a number of stages. The full reoptimization strategy was applied to fusion peptides 1 and 5 from the initial selections (Fig. 4). Peptide 3, which also was reoptimized, yielded variants that appeared to form higher-order oligomers and therefore was not studied in detail (data not shown). We typically used six to nine selection–amplification cycles for the reoptimization of any particular five-residue block. One of the final sequences—resembling the consensus for the set—was then chosen for use in the next step. In some cases, choosing a consensus sequence was complicated somewhat by the presence of spurious mutations (Fig. 4) in nonrandomized portions of the peptide. Because such mutations probably conferred some other selective advantage (independent of the peptide block targeted for reoptimization), clones carrying them were not used in assigning a consensus. The final selected peptides—with all three five-residue blocks reoptimized—were designated as 1* and 5* to indicate that they had been obtained by sequentially reoptimizing peptides 1 and 5. A variant form of peptide 1* also was chosen for further analysis (Fig. 4).
Figure 4.
Evolution of peptides 1 and 5 by sequential block reoptimization. The sequences selected from each reoptimization step are shown in bold, with the number of selection and amplification cycles given in parentheses. Sequences roughly matching the consensus that were used in later steps have been boxed. In some cases, such as in reoptimization step 3 for peptide 1 and reoptimization step 1 for peptide 5, we isolated clones that carried spurious mutations at a nondegenerate position of the peptide extension. In addition, the E21D mutation in the zinc finger region (which also was seen in the original peptide 2 sequence) arose several times; this mutation may stabilize complex formation by improving contacts at the protein–DNA interface. [Note: some confusion was caused by this E21D mutation, which occurred in the first reoptimization step for peptide 1, but was discovered only after reoptimization step 2. Thus, the “consensus” sequence from reoptimization step 1 (VPKQR), chosen after selection–amplification cycle 9, had a glutamine that did not occur in sequences isolated after cycle 6. To double-check this position of peptide 1, it was randomized again during reoptimization step 3. The corresponding position was allowed to vary as Q, M, I, or L, along with the complete randomization of the third block. The reselections showed that methionine or leucine is preferred at this position.]
The reoptimized fusion peptides were expressed and purified, and their DNA-binding properties were assessed with quantitative gel-shift assays (Table 1). For fusion peptides 1* and 5*, half-maximal DNA binding was observed with peptide concentrations in the nanomolar range, demonstrating that the reoptimization process produced peptides with significantly higher affinity. The variant form of peptide 1* also bound very tightly, but appeared to form higher-order complexes with the DNA, and this peptide was not studied further.
Sedimentation equilibrium experiments were conducted with several fusion peptides to determine their oligomeric state in solution. Peptide 1*, peptide 5, and the isolated Zif12 domain were monomeric at all concentrations tested (up to ≈100 μM). Peptide 5* was monomeric at concentrations up to 50 μM, above which the peptide appeared to form higher-order aggregates (apparently tetramers). These results confirmed that our selected peptides exist as monomers in solution at the concentrations used in the DNA-binding assays.
DISCUSSION
Protein–protein interactions can play important roles in protein–DNA recognition by facilitating cooperative binding. In this project, we sought to “evolve” stable zinc finger dimerization elements starting from libraries of random polypeptides. Broadly speaking, our goals in this study were 3-fold: (i) to gain some impression about the frequency of functional dimerization elements in a pool of random polypeptides; (ii) to explore the utility of a sequential block-reoptimization strategy to improve the activity of selected peptides; and (iii) to generate dimerization elements for zinc finger proteins that may be useful in future efforts to create designer DNA-binding proteins for applications in gene therapy.
Given the length of the random peptides tested in this study, phage display allows one to search only a tiny fraction of the relevant sequence space. We screened about 108 sequences from each library, but there are 1019 possible 15-mers and 1039 possible 30-mers. The success of the initial screen, which yielded several different peptides that mediate dimerization, suggests that such peptides are relatively “common” in sequence space. Zhang et al. (19) have isolated dimerization elements by fusing random fragments of the yeast genome to the DNA-binding domain of lambda repressor and selecting fusion proteins that reconstitute repressor activity. This group reached similar conclusions regarding the frequency of functional dimerization domains. Our findings may help explain why dimerization elements are so common and have such diverse sequences in natural DNA-binding proteins. The peptides that we have isolated may be analogous—in an evolutionary and functional sense—to the peptide extensions that are responsible for heterodimerization of certain homeodomain proteins (20–22).
It is interesting that we obtained only 15-mer extensions in our initial selection, although the starting library consisted of equal numbers of fusion proteins with 15- and 30-residue N-terminal extensions. At this stage, the significance of this observation remains unclear. Our sample may be too small to determine the relative effectiveness of 15-mers and 30-mers as dimerization units, and it is possible that problems with processing and display on the phage surface become more severe with large random peptides. Considering the number of selection–amplification cycles used, even a modest difference in propagation efficiency between 15-mers and 30-mers could have resulted in a substantial bias in the final pool.
During the natural evolution of a protein, many sequence variants are tested for improved activity. We adopted a generally similar strategy, searching sequences related to the initial peptides, but we generated variants in a distinctive way. The peptides were reoptimized in three steps, with each step involving an exhaustive search of a five-residue sequence block. Envisioning that the zinc fingers would provide a relatively rigid structural framework, we began reoptimization with the five-residue block closest to the fingers and then proceeded outward. Because we completely randomize each block when it is reoptimized, our procedure systematically searches a large number of sequence variants that can differ dramatically from the initial peptide, and the final sequence may be altered (potentially) at every position. In this respect, our strategy encompasses a broader search than more traditional mutagenesis schemes, which often involve creating variants of the initial sequence with a limited number of changes (see ref. 2 for a theoretical discussion of different mutagenesis strategies). Given the number of residues that are randomized in a reoptimization step, it is even possible that the overall fold of a reoptimized peptide will be different from the fold of the original peptide.
Our “sequential block reoptimization” strategy was applied successfully to several fusion peptides and yielded variants with high DNA-binding affinity (Table 1). Assuming that the binding of the isolated Zif12 domain reflects the binding of the Zif12 moiety in the selected fusion peptides, the binding energy contributed by each peptide extension is represented by the free energy of binding for the fusion peptide minus that for Zif12 alone. The contribution of the peptide extension includes the energy of dimerization as well as any energy derived from contacts between the peptide extension and the DNA. For peptides 1* and 5*, this value is about 7.3 kcal/mol (i.e., 20.0 kcal/mol − 12.7 kcal/mol). This is more than twice that contributed by peptides 1 and 5 (≈3.5 kcal/mol), which had been obtained in the initial selections. The DNA-binding affinities of our optimized peptides (1* and 5*) are roughly comparable to those of ZFGD1, a rationally designed chimeric protein composed of Zif12 fused to a 60-residue linker and coiled-coil dimerization domain from Gal4 (1). It appears that, in some situations, such small peptides may be able to functionally replace larger protein domains.
In the course of our reoptimizations, we may have approached a practical limit of our selection system. Although our selections employed a “monovalent” display format (23), we assume that most of the phage that bound in a given cycle actually were bivalent. (Presumably, monovalent phage predominate in the sample, but the fraction of retained phage was always less than 1%, and it is certainly possible that these are predominantly bivalent phage.) Phage displaying two copies of the peptide–zinc finger fusion would bind more tightly to the dimeric site than their monovalent counterparts, because the peptides attached to the same phage would be tethered at a high “effective concentration.” The opportunity for bivalent binding presumably aided the initial selections but may complicate reoptimization. As higher-affinity dimers arise, peptides on bivalent phage may, aided by the high “effective concentration,” form dimers even in the absence of DNA, and this would eliminate any basis for selecting tighter dimers. In addition, as dimer interfaces become more stable, and as members of the phage pool become more similar (in the reoptimizations, all sequences in a pool share at least 10 residues), there also is an increased probability of two slightly different monovalent phage binding to a single DNA target molecule. Such phage heterodimers could seriously complicate the selection process. (Lowering the concentration of phage might minimize this possibility.) Finally, selection for high-affinity dimers may inadvertently isolate peptides with alternative oligomerization states. Several of our reoptimized sequences appeared to form higher-order complexes, and it is interesting in this context that design studies with self-associating amphipathic helices have shown that subtle sequence changes can dramatically alter the oligomeric state (24). Similar adventitious effects may have occurred with some of our selected peptides, or there may be binding modes that somehow permit two bivalent phage to occupy the same DNA site.
Investigating the structural details of the complexes presented here should yield basic insights into how dimerization can be achieved with short peptides. The different peptide “classes” uncovered in this study share no obvious sequence similarity with each other or with natural dimerization elements, suggesting that we have isolated several distinct motifs that may represent novel dimerization units. Furthermore, secondary structure predictions (25) for our sequences indicate very different structural propensities for the different peptides. The selected peptides might fold into different structures or pack in different ways at the dimer interface. Although we tend to envision dimerization in terms of peptide–peptide interactions, cooperative binding also would be obtained if the peptide extension from one fusion protein reached across the center of the DNA site and bound to the zinc finger domain of a symmetry-related molecule. In principle, a peptide also might induce dimer formation by promoting domain swapping (26) between substructures of two Zif12 monomers when they are bound to adjacent sites on the DNA. Finally, we note that improved DNA-binding affinity could result from additional peptide–DNA contacts (the Lys or Arg residues that are preferred at the position immediately preceding the fingers may play some such role), but these contacts would not be expected to contribute to the observed cooperativity of binding.
The selection of dimerization elements for zinc fingers demonstrates that these elements are relatively common in sequence space and reemphasizes the utility of screening random polypeptide libraries when developing proteins with desired activities. We have shown that a sequential reoptimization strategy can generate peptides with significantly higher activity, and peptide sequences such as those described here may prove useful for other zinc finger and DNA-binding protein design studies. There may be practical advantages to using these selected peptides (as opposed to known dimerization motifs such as coiled coils), because it seems less likely that these peptides will “crossreact” by heterodimerizing with natural dimerization interfaces presented by proteins in the cell. Further characterization of these novel motifs should broaden our understanding of macromolecular recognition and protein evolution by providing interesting comparisons to natural polypeptide sequences involved in dimerization and cooperative binding.
Acknowledgments
We thank R. Sauer, P. Kim, S. Wolfe, J. Szostak, and D. Akey for helpful discussions; R. Sauer, N. Clarke, S. Wolfe, K. Joung, and J. Miller for comments on the manuscript; and R. Sauer and P. Kim for the use of equipment. B.S.W. is a Howard Hughes Medical Institute predoctoral fellow. This work was supported by the Howard Hughes Medical Institute.
References
- 1.Pomerantz J L, Wolfe S A, Pabo C O. Biochemistry. 1998;37:965–970. doi: 10.1021/bi972464o. [DOI] [PubMed] [Google Scholar]
- 2.Smith G P, Petrenko V A. Chem Rev. 1997;97:391–410. doi: 10.1021/cr960065d. [DOI] [PubMed] [Google Scholar]
- 3.Lowman H B. Annu Rev Biophys Biomol Struct. 1997;26:401–424. doi: 10.1146/annurev.biophys.26.1.401. [DOI] [PubMed] [Google Scholar]
- 4.Cunningham B C, Wells J A. Curr Opin Struct Biol. 1997;7:457–462. doi: 10.1016/s0959-440x(97)80107-8. [DOI] [PubMed] [Google Scholar]
- 5.Wrighton N C, Farrell F X, Chang R, Kashyap A K, Barbone F P, Mulcahy L S, Johnson D L, Barrett R W, Jolliffe L K, Dower W J. Science. 1996;273:458–463. doi: 10.1126/science.273.5274.458. [DOI] [PubMed] [Google Scholar]
- 6.Livnah O, Stura E A, Johnson D L, Middleton S A, Mulcahy L S, Wrighton N C, Dower W J, Jolliffe L K, Wilson I A. Science. 1996;273:464–471. doi: 10.1126/science.273.5274.464. [DOI] [PubMed] [Google Scholar]
- 7.Rebar E J, Greisman H A, Pabo C O. Methods Enzymol. 1996;267:129–149. doi: 10.1016/s0076-6879(96)67010-4. [DOI] [PubMed] [Google Scholar]
- 8.Pavletich N P, Pabo C O. Science. 1991;252:809–817. doi: 10.1126/science.2028256. [DOI] [PubMed] [Google Scholar]
- 9.Rebar E J, Pabo C O. Science. 1994;263:671–673. doi: 10.1126/science.8303274. [DOI] [PubMed] [Google Scholar]
- 10.Carey J. Methods Enzymol. 1991;208:103–117. doi: 10.1016/0076-6879(91)08010-f. [DOI] [PubMed] [Google Scholar]
- 11.Brenowitz M, Senear D F, Shea M A, Ackers G K. Methods Enzymol. 1986;130:132–181. doi: 10.1016/0076-6879(86)30011-9. [DOI] [PubMed] [Google Scholar]
- 12.Klemm J D, Pabo C O. Genes Dev. 1996;10:27–36. doi: 10.1101/gad.10.1.27. [DOI] [PubMed] [Google Scholar]
- 13.Laue T M, Shah B D, Ridgeway T M, Pelletier S L. In: Analytical Ultracentrifugation in Biochemistry and Polymer Science. Harding S E, Rowe A J, Horton J C, editors. Fullerton, CA: R. Soc. Chem.Cambridge, U.K.90125Laue, T. M., Shah, B. D., Ridgeway, T. M. & Pelletier, S. L. (1992) in Analytical Ultracentrifugation in Biochemistry and Polymer Science, eds. Harding, S. E., Rowe, A. J. & Horton, J. C. (R. Soc. Chem., Cambridge, U.K.), pp. 90–125.McRorieD. K.VoelkerP. J.1993Self-Associating Systems in the Analytical UltracentrifugeBeckman Instruments; 1992. [Google Scholar]
- 15.Laue T M. Methods Enzymol. 1995;259:427–452. doi: 10.1016/0076-6879(95)59055-2. [DOI] [PubMed] [Google Scholar]
- 16.Johnson M L, Correia J J, Yphantis D A, Halvorson H R. Biophys J. 1981;36:575–588. doi: 10.1016/S0006-3495(81)84753-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Christy B A, Lau L F, Nathans D. Proc Natl Acad Sci USA. 1988;85:7857–7861. doi: 10.1073/pnas.85.21.7857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 19.Zhang Z, Murphy A, Hu J C, Kodadek T. Curr Biol. 1999;9:417–420. doi: 10.1016/s0960-9822(99)80188-2. [DOI] [PubMed] [Google Scholar]
- 20.Li T, Stark M R, Johnson A D, Wolberger C. Science. 1995;270:262–269. doi: 10.1126/science.270.5234.262. [DOI] [PubMed] [Google Scholar]
- 21.Tan S, Richmond T J. Nature (London) 1998;391:660–666. doi: 10.1038/35563. [DOI] [PubMed] [Google Scholar]
- 22.Passner J M, Ryoo H D, Shen L, Mann R S, Aggarwal A K. Nature (London) 1999;397:714–719. doi: 10.1038/17833. [DOI] [PubMed] [Google Scholar]
- 23.Bass S, Greene R, Wells J A. Proteins Struct Funct Genet. 1990;8:309–314. doi: 10.1002/prot.340080405. [DOI] [PubMed] [Google Scholar]
- 24.Harbury P B, Zhang T, Kim P S, Alber T. Science. 1993;262:1401–1407. doi: 10.1126/science.8248779. [DOI] [PubMed] [Google Scholar]
- 25.Rost B, Sander C. J Mol Biol. 1993;232:584–599. doi: 10.1006/jmbi.1993.1413. [DOI] [PubMed] [Google Scholar]
- 26.Bennett M J, Schlunegger M P, Eisenberg D. Protein Sci. 1995;4:2455–2468. doi: 10.1002/pro.5560041202. [DOI] [PMC free article] [PubMed] [Google Scholar]