Abstract
Previous work has demonstrated the critical role for transcription repression in quiescent cells through the action of E2F-Rb or E2F-p130 complexes. Recent studies have shown that at least one mechanism for this repression involves the recruitment of histone deacetylase. Nevertheless, these studies also suggest that other events likely contribute to E2F/Rb-mediated repression. Using a yeast two-hybrid screen to identify proteins that specifically interact with the Rb-related p130 protein, we demonstrate that p130, as well as Rb, interacts with a protein known as CtIP. This interaction depends on the p130 pocket domain, which is important for repression activity, as well as an LXCXE sequence within CtIP, a motif previously shown to mediate interactions of viral proteins with Rb. CtIP interacts with CtBP, a protein named for its ability to interact with the C-terminal sequences of adenovirus E1A. Recent work has demonstrated that the Drosophila homologue of CtBP is a transcriptional corepressor for Hairy, Knirps, and Snail. We now show that both CtIP and CtBP can efficiently repress transcription when recruited to a promoter by the Gal4 DNA binding domain, thereby identifying them as corepressor proteins. Moreover, the full repression activity of CtIP requires a PLDLS domain that is also necessary for the interaction with CtBP. We propose that E2F-mediated repression involves at least two events, either the recruitment of a histone deacetylase or the recruitment of the CtIP/CtBP corepressor complex.
The control of the early events of cell proliferation through the action of the G1 cyclin-dependent kinases, leading to the phosphorylation of Rb and related proteins, and the subsequent accumulation of E2F transcription factor activity is now well established (for reviews, see refs. 1–6). It is also evident that most, if not all, human cancers arise as a result of the disruption of this pathway, either through the activation of positive acting components such as the G1 cyclins or the inactivation of negative-acting components such as p53, Rb, and the cyclin kinase inhibitors (6, 7).
E2F transcription activity is now recognized to be a complex array of DNA binding activities that function both as transcriptional activating proteins as well as transcription repressors (8, 9). The E2F4 and E2F5 proteins, which specifically associate with the Rb-related p130 protein in quiescent cells (10), function to repress transcription of various genes encoding proteins important for cell growth. In contrast, the E2F1, E2F2, and E2F3 proteins are tightly regulated by cell proliferation, accumulate as cells progress through mid- to late G1, and appear to function as positive regulators of transcription. The complexity of E2F transcription control is illustrated by the fact that the E2F1, E2F2, and E2F3 genes are repressed in quiescent cells through the action of E2F4 or E2F5 complexes containing Rb or p130. In addition to the E2F1, E2F2, and E2F3 genes, the targets for E2F-mediated repression include a very large number of genes that encode proteins that guide cell cycle progression and that participate directly in DNA replication (8, 9).
Initial studies of Dean and colleagues clearly demonstrated that the role of Rb in controlling E2F-dependent transcription was not merely an inhibition of positive activation of transcription but, rather, that E2F/Rb-mediated repression was a dominant event, capable of shutting off an otherwise active promoter (11, 12). Indeed, a series of recent reports has provided evidence that one mechanism for this Rb-mediated repression involves an ability of Rb to recruit histone deacetylase to E2F-site containing promoters, presumably resulting in an alteration of chromatin conformation that hinders transcription (13–15).
Nevertheless, despite the evidence implicating histone deacetylase recruitment as a mechanism for Rb-mediated repression, several observations suggest that additional events may contribute to the repression. For example, many genes subject to E2F/Rb-mediated repression are not derepressed by treatment with the histone deacetylase inhibitor trichostatin A (13). Moreover, although the recruitment of histone deacetylase is effective in repressing some promoters, others appear to be unaffected. Based on these observations, it would appear that a histone deacetylase-independent mechanism of transcriptional repression contributes to the Rb control of transcription.
To further explore the mechanistic basis for Rb-mediated repression, we have used a yeast two-hybrid screen to identify proteins that specifically interact with the p130 protein. In so doing, we have identified a protein known as CtIP that interacts with p130 dependent on the p130 pocket domain. CtIP has previously been described (16) as a protein that interacts with CtBP, an adenovirus E1A-interacting protein (17), and CtBP has recently been shown to function as a corepressor in Drosophila. We show here that the CtIP protein can itself repress transcription and that this repression is caused, at least in part, by its ability to recruit the CtBP corepressor. Rb/p130-mediated repression therefore functions not only through histone deacetylase activity but also through a CtIP/CtBP repressor complex.
MATERIALS AND METHODS
Cell Culture.
C33A cells were grown in DMEM containing 10% fetal bovine serum.
Plasmids and Reagents.
The SV40 promoter containing upstream Gal4 sites (pSVECG) was a kind gift from D. Dean (Washington University, St. Louis) (12). The MLP with Gal4 sites was a kind gift from D. Dean and R. Eisenman (Fred Hutchinson Cancer Research Center, Seattle), the Gal4-HDAC expression plasmid was a kind gift from D. Reinberg (Robert Wood Johnson Medical School, Piscataway, NJ) (18), and the RbΔp34 plasmid was a kind gift from R. Bremner (University of Toronto) (19). The Gal4Rb plasmids were created in several steps. The PvuII Fragment of Rb was first subcloned into the SmaI site of the pGBT9 vector. The BsaH-BglII fragment of Rb then was cloned into the ClaI and BglII sites of the pSP70 cloning vector. An EcoRI fragment was isolated from Rb-pSP70 (including the upstream EcoRI site from the polylinker) and was cloned into the Rb-pGBT9 vector, creating a full length Gal4Rb-pGBT9 plasmid used for expression in yeast. A HindIII fragment was isolated from the Gal4Rb-pGBT9 plasmid and was cloned into the same site in pCDNA3 to create the Gal4Rb-pCDNA3 plasmid used for expression in mammalian cells. The Gal4p130 plasmid was created by digesting pBluescript SK(+) (Stratagene), which contained full length p130 in the HindIII site, with BamHI and SalI. This fragment then was cloned into the BamHI and SalI sites of the pGBT9 vector. To put the fragment in frame with the Gal4 DNA binding domain, the p130-pGBT9 plasmid was cut with EagI and SmaI and was religated, creating Gal4p130-pGBT9 that was used for expression in yeast. To create Gal4p130 for expression in mammalian cells, a HindIII fragment of Gal4p130-pGBT9, including the Gal4DBD, was cloned into the HindIII site of pCDNA3. The p130C894F and RbC706F mutants were made by using the CLONTECH Transformer Site-Directed Mutagenesis Kit and the primers 5′ CAA ATT ATG ATG TTT TCC ATG TAT GG 3′ for Rb and 5′ CAG TTA TTA ATG TTT GCC ATT TAT GTG 3′ for p130. The p130 pocket domain-containing plasmid was made by PCR using primers with BamHI sites followed by 5′ CCA GTT TCT ACA GCT ACG CAT 3′ and 5′ TTA ATG TGG GGA AAT GTA GAC 3′. The BamHI fragment was cloned into the BamHI site of a pCDNA3-Gal4DBD plasmid. The pCDNA3-Gal4DBD plasmid was made by cloning the HindIII-SalI fragment from pGBT9 into the HindIII and XhoI sites of pCDNA3. The Gal4AD CtIP clone was isolated from the two-hybrid screen. Full length Gal4AD CtIP was created by PCR using the Gal4AD human fetal liver library as the template with the primers 5′ GTT ACT GTA ATA GAT ACA AA 3′ and 5′ AAA AGG GCC CCT ATG TCT TCT GCT CCT TGC 3′. The PCR product was cut with BsrGI and ApaI and was subcloned into the same sites in Gal4AD CtIP. The resulting full length Gal4AD CtIP was sequenced to confirm that no mutations were introduced. Myc-CtIP was created by subcloning the BglII-ApaI fragment of Gal4AD CtIP into the BamHI and ApaI sites of the pCDNA3-Myc vector. The Myc-CtIP ΔLXCXE was created by cutting the Myc-CtIP vector with BamHI and HpaI, treating with Klenow to fill the DNA ends, and then religating with DNA ligase. The MycCtIP ΔPLDLS vector was made in several steps. By using PCR with the primers 5′ TTT AGC AAC ACT TGT 3′ and 5′ AAA AGG ATC CTT TAT CCA TCA CAC 3′, an N-terminal fragment of CtIP was made. This fragment was subcloned into pBluescript SK(+) at the XbaI and BamHI sites. A second fragment was created by PCR using the primers 5′ AAA AGG ATC CGA TCG ATT TTC AGC 3′ and 5′ AAA AGG GCC CCT ATG TCT TCT GCT CCT TGC 3′. This fragment was subcloned behind the first fragment in the pBluescript SK(+) BamHI and ApaI sites. The whole fragment of CtIP, which now contained a deletion of the PLDLS motif, was cut out of pBluescript SK(+) by using XbaI and ApaI and was subcloned into the same sites of the Myc-CtIP vector. Gal4BD CtIP was made by subcloning the BamHI-ApaI fragment from Myc-CtIP into the BglII and ApaI sites of the pCDNA3 Gal4DBD plasmid. Gal4BD CtIP ΔPLDLS was generated by subcloning the BamHI-ApaI fragment from Myc-CtIP ΔPLDLS into the BglII and ApaI sites of the pCDNA3 Gal4DBD plasmid. Gal4BD CtBP was made by PCR using the primers 5′ AAA AGA ATT CAT GGG CAG CTC GCA CTT GCT 3′ and 5′ AAA ATC TAG ACT ACA ACT GGT CAC TGG CGT 3′ and using the CtBP clone that was a kind gift from G. Chinnadurai (Saint Louis University) as a template. The PCR product was digested with EcoRI and XbaI and was cloned into the same sites of the pCDNA3 Gal4DBD plasmid.
Repression Assays.
C33A cells were transiently transfected by the calcium phosphate method with the pSVECG reporter and Gal4DBD fusion proteins. One microgram of the β-galactosidase (β-gal) plasmid was cotransfected as a control for transfection efficiency. After 15 hours, cells were washed twice with PBS and were allowed to recover in DMEM with 10% serum. Forty hours posttransfection, cells were harvested, and chloramphenicol acetyltransferase (CAT) assays were performed as described (20), although extracts were not heat inactivated. CAT assay reaction mixture included 75 μl of extract, 75 μl of 1M Tris⋅Cl (7.8) 1 μl of C14-labeled chloramphenicol (1 mCi/ml), and 30 μl of acetyl CoA (3.5 mg/ml in H2O). Reactions were incubated at 37° for 3.5 hours. CAT values were normalized relative to the vector alone control β-gal values. β-gal activity was measured by adding 10 μl of the extract prepared for the CAT assays to 590 μl of 0.1 mg/ml chlorophenol red-β-d-galactopyranoside in lac Z buffer (60 mM Na2HPO4/40 mM NaH2PO4/10 mM KCl/1 mM MgSO4/38 mM 2-mercaptoethanol, pH 7.0). The absorbance of each sample was measured at 570 nm. In all instances of comparison, Western blot analyses were performed to determine that equal protein was expressed.
Yeast Two-Hybrid Screen.
The yeast two-hybrid screen was performed as recommended in the CLONTECH protocol. Inserts from positive clones were sequenced according to Sequenase Kit (United States Biochemical) instructions.
Immunoprecipitations.
C33A cells were transiently transfected by the calcium phosphate method. After 15 hours of transfection, cells were washed twice with DMEM, and then complete media was replaced. Forty hours posttransfection, cells were harvested and lysed in IP buffer containing 50 mM Tris⋅HCl (ph 7.4), 250 mM NaCl, 5 mM EDTA, 0.1% NP40, and the protease inhibitors Leupeptin at 1 μg/ml, Aprotinin at 1 μg/ml, at Pepstatin 1 μg/ml, and PMSF or Perfablock (Boehringer Mannheim) at 1 mM. Extracts were precleared by incubating with protein A agarose beads (Calbiochem) for 1 hour and then were centrifuged at 20,000 × g for 10 minutes. An aliquot of the sample (10%) was used as input, and 10% was used in a β-gal assay. The amount of extract used in the immunoprecipitation was normalized based on the β-gal values. One microgram of the appropriate antibody was added to precleared extracts and was allowed to mix at 4°C for 3 hours. Protein A agarose beads then were added and allowed to mix at 4°C for 1.5 hours. Samples then were washed 4 times at 4°C with 1 ml of IP buffer and were run on an SDS polyacrylamide gel.
RESULTS
Recent work has provided evidence for a mechanism for E2F/Rb-mediated repression that involves the recruitment of histone deacetylase (13–15). In particular, these studies demonstrated an ability of Rb to physically interact with HDAC that coincided with the ability of Rb to repress transcription. Nevertheless, this work also suggested that additional events may contribute to the ability of Rb to repress transcription. For instance, whereas the addition of the HDAC inhibitor trichostatin A reversed the repression of the adenovirus major late promoter, trichostatin A had little effect on the ability of Rb to repress transcription of the SV40 or TK promoter (13). Moreover, although the MAD protein, which is known to repress transcription through the recruitment of HDAC (21), could efficiently repress the major late promoter, it had no effect on the SV40 promoter (13). Although the distinction between these promoters remains unclear, the apparent insensitivity of the SV40 promoter to the recruitment of HDAC provides an assay to examine mechanisms of Rb-mediated repression that are independent of HDAC recruitment.
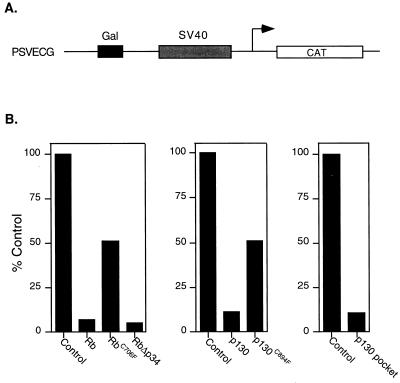
To further explore the basis for HDAC-independent E2F/Rb-mediated repression, we assayed wild-type and mutant versions of the Rb family proteins for their ability to repress transcription driven by an SV40 promoter that also contained upstream Gal4 sites (Fig. 1A). As shown in Fig. 1B, fusion proteins linking either Rb or p130 to the Gal4 DNA binding domain were capable of repressing transcription of the reporter in C33A cells. Similarly, a mutant of Rb that deletes eight sites for phosphorylation by cdc2 (RbΔp34) is an even more efficient repressor of transcription than wild-type Rb (19). As previously published for Rb (12), the pocket domain of p130, when tethered to the Gal4 DNA binding domain, repressed transcription, indicating that the pocket domain is sufficient for repression of the SV40 promoter. An Rb mutation found in human tumors, involving a Cys to Phe change at position 706 within the pocket domain (RbC706F), has been shown to disrupt the structure of the pocket domain and therefore to abolish the interaction of Rb with the viral oncoproteins E1A and T antigen (22). As previously shown by Dean and colleagues (12), the RbC706F mutant failed to repress transcription of the SV40 promoter (Fig. 1B). Likewise, a p130 mutant constructed to contain the equivalent alteration in the homologous sequence (p130C894F) also failed to repress transcription (Fig. 1B).
Figure 1.
Rb/p130-mediated repression independent of histone deacetylase recruitment. (A) Schematic representation of the pSVECG reporter. Constitutive CAT expression is driven by the SV40 promoter/enhancer. Upstream Gal4 sites provide a binding site for Gal4 DNA biding domain fusion proteins. (B) C33A cells were transiently transfected with 1 μg of β-gal, 0.5 μg of the pSVECG reporter, and 2 μg of Gal4-Rb, Gal4-RbΔp34, Gal4-p130, or Gal4-p130C894F or 5 μg of Gal4-RbC706F. Western blotting was performed to verify that transfected constructs expressed equal protein. For the repression assay using the pocket domain, 0.5 μg of pSVECG was transfected with 3 μg of Gal4-p130 pocket. As controls, C33A cells were transfected with 1 μg of β-gal, 0.5 μg of pSVECG reporter, and 2 or 3 μg of a vector encoding a Gal4 DNA binding domain (control vector). Cells were harvested 40 hours posttransfection, and CAT activity was assayed. β-gal values were used to normalize for transfection efficiency. Results of typical experiments are shown.
Rb and p130 Interact with CtIP.
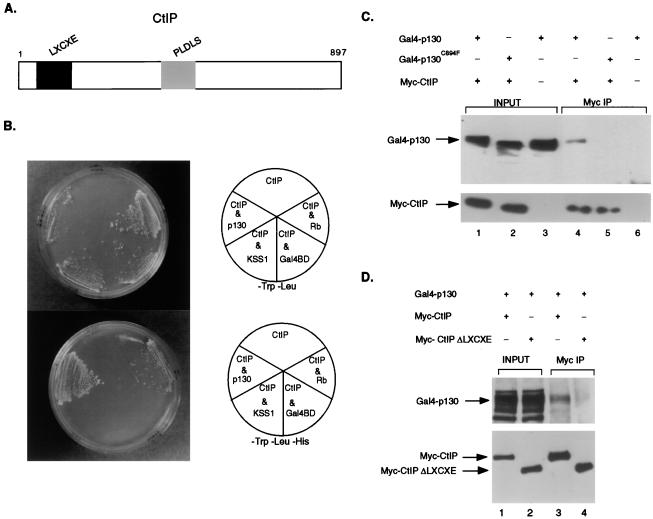
Given the indication from previous work that E2F/Rb-mediated repression could not be fully explained by recruitment of histone deacetylase, we initiated a search for other proteins that might be involved in an HDAC-independent transcriptional repression by Rb or p130. We used a full length p130 protein in a yeast two-hybrid assay to screen for potential protein partners of p130 that could mediate this HDAC-independent repression. The HF7C yeast strain was transformed with a plasmid encoding a Gal4 DNA binding domain-p130 fusion protein together with a human fetal liver cDNA library that incorporated the Gal4 activation domain. Forty positive transformants yielded 11 different clones encoding proteins that interacted with p130. Among the positive clones were cyclin D1, cyclin D3, E2F4, and E2F5, proteins known to specifically interact with Rb/p130.
In addition to these anticipated interacting proteins, one clone was found to encode the first 800 amino acids of a protein previously identified as CtIP. CtIP (C-terminal interacting protein) was originally recovered in a yeast two-hybrid screen as a partner of a protein known as CtBP (C-terminal binding protein) (16), a protein that binds to the C terminus of adenovirus E1A (23). The N-terminal portion of CtIP contains an LXCXE sequence (Fig. 2A), a motif found in the viral oncoproteins E1A, T antigen, and E7, as well as the D type cyclins, and that mediates the interaction with the Rb family proteins. Given the presence of the LXCXE motif in CtIP, we tested the ability of CtIP to interact with Rb in the two-hybrid assay. Fig. 2B shows that CtIP can specifically interact with p130 and Rb but not with an unrelated yeast protein, KSS1.
Figure 2.
Rb/p130 Interaction with CtIP. (A) Schematic representation of the CtIP protein. The position of an LXCXE sequence motif and a PLDLS sequence motif within the 897-aa CtIP protein are indicated. (B) HF7C yeast were transformed with plasmid encoding the Gal4AD-CtIP fusion protein alone or with a Gal4BD-p130, Gal4BD-Rb, Gal4BD-KSS1, or the empty Gal4BD vector. Yeast were streaked on nonselective media lacking Trp and Leu and on media that lacks Trp, Leu, and His that is selective for protein/protein interactions. (C) C33A cells were transfected with 10 μg of Myc-CtIP and 10 μg of either Gal4-p130 or Gal4-p130C894F. Cells were harvested 40 hours posttransfection and were lysed in IP buffer. Ten percent of the extract was loaded in input lanes 1–3. Myc antibody (Santa Cruz Biotechnology, 9E10) was used to immunoprecipitate Myc-CtIP. p130 antibody (Santa Cruz Biotechnology) was used in Western blotting to detect p130 in the immunoprecipitates (lanes 4–6). The blot was stripped and reprobed with Myc antibody to verify that equal amounts of Myc-CtIP were immunoprecipitated (lanes 4–6). (D) C33A cells were transfected as in C with 10 μg of Gal4-p130 and 10 μg of either Myc-CtIP or Myc-CtIP ΔLXCXE. Ten percent of the extract was loaded in input lanes 1 and 2. Myc antibody was used to immunoprecipitate Myc-CtIP. p130 antibody was used in Western blotting to detect p130 in the immunoprecipitates (lanes 3 and 4). The blot was stripped and reprobed with Myc antibody to verify that equal amounts of Myc-CtIP were immunoprecipitated (lanes 3 and 4).
We also used a coimmunoprecipitation assay to measure the ability of CtIP to interact with p130, as well as to define the sequences important in each protein for the interaction. As shown in Fig. 2C, wild-type p130 could be recovered in an immunoprecipitate with the CtIP protein, but a pocket disrupting mutant p130 protein (p130C894F) could not. In addition, p130 could be found to associate with the wild-type CtIP but not with a mutant of CtIP in which the first 170 amino acids, including the LXCXE domain, was deleted (Fig. 2D). It thus appears clear that CtIP interacts with p130, as well as Rb, and does so via an LXCXE-pocket domain interaction.
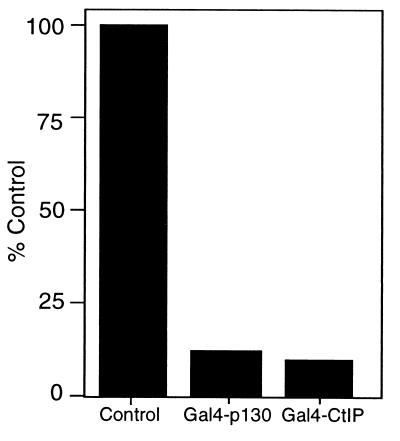
The observation that p130 interacts with CtIP, dependent on the pocket domain that is also required for p130-mediated repression, suggested a possible role for CtIP in transcriptional repression. To explore such a function, a fusion protein containing the Gal4 DNA binding domain linked to CtIP was created and assayed for its ability to repress the SV40 promoter reporter construct. As shown in Fig. 3, the Gal4-CtIP fusion protein was indeed active as a repressor; in fact, the Gal4-CtIP protein was as efficient as the Gal4-p130 fusion protein in the repression of the SV40 promoter. Based on all of these results, we conclude that recruitment of the CtIP protein represents an alternative mechanism, in addition to the recruitment of histone deacetylase, for p130-mediated repression.
Figure 3.
CtIP is a transcriptional repressor. C33A cells were transiently transfected with 1 μg of β-gal, 0.5 μg of the pSVECG reporter, and 2 μg of Gal4-p130 or 20 μg of Gal4-CtIP. Western blotting was performed to verify that transfected constructs expressed equal protein. As controls, C33A cells were transfected with 1 μg of β-gal, 0.5 μg of pSVECG reporter, and 2 μg of a vector encoding a Gal4 DNA binding domain (control vector). Cells were harvested 40 hours posttransfection, and CAT activity was assayed. β-gal values were used to normalize for transfection efficiency. Results of a typical experiment are shown.
CtIP Recruits the CtBP CoRepressor.
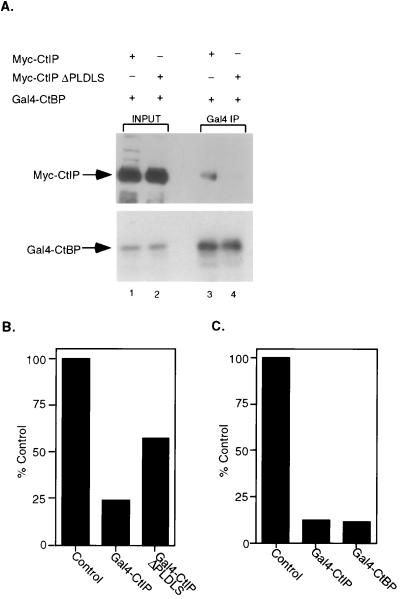
CtIP was isolated based on its interaction with CtBP, a protein identified as an adenovirus E1A-binding protein (16). More recently, a Drosophila homologue of CtBP has been identified and shown to function as a corepressor for the transcriptional regulatory proteins Hairy, Knirps, and Snail (24–26). These observations thus suggested the possibility that CtIP might function in E2F/p130-mediated repression by recruiting the CtBP protein. To investigate this possibility, we first examined the ability of CtIP to interact with CtBP. C33A cells were cotransfected with a plasmid encoding a Myc-tagged CtIP protein and a plasmid encoding Gal4-CtBP. Cells then were assayed for an interaction of the two proteins by immunoprecipitating Gal4-CtBP and then assaying for the presence of CtIP in the immunoprecipitates by Western blotting. As shown in Fig. 4A, wild-type CtIP was indeed recovered in the CtBP immunoprecipitate.
Figure 4.
p130/CtIP-mediated repression involves the recruitment of the CtBP corepressor. (A) C33A cells were transfected with 10 μg of Gal4-CtBP and 10 μg of either Myc-CtIP or Myc-CtIP ΔPLDLS. Cells were harvested 40 hours posttransfection and were lysed in IP buffer. Ten percent of the extract was loaded in input lanes 1 and 2. Gal4 DNA binding domain antibody (Santa Cruz Biotechnology, monoclonal) was used to immunoprecipitate Gal4-CtBP. Myc antibody (Santa Cruz Biotechnology, 9E10) was used in Western blotting to detect CtIP in the immunoprecipitates (lanes 3 and 4). The blot was stripped and reprobed with Gal4 DNA binding domain antibody to verify that equal amounts of Gal4-CtBP were immunoprecipitated (lanes 3 and 4). (B) C33A cells were transiently transfected with 1 μg of β-gal, 0.5 μg of the pSVECG reporter, and 2 μg of Gal4-CtIP or Gal4-CtIP ΔPLDLS. Western blotting was performed to verify that transfected constructs expressed equal protein. As controls, C33A cells were transfected with 1 μg of β-gal, 0.5 μg of pSVECG reporter, and 2 μg of control vector. Cells were harvested 40 hours postinfection, and CAT activity was assayed. β-gal values were used to normalize for transfection efficiency. Results of typical experiments are shown. (C) Same as in B except that cells were transfected with 1 μg of β-gal, 0.5 μg of pSVECG reporter, and 2 μg of Gal4-CtBP or 20 μg of Gal4 CtIP.
Previous work has demonstrated that the interaction of CtBP with E1A, or the interaction of the Drosophila CtBP with Hairy, Knirps, and Snail, depends on a PLDLS sequence found within these interacting proteins. Examination of the CtIP sequence reveals a PLDLS motif within the C-terminal region of the protein (see Fig. 2A). As such, we have generated a deletion mutant lacking this sequence and have tested the ability of the mutant to interact with CtBP. As shown in Fig. 4A, deletion of the PLDLS sequence in CtIP abolished the interaction with CtBP. We thus conclude that CtIP and CtBP do indeed interact and, like the interaction of the Drosophila proteins, the interaction depends on the PLDLS domain of CtIP.
Finally, to explore the role of CtBP in E2F/p130/CtIP-mediated repression, we assayed the effect of the CtIP PLDLS mutation on CtIP-mediated repression. As shown in Fig. 4B, the repressing activity of CtIP was clearly impaired by the PLDLS mutation, coincident with the role of the PLDLS sequence in mediating the CtBP interaction. Given the indication that CtIP has the ability to repress transcription when recruited to a promoter, together with the evidence that CtIP can interact with CtBP, we assayed the ability of CtBP alone to function as a transcriptional repressor. A Gal4 DNA binding domain-CtBP fusion was assayed for its ability to repress the SV40 promoter. As shown in Fig. 4C, the Gal4-CtBP fusion was equally effective as the Gal4-CtIP fusion protein in repressing the SV40 promoter. Based on these results, we conclude that the CtBP protein does possess transcriptional repressing activity and that the recruitment of CtBP via an interaction with CtIP represents an alternate mechanism for E2F/Rb-mediated repression of transcription.
DISCUSSION
The role of histone deacetylase recruitment in transcription repression, including E2F/Rb-mediated transcription repression, has now been shown in multiple instances (13–15, 27). Nevertheless, it is also clear that other mechanisms, functioning independently of HDAC recruitment, must play a role in repression. The data we present here now describe at least one additional mechanism for E2F/Rb-mediated repression that involves the recruitment of the CtIP/CtBP corepressor complex.
Alternate Mechanisms of E2F/Rb-Mediated Repression.
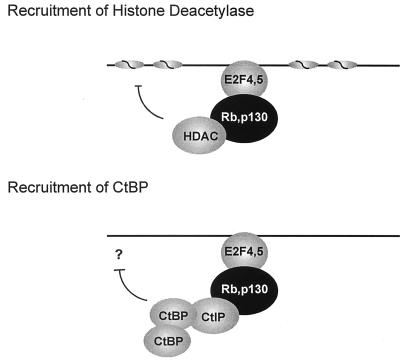
The CtBP protein has been implicated in several forms of transcription repression including the recent studies of the Drosophila proteins Hairy, Knirps, and Snail (24–26). In each case, CtBP is recruited to a promoter through the interaction with a PLDLS-containing protein. The results we now present here demonstrate that the mammalian CtBP protein can be recruited to a target promoter through an interaction with CtIP, which in turn interacts with Rb or p130. Although the majority of the PLDLS-containing proteins that are known to interact with CtBP are DNA binding proteins, including Hairy, Knirps, and Snail, there is no evidence to suggest that CtIP has intrinsic DNA binding activity. Rather, CtIP appears to act as a bridging protein, bringing the CtBP corepressor to a promoter through the interaction with Rb or p130 and then E2F (Fig. 5). The observation that CtIP contains distinct motifs that can mediate Rb/p130 binding (LXCXE) as well as CtBP binding (PLDLS) provides a mechanism by which CtIP could serve to bridge the two sets of proteins.
Figure 5.
Alternative mechanisms for Rb/p130-mediated repression. E2F target promoters can be repressed in two fashions. Histone deacetylase is recruited to promoters that contain E2F/Rb or E2F/p130 complexes through an interaction with Rb or p130. Histone deacetylase then modifies the histones proximal to the promoter, causing transcriptional silencing. Rb and p130 recruit CtIP/CtBP to E2F complexes. CtIP bridges the interaction between CtBP and the E2F/Rb complex. CtBP, most likely acting as a dimer, then functions by an undetermined mechanism to mediate repression.
Although CtIP does recruit CtBP, and this can serve as a mechanism for transcriptional repression, it is also possible that other proteins interact with CtIP, possibly leading to other events of transcriptional repression. In this regard, it is of interest to note that, although the recruitment of CtBP coincides with repression by Hairy as well as BKLF, each of these proteins appears to interact with other factors to establish a more complete repression (26, 28). Possibly, the fact that the CtIP PLDLS mutation did not completely abolish repression might suggest that other activities of CtIP could contribute to full repression.
Yet to be determined is the precise mechanism by which CtBP might effect a repression of transcription. Although there has been one report suggesting an interaction of CtBP with histone deacetylase (29), this is unlikely to be the primary mechanism of CtBP-mediated repression given our observations and the observations of others that the SV40 promoter, shown to be repressed by CtBP, is relatively insensitive to histone deacetylase (13). An alternative possibility stems from recent observations that CtBP interacts with the human polycomb proteins (30). Polycomb proteins have been shown in Drosophila to be important in repression of certain homeotic genes. Although the mechanisms of this repression are unclear, current models speculate that the PcG proteins can package regions of DNA into heterochromatin-like structures (30).
Multiple Roles for E1A in Affecting Cellular Transcription.
Human CtBP was originally identified as a phosphoprotein that associates with the C terminus of E1A (17). The C-terminal E1A sequences that are involved in the CtBP interaction are well conserved among adenovirus serotypes, implying a functional importance. Nevertheless, the analysis of function of these sequences has been somewhat confusing. Some experiments suggest that the C-terminal E1A domain functions to suppress cell transformation in conjunction with an activated Ras protein (23). That is, mutation of the E1A C terminus, which include the domain responsible for binding to CtBP, leads to enhanced oncogenicity in conjunction with Ras. In contrast, other experiments have provided evidence for a role for these sequences in the immortalizing function of E1A as well as to collaborate with adenovirus E1B in transformation (31). Although the basis for the apparent discrepancy in these results is unclear, the latter findings, indicating a requirement for the C-terminal domain in E1A function in immortalization and transformation with E1B, are certainly consistent with the findings that this domain interacts with CtBP and thus would disrupt the formation of the E2F-p130-CtIP-CtBP repressor complex. It is interesting to note that both E1A and CtIP contain LXCXE and PLDLS motifs, implying that the two proteins target the same factors, Rb and CtBP, and perhaps compete for their binding. In this way, E1A would be seen to disrupt transcriptional repression in three complementary fashions—either the inhibition of Rb family protein interaction with E2F, the inhibition of the interaction of Rb with the CtIP/CtBP complex, or the inhibition of the CtBP repressor with the E2F complex. Interestingly, two recent reports describe yet another mechanism for E1A action that involves a direct inhibition of histone acetyl transferase activity (32, 33). In addition to its ability to disrupt complexes involving the p300 protein, these two reports demonstrate that the direct interaction of E1A with either p300/CBP or PCAF leads to an inhibition of histone acetylase activity. As such, it appears that the E1A protein has evolved a series of distinct activities to affect transcription through an alteration of chromatin structure.
Acknowledgments
We are grateful to Jeanette Cook, Rosalie Sears, Chi-Hyun Park, and Nicole Liberatti for invaluable advice, discussion, and technical assistance. We are grateful to D. Dean, R. Eisenman, D. Reinberg, G. Chinnadurai, and R. Bremner for the generous gift of reagents. We also thank Kaye Culler for assistance in the preparation of the manuscript. This work was supported by the Howard Hughes Medical Institute. A.R.M. was supported by a fellowship from the Department of the Army (DAMD17-98-8074), and J.R.N. is an Investigator of the Howard Hughes Medical Institute.
ABBREVIATIONS
- β-gal
β-galactosidase
- CAT
chloramphenicol acetyltransferase
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
References
- 1.Sherr C J. Cell. 1993;73:1059–1065. doi: 10.1016/0092-8674(93)90636-5. [DOI] [PubMed] [Google Scholar]
- 2.Hunter T. Cell. 1993;75:839–841. doi: 10.1016/0092-8674(93)90528-x. [DOI] [PubMed] [Google Scholar]
- 3.Nevins J R. Science. 1992;258:424–429. doi: 10.1126/science.1411535. [DOI] [PubMed] [Google Scholar]
- 4.Helin K, Harlow E. Trends Cell Biol. 1993;3:43–46. doi: 10.1016/0962-8924(93)90150-y. [DOI] [PubMed] [Google Scholar]
- 5.Sherr C J, Roberts J M. Genes Dev. 1995;9:1149–1163. doi: 10.1101/gad.9.10.1149. [DOI] [PubMed] [Google Scholar]
- 6.Weinberg R A. Cell. 1995;81:323–330. doi: 10.1016/0092-8674(95)90385-2. [DOI] [PubMed] [Google Scholar]
- 7.Hunter T, Pines J. Cell. 1994;79:573–582. doi: 10.1016/0092-8674(94)90543-6. [DOI] [PubMed] [Google Scholar]
- 8.Dyson N. Genes Dev. 1998;12:2245–2262. doi: 10.1101/gad.12.15.2245. [DOI] [PubMed] [Google Scholar]
- 9.Nevins J R. Cell Growth Differ. 1998;9:585–593. [PubMed] [Google Scholar]
- 10.Smith E J, Leone G, DeGregori J, Jakoi L, Nevins J R. Mol Cell Biol. 1996;16:6965–6976. doi: 10.1128/mcb.16.12.6965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weintraub S J, Prater C A, Dean D C. Nature (London) 1992;358:259–261. doi: 10.1038/358259a0. [DOI] [PubMed] [Google Scholar]
- 12.Weintraub S J, Chow K N B, Luo R X, Zhang S H, He S, Dean D C. Nature (London) 1995;375:812–815. doi: 10.1038/375812a0. [DOI] [PubMed] [Google Scholar]
- 13.Luo R X, Postigo A A, Dean D C. Cell. 1998;92:463–473. doi: 10.1016/s0092-8674(00)80940-x. [DOI] [PubMed] [Google Scholar]
- 14.Magnaghi-Jaulin L, Groisman R, Naguibneva I, Robin P, Lorain S, LeVillain J P, Troalen F, Trouche D, Harel-Bellan A. Nature (London) 1998;391:601–605. doi: 10.1038/35410. [DOI] [PubMed] [Google Scholar]
- 15.Brehm A, Miska E A, McCance D J, Reid J L, Bannister A J, Kouzarides T. Nature (London) 1998;391:597–601. doi: 10.1038/35404. [DOI] [PubMed] [Google Scholar]
- 16.Schaeper U, Subramanian T, Lim L, Boyd J M, Chinnadurai G. J Biol Chem. 1998;273:8549–8552. doi: 10.1074/jbc.273.15.8549. [DOI] [PubMed] [Google Scholar]
- 17.Schaeper U, Boyd J M, Verma S, Uhlmann E, Subramanian T, Chinnadurai G. Proc Natl Acad Sci USA. 1995;92:10467–10471. doi: 10.1073/pnas.92.23.10467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang Y, Iratni R, Erdjument-Bromage H, Tempst P, Reinberg D. Cell. 1997;89:357–364. doi: 10.1016/s0092-8674(00)80216-0. [DOI] [PubMed] [Google Scholar]
- 19.Bremner R, Cohen B L, Sopta M, Hamel P A, Ingles C J, Gallie B L, Phillips R A. Mol Cell Biol. 1995;15:3256–3265. doi: 10.1128/mcb.15.6.3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smith E J, Nevins J R. Mol Cell Biol. 1995;15:338–344. doi: 10.1128/mcb.15.1.338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Laherty C D, Yang W-M, Sun J-M, Davie J R, Seto E, Eisenman R N. Cell. 1997;89:349–356. doi: 10.1016/s0092-8674(00)80215-9. [DOI] [PubMed] [Google Scholar]
- 22.Kaye F J, Kratzke R A, Gerster J L, Horowitz J M. Proc Natl Acad Sci USA. 1990;87:6922–6926. doi: 10.1073/pnas.87.17.6922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boyd J M, Subramanian T, Schaeper U, Regina M, Bayley S, Chinnadurai G. EMBO J. 1993;12:469–478. doi: 10.1002/j.1460-2075.1993.tb05679.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nibu Y, Zhang H, Bajor E, Barolo S, Small S, Levine M. EMBO J. 1998;17:7009–7020. doi: 10.1093/emboj/17.23.7009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nibu Y, Zhang H, Levine M. Science. 1998;280:101–104. doi: 10.1126/science.280.5360.101. [DOI] [PubMed] [Google Scholar]
- 26.Poortinga G, Watanabe M, Parkhurst S M. EMBO J. 1998;17:2067–2078. doi: 10.1093/emboj/17.7.2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ferreira R, Magnaghi-Jaulin L, Robin P, Harel-Bellan A, Trouche D. Proc Natl Acad Sci USA. 1998;95:10493–10498. doi: 10.1073/pnas.95.18.10493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Turner J, Crossley M. EMBO J. 1998;17:5129–5140. doi: 10.1093/emboj/17.17.5129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sundqvist A, Sollerbrant K, Svensson C. FEBS Lett. 1998;429:183–188. doi: 10.1016/s0014-5793(98)00588-2. [DOI] [PubMed] [Google Scholar]
- 30.Sewalt R G A B, Gunster M J, van der Vlag J, Satijn D P E, Otte A P. Mol Cell Biol. 1999;19:777–787. doi: 10.1128/mcb.19.1.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Quinlan M P, Whyte P, Grodzicker T. Mol Cell Biol. 1988;8:3191–3203. doi: 10.1128/mcb.8.8.3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chakravarti D, Ogryzko V, Kao H-Y, Nash A, Chen H, Nakatani Y, Evans R M. Cell. 1999;96:393–403. doi: 10.1016/s0092-8674(00)80552-8. [DOI] [PubMed] [Google Scholar]
- 33.Hamamori Y, Sartorelli V, Ogryzko V, Puri P L, Wu H-Y, Wang J Y J, Nakatani Y, Kedes L. Cell. 1999;96:405–413. doi: 10.1016/s0092-8674(00)80553-x. [DOI] [PubMed] [Google Scholar]