S. cerevisiae Atg5 in complex with the N-terminal regions of Atg16 was expressed, purified and crystallized in four crystal forms.
Keywords: Atg5, Atg16, autophagy
Abstract
Atg5 is a novel 34 kDa protein that is covalently modified by Atg12, a ubiquitin-like modifier, and forms a complex with Atg16. The Atg12–Atg5–Atg16 complex localizes to autophagosome precursors and plays an essential role in autophagosome formation. Saccharomyces cerevisiae Atg5 in complex with the N-terminal regions of Atg16 was expressed, purified and crystallized in four crystal forms. Forms I, II and III belong to space group P21, with unit-cell parameters a = 66.3, b = 104.4, c = 112.1 Å, β = 92.1° (form I), a = 79.5, b = 101.4, c = 95.1 Å, β = 98.6° (form II) or a = 56.9, b = 101.2, c = 66.5 Å, β = 100.6° (form III). Form IV belongs to space group P42212, with unit-cell parameters a = 73.3, c = 148.1 Å. Diffraction data were collected from all crystal forms and high-resolution data to beyond 2.0 Å resolution were obtained from a form IV crystal.
1. Introduction
Autophagy is a starvation-induced response that mediates the bulk degradation of cytoplasmic components in lysosomes/vacuoles (Seglen & Bohley, 1992 ▶; Takeshige et al., 1992 ▶) and plays a critical role in numerous biological processes such as neurodegeneration and pathogen infection as well as survival response during neonatal starvation (Hara et al., 2006 ▶; Komatsu et al., 2006 ▶; Ogawa et al., 2005 ▶; Nakagawa et al., 2004 ▶; Kuma et al., 2004 ▶). In autophagy, a double-membrane structure called an autophagosome sequesters a portion of cytoplasm and fuses with the lysosome/vacuole to deliver its contents into the organelle lumen.
16 autophagy genes involved in autophagosome formation, named ATG genes, have been identified using genetic approaches in Saccharomyces cerevisiae (Klionsky et al., 2003 ▶). Among these, five Atg proteins were shown to be involved in a novel ubiquitin-like conjugation system named the Atg12 system (Mizushima et al., 1998 ▶). In the Atg12 system, the carboxyl-terminal glycine of Atg12 is activated by Atg7, an E1-like enzyme (Tanida et al., 1999 ▶), and is then transferred to Atg10, an E2-like enzyme (Shintani et al., 1999 ▶). Finally, Atg12 is conjugated to its sole target, Atg5 (Mizushima et al., 1998 ▶). In addition to the covalent interaction with Atg12, Atg5 interacts with the N-terminal region of a multimeric protein, Atg16, non-covalently (Mizushima et al., 1999 ▶). Using this interaction, the Atg12–Atg5 conjugate forms a multimeric complex with Atg16 (Kuma et al., 2002 ▶; Mizushima et al., 1999 ▶). The Atg12–Atg5–Atg16 complex localizes to autophagosome precursors and plays an essential role in autophagosome formation (Kim et al., 2001 ▶; Suzuki et al., 2001 ▶).
Recently, we reported the crystal structure of plant Atg12 and revealed that Atg12 is a ubiquitin-fold protein (Suzuki et al., 2005 ▶). However, structural information on Atg5 and Atg16 is completely lacking, preventing us from elucidating the molecular role of the Atg12–Atg5–Atg16 complex. In this report, we describe the expression, purification and crystallization of Atg5 in complex with the N-terminal region of Atg16.
2. Expression and purification
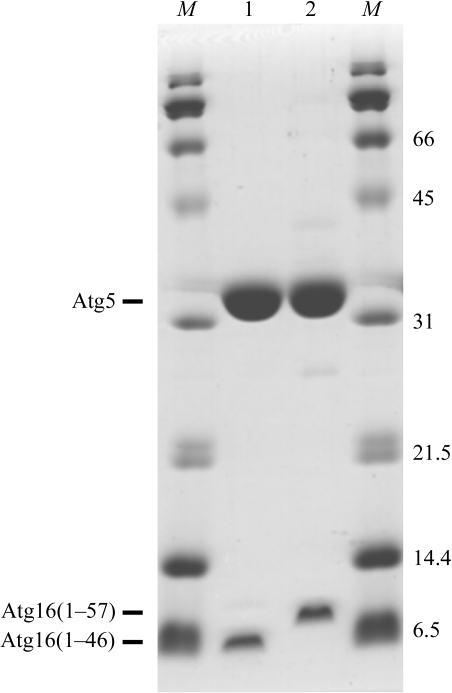
The full-length gene of S. cerevisiae Atg5 was inserted into a pHT1 vector [a pET28a(+) vector (Novagen) modified by insertion of sequences encoding a hexahistidine tag and a TEV protease-cleavage site] using NdeI/BamHI restriction sites. Residues 1–46 and 1–57 of S. cerevisiae Atg16 were inserted into a pET-11a vector (Novagen) using NdeI/BamHI restriction sites. Each Atg16 construct was co-expressed with N-terminally hexahistidine-tagged Atg5 in Escherichia coli BL21 (DE3). After cell lysis, Atg5 was purified by affinity chromatography using an Ni–NTA column (Qiagen). Throughout the purification steps, both Atg16(1–46) and Atg16(1–57) co-migrated with Atg5, indicating that they form a stable complex with Atg5. After affinity chromatography, the protein complexes were purified on a HiTrap DEAE FF column (GE Healthcare Biosciences) equilibrated with 20 mM Tris–HCl pH 8.5 and were eluted with a 0–500 mM NaCl gradient in the same buffer. The hexahistidine tag was then cleaved from Atg5 with TEV protease (GE Healthcare Biosciences; a Gly-Ala-His sequence remained on the N-terminus of Atg5). Further purification was performed using a HiTrap CM FF column (GE Healthcare Biosciences) equilibrated with 20 mM HEPES buffer pH 6.8 and product was eluted with a 0–500 mM NaCl gradient in the same buffer. Final purification was carried out on a Superdex75 column (GE Healthcare Biosciences) eluted with 20 mM HEPES buffer pH 6.8 and 150 mM NaCl for the Atg5–Atg16(1–46) complex and on a Superdex200 column (GE Healthcare Biosciences) eluted with 20 mM Tris buffer pH 7.4 and 150 mM NaCl for the Atg5–Atg16(1–57) complex. The purified Atg5–Atg16(1–46) and Atg5–Atg16(1–57) complexes (Fig. 1 ▶) were concentrated to 8 and 12.5 mg ml−1, respectively, and used for crystallization.
Figure 1.
SDS–PAGE of purified Atg5–Atg16(1–46) and Atg5–Atg16(1–57) complexes on a 15% gel. Lanes 1 and 2 contain Atg5–Atg16(1–46) and Atg5–Atg16(1–57) complexes, respectively, and lanes M contain molecular-weight markers (labelled in kDa). Proteins were stained with Coomassie Brilliant Blue.
3. Crystallization
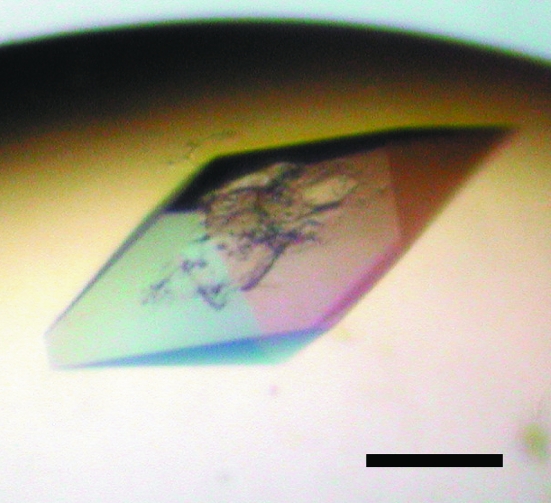
Crystallization trials were performed using the sitting-drop vapour-diffusion method at 293 K. Initial screening was performed using Crystal Screen and Crystal Screen 2 from Hampton Research and Wizard I and II from Emerald Biostructures as reservoir solutions. 0.3 µl drops of 8 mg ml−1 Atg5–Atg16(1–46) complex in 20 mM HEPES buffer pH 6.8 and 150 mM NaCl were mixed with equal amounts of reservoir solution and were equilibrated against 100 µl of the same reservoir solution by vapour diffusion. In the same way, 0.3 µl drops of 12.5 mg ml−1 Atg5–Atg16(1–57) complex in 20 mM Tris buffer pH 7.4 and 150 mM NaCl were mixed with equal amounts of reservoir solution and were equilibrated against 100 µl of the same reservoir solution by vapour diffusion. The Atg5–Atg16(1–46) complex was crystallized with a reservoir solution consisting of 24% PEG 3350, 0.1 M CAPS pH 10.0 (form I crystal). The Atg5–Atg16(1–57) complex was crystallized in three different crystal forms (forms II, III and IV). Only a photograph of form IV is shown in Fig. 2 ▶, as we judge the photographs we have of the other forms to not be good enough for publication. These three crystal forms were obtained with the same reservoir solution consisting of 15% PEG 3350, 0.1 M HEPES pH 6.8. All crystals were obtained within a week.
Figure 2.
Crystal of Atg5–Atg16(1–57) complex (form IV). The black scale bar is 100 µm in length.
4. Preliminary X-ray analysis
Crystals were immersed into reservoir solution supplemented with 11–20% glycerol as a cryoprotectant for several seconds and then flash-cooled and kept in a stream of nitrogen gas at 90–120 K during data collection. Diffraction data were collected from a form I crystal using an ADSC Quantum 315 charge-coupled device detector on SPring-8 beamline BL41XU at a wavelength of 1.00 Å. Diffraction data from form II, III and IV crystals were collected on a Rigaku R-AXIS VII imaging-plate detector using Cu Kα radiation. All diffraction data were processed using the HKL-2000 program suite (Otwinowski & Minor, 1997 ▶). The data-collection statistics are summarized in Table 1 ▶. The acceptable range of the volume-to-weight ratio (V M) values (Matthews, 1968 ▶) indicates that form I contains three, four or five Atg5–Atg16(1–46) complexes (the corresponding V M values are 3.31, 2.48 and 1.99 Å3 Da−1, respectively), form II contains three, four or five Atg5–Atg16(1–57) complexes (V M values of 3.15, 2.36 and 1.89 Å3 Da−1, respectively), form III contains two Atg5–Atg16(1–57) complexes (V M = 2.35 Å3 Da−1) and form IV contains one Atg5–Atg16(1–57) complex (V M = 2.48 Å3 Da−1) per asymmetric unit. We calculated self-rotation functions using form I and II diffraction data, but failed to find any obvious noncrystallographic symmetry axes. Therefore, the numbers of complexes contained in the asymmetric units of form I and II crystals remain to be determined. Phasing experiments are now in progress by a combination of the multiple isomorphous replacement and multiwavelength anomalous dispersion methods.
Table 1. Diffraction data statistics of four crystal forms of the Atg5–Atg16 complex.
Values in parentheses refer to the outer shell.
| Crystal form | I | II | III | IV |
|---|---|---|---|---|
| Protein complex | Atg5–Atg16(1–46) | Atg5–Atg16(1–57) | Atg5–Atg16(1–57) | Atg5–Atg16(1–57) |
| Space group | P21 | P21 | P21 | P42212 |
| Unit-cell parameters | ||||
| a (Å) | 66.3 | 79.5 | 56.9 | 73.3 |
| b (Å) | 104.4 | 101.4 | 101.2 | 73.3 |
| c (Å) | 112.2 | 95.1 | 66.5 | 148.1 |
| β (°) | 92.1 | 98.6 | 100.6 | 90 |
| Resolution range (Å) | 50–2.1 (2.18–2.10) | 50–2.95 (3.06–2.95) | 50–3.0 (3.11–3.00) | 50–1.97 (2.04–1.97) |
| Observed reflections | 317606 | 169957 | 74216 | 257319 |
| Unique reflections | 88015 | 31125 | 14118 | 29390 |
| Completeness (%) | 98.6 (95.7) | 98.8 (91.5) | 94.3 (77.8) | 99.9 (100.0) |
| Rmerge(I)† | 0.048 (0.306) | 0.092 (0.312) | 0.090 (0.304) | 0.055 (0.304) |
| I/σ(I) | 15.0 (3.8) | 8.7 (3.9) | 10.9 (4.0) | 28.9 (11.1) |
R
merge(I) = 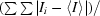
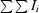 , where I
i is the intensity of the ith observation and 〈I〉 is the mean intensity.
, where I
i is the intensity of the ith observation and 〈I〉 is the mean intensity.
Acknowledgments
We thank Dr M. Kawamoto, Dr H. Sakai and the staff at beamline BL41XU, SPring-8, Japan for data-collection support. This work was supported by a Grant-in-Aid for Young Scientists (B) 17790048 and by National Project on Protein Structural and Functional Analyses from the Ministry of Education, Culture, Sports, Science and Technology, Japan. This work was carried out under the NIBB Cooperative Research Program (4-148).
References
- Hara, T., Nakamura, K., Matsui, M., Yamamoto, A., Nakahara, Y., Suzuki-Migishima, R., Yokoyama, M., Mishima, K., Saito, I., Okano, H. & Mizushima, N. (2006). Nature (London), 441, 885–889. [DOI] [PubMed] [Google Scholar]
- Kim, J., Huang, W. P. & Klionsky, D. J. (2001). J. Cell Biol.152, 51–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky, D. J., Cregg, J. M., Dunn, W. A. Jr, Emr, S. D., Sakai, Y., Sandoval, I. V., Sibirny, A., Subramani, S., Thumm, M., Veenhuis, M. & Ohsumi, Y. (2003). Dev. Cell, 5, 539–545. [DOI] [PubMed] [Google Scholar]
- Komatsu, M., Waguri, S., Chiba, T., Murata, S., Iwata, J., Tanida, I., Ueno, T., Koike, M., Uchiyama, Y., Kominami, E. & Tanaka, K. (2006). Nature (London), 441, 880–884. [DOI] [PubMed] [Google Scholar]
- Kuma, A., Hatano, M., Matsui, M., Yamamoto, A., Nakaya, H., Yoshimori, T., Ohsumi, Y., Tokuhisa, T. & Mizushima, N. (2004). Nature (London), 432, 1032–1036. [DOI] [PubMed] [Google Scholar]
- Kuma, A., Mizushima, N., Ishihara, N. & Ohsumi, Y. (2002). J. Biol. Chem.277, 18619–18625. [DOI] [PubMed] [Google Scholar]
- Matthews, B. W. (1968). J. Mol. Biol.33, 491–497. [DOI] [PubMed] [Google Scholar]
- Mizushima, N., Noda, T. & Ohsumi, Y. (1999). EMBO J.18, 3888–3896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima, N., Noda, T., Yoshimori, T., Tanaka, Y., Ishii, T., George, M. D., Klionsky, D. J., Ohsumi, M. & Ohsumi, Y. (1998). Nature (London), 395, 395–398. [DOI] [PubMed] [Google Scholar]
- Nakagawa, I., Amano, A., Mizushima, N., Yamamoto, A., Yamaguchi, H., Kamimoto, T., Nara, A., Funao, J., Nakata, M., Tsuda, K., Hamada, S. & Yoshimori, T. (2004). Science, 306, 1037–1040. [DOI] [PubMed] [Google Scholar]
- Ogawa, M., Yoshimori, T., Suzuki, T., Sagara, H., Mizushima, N. & Sasakawa, C. (2005). Science, 307, 727–731. [DOI] [PubMed] [Google Scholar]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol.276, 307–326. [DOI] [PubMed]
- Seglen, P. O. & Bohley, P. (1992). Experientia, 48, 158–172. [DOI] [PubMed] [Google Scholar]
- Shintani, T., Mizushima, N., Ogawa, Y., Matsuura, A., Noda, T. & Ohsumi, Y. (1999). EMBO J.18, 5234–5241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki, K., Kirisako, T., Kamada, Y., Mizushima, N., Noda, T. & Ohsumi, Y. (2001). EMBO J.20, 5971–5981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki, N. N., Yoshimoto, K., Fujioka, Y., Ohsumi, Y. & Inagaki, F. (2005). Autophagy, 1, 119–126. [DOI] [PubMed] [Google Scholar]
- Takeshige, K., Baba, M., Tsuboi, S., Noda, T. & Ohsumi, Y. (1992). J. Cell Biol.119, 301–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanida, I., Mizushima, N., Kiyooka, M., Ohsumi, M., Ueno, T., Ohsumi, Y. & Kominami, E. (1999). Mol. Biol. Cell, 10, 1367–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]