Crystals of an N-terminally truncated variant of the Salmonella flagellar ATPase FliI, which exports substrate proteins into the central channel of the growing flagellar structure by utilizing the energy of ATP hydrolysis, have been obtained and characterized by X-ray diffraction.
Keywords: type III flagellar protein export, FliI ATPase
Abstract
Most of the structural components making up the bacterial flagellum are translocated through the central channel of the growing flagellar structure by the type III flagellar protein-export apparatus in an ATPase-driven manner and are assembled at the growing end. FliI is the ATPase that drives flagellar protein export using the energy of ATP hydrolysis. FliI forms an oligomeric ring structure in order to attain maximum ATPase activity. In this study, FliI(Δ1–18), an N-terminally truncated variant of FliI lacking the first 18 residues, was purified and crystallized. Crystals were obtained using the hanging-drop vapour-diffusion technique with PEG 8000 as a precipitant. FliI(Δ1–18) crystals grew in the monoclinic space group P21, with unit-cell parameters a = 48, b = 73, c = 126 Å, β = 94°, and diffracted to 2.4 Å resolution. Anomalous difference Patterson maps of Os-derivative and Pt-derivative crystals showed significant peaks in their Harker sections, indicating that both derivatives are suitable for structure determination.
1. Introduction
The bacterial flagellum is a locomotive organelle consisting of a basal body, a hook and a filament. Flagellar assembly begins with the basal body, followed by the hook and finally the filament. The axial structure of the flagellum extends from the MS ring, which spans the cytoplasmic membrane. Therefore, flagellar axial component proteins must be exported from the cytoplasm to their final destination, the distal end of the growing flagellum (Iino, 1969 ▶; Emerson et al., 1970 ▶), where their self-assembly occurs (Yonekura et al., 2000 ▶). Several other proteins involved in flagellar assembly are also exported. All these proteins are translocated through the central narrow channel of the growing structure by the flagellar type III protein-export apparatus in an ATPase-driven manner (Macnab, 2003 ▶, 2004 ▶; Minamino & Namba, 2004 ▶). The export apparatus consists of six integral membrane components (FlhA, FlhB, FliO, FliP, FliQ and FliR) and three cytoplasmic components (FliH, FliI and FliJ) and its membrane-spanning portion is postulated to be inserted into the putative central pore of the MS ring (Minamino & Macnab, 1999 ▶, 2000a ▶).
FliI is the flagellar ATPase, the enzymatic activity of which is essential for flagellar protein export (Dreyfus et al., 1993 ▶; Fan & Macnab, 1996 ▶). The middle region of FliI shows a significant sequence similarity to the catalytic β-subunit of F1-ATPase, including well conserved residues within the Walker A and B boxes (Vogler et al., 1991 ▶). Unlike the β-subunit, however, FliI can self-assemble into a homohexameric ring structure in vitro and attain its full ATPase activity (Claret et al., 2003 ▶), suggesting that FliI oligomerization could occur at some stage of the type III flagellar protein-export process. The extreme N-terminal region of FliI controls its oligomerization and thus regulates its catalytic activity (Minamino et al., 2006 ▶).
The ATPase activity of FliI is also controlled by its regulator FliH (Auvray et al., 2002 ▶; González-Pedrajo et al., 2002 ▶; Minamino & Macnab, 2000b ▶). FliH is also required for the effective docking of FliI to FlhA and FlhB and is thought to play an important role in the energy coupling of ATP hydrolysis by FliI with flagellar protein export (Dreyfus et al., 1993 ▶; Fan & Macnab, 1996 ▶; Minamino & Macnab, 2000b ▶; Minamino et al., 2003 ▶). The FliH–FliI complex interacts with FliJ and/or export substrate–chaperone complexes such as the FlgK–FlgN complex (González-Pedrajo et al., 2002 ▶; Minamino et al., 2000 ▶; Thomas et al., 2004 ▶). Salmonella InvC, which is a FliI homolog in the virulence type III protein-secretion system, has been shown to induce chaperone release from and unfolding of the cognate secreted protein in an ATP-dependent manner (Akeda & Galán, 2005 ▶). However, it remains unknown how FliI drives the translocation of export substrates into the central channel of the flagellum by utilizing the energy of ATP hydrolysis.
In order to understand the mechanism of flagellar protein export driven by ATP hydrolysis, the atomic structure of FliI is essential. Here, we report an X-ray crystallographic study of FliI. Because of the low solubility of native FliI, we prepared a highly soluble variant lacking the first 18 residues, FliI(Δ1–18), for the following crystallization trial.
2. Materials and methods
2.1. Protein purification
FliI(Δ1–18) was produced from N-terminally His-tagged FliI(Δ1–7), a variant of FliI lacking the first seven residues. A 50 ml overnight culture of BL21(DE3)pLysS carrying pBGeI(Δ1–7), which encodes His-FliI(Δ1–7) on pET19b (Minamino et al., 2003 ▶), was inoculated into 5 l LB medium containing 100 µg ml−1 ampicillin and 30 µg ml−1 chloramphenicol. The cells were grown at 303 K until the culture density reached an OD600 of 0.6. IPTG was then added to a final concentration of 1 mM and growth continued for another 5 h. The cells were collected by centrifugation (8000g, 10 min, 277 K) and stored at 193 K. The cells were thawed, resuspended in 300 ml binding buffer (20 mM Tris–HCl pH 8.0, 500 mM NaCl) containing six tablets of Complete protease-inhibitor cocktail (Boehringer Mannheim) and sonicated (ASTRASON model XL2020 sonicator, Misonix Inc.). The cell lysate was centrifuged (13 000g, 10 min, 277 K) to remove cell debris and the supernatant fraction centrifuged at 130 000g for 1 h at 277 K.
The soluble fraction was subjected to a HiTrap chelating column (Amersham Pharmacia Biotech) equilibrated with binding buffer. The column was washed with binding buffer containing 50 mM imidazole and proteins were eluted using a linear gradient of 50–500 mM imidazole. Fractions containing His-FliI(Δ1–7) were identified by SDS–PAGE, concentrated by Centriprep YM30 (Amicon), and dialyzed overnight against two changes of buffer containing 50 mM Tris–HCl pH 8.0, 100 mM NaCl, 1 mM EDTA, 1 mM DTT at 277 K. The dialyzed sample was loaded onto a HiLoad 26/60 Superdex 200 prep-grade column (Amersham Biosciences) equilibrated with the buffer used for dialysis and 5 ml fractions were collected at a flow rate of 2.5 ml min−1. Fractions containing His-FliI(Δ1–7) were pooled, concentrated and dialyzed overnight against two changes of buffer containing 50 mM Tris–HCl pH 8.0, 50 mM NaCl.
FliI(Δ1–18) was then prepared from purified His-FliI(Δ1–7) by limited proteolysis. His-FliI(Δ1–7) was mixed with recombinant enterokinase (Novagen) and incubated overnight at room temperature. FliI(Δ1–18) was produced by removal of the His tag together with the first 11 residues of FliI(Δ1–7) as judged by N-terminal amino-acid sequencing and MALDI–TOF mass spectrometry (Voyager DE/PRO). The proteolytic product was then purified by affinity chromatography to remove N-terminally His-tagged peptide as well as intact N-His-FliI(Δ1–7) followed by gel-filtration chromatography on a HiLoad 26/60 Superdex 200 prep-grade column. The purity of the product was examined by SDS–PAGE and MALDI–TOF mass spectrometry. Purified proteins were dialyzed overnight against a buffer containing 10 mM Tris–HCl pH 8.5, 10 mM NaCl, 1 mM DTT and were concentrated to 3.0–3.5 mg ml−1.
2.2. Crystallization
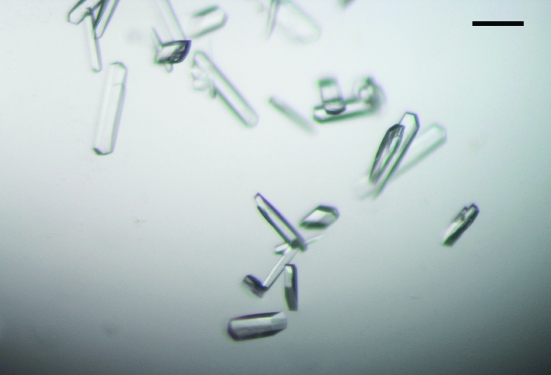
Initial crystallization screening of FliI(Δ1–18) was performed at 293 K by the sitting-drop vapour-diffusion technique using the following screening kits: Wizard I and II, Cryo I and II (Emerald Biostructures) and Crystal Screen I and II (Hampton Research). Drops were prepared by mixing 2 µl protein solution containing 1 mM ADP and 1 mM MgCl2 with 2 µl reservoir solution and were equilibrated against 1 ml reservoir solution. Small crystals of FliI(Δ1–18) were grown from various conditions containing PEG 8000 as a precipitant at pH 8. We optimized the conditions by varying the additives and the concentration of precipitant using the hanging-drop method. Finally, we obtained elongated hexagonal crystals (Fig. 1 ▶) with typical dimensions of 0.2 × 0.05 × 0.02 mm from a solution containing 0.1 M imidazole pH 8.0, 50–100 mM calcium acetate, 4–8%(v/v) PEG 8000 and 4–8%(v/v) 2-propanol at 293 K using the seeding technique.
Figure 1.
Crystals of Salmonella FliI(Δ1–18). Scale bar, 0.1 mm.
Heavy-atom derivatives were prepared by soaking the crystals in a solution containing K2OsCl6 at 50% saturation for 1 d or 10 mM K2PtCl4 for 8 h. The soaking solution included the same precipitant and additives at the same concentration as the reservoir solution but in a different buffer, 0.1 M HEPES pH 8.0, to reduce aggregation of heavy-atom compounds. We also tried to prepare SeMet-labeled crystals, but the purified protein sample was unusable owing to its low solubility.
2.3. Data collection and processing
All X-ray diffraction data were collected at SPring-8 beamline BL41XU. Crystals used for the diffraction experiments were frozen in liquid nitrogen and mounted in a cryo-gas flow. The diffraction patterns from native crystals were recorded on a MAR CCD (MAR Research) detector under a 100 K nitrogen-gas flow. The anomalous diffraction data of derivative crystals were collected on ADSC Quantum 4R CCD detectors (Area Detector Systems Corporation) at 35 or 100 K using a helium-cryocooling device (Rigaku). The diffraction data were indexed, integrated and scaled using the programs MOSFLM (Leslie, 1992 ▶) and SCALA from the CCP4 program suite (Collaborative Computational Project, Number 4, 1994 ▶). The statistics of data collection are summarized in Table 1 ▶.
Table 1. Summary of the data statistics.
Values in parentheses indicate the statistics for the highest resolution shell.
| Native | Pt derivative | Os derivative | |
|---|---|---|---|
| Space group | P21 | P21 | P21 |
| Unit-cell parameters (Å, °) | a = 48.16, b = 72.75, c = 125.74, β = 94.13 | a = 47.68, b = 72.85, c = 126.39, β = 94.04 | a = 47.90, b = 72.86, c = 126.46, β = 93.17 |
| Wavelength (Å) | 1.0000 | 1.0718 | 1.1399 |
| Resolution range (Å) | 2.4 (2.53–2.4) | 2.8 (2.95–2.8) | 2.8 (2.95–2.8) |
| Observations | 128271 (17594) | 81089 (11863) | 77487 (11489) |
| Unique reflections | 33960 (4885) | 21398 (3069) | 21515 (3108) |
| Completeness (%) | 99.6 (98.8) | 99.7 (99.9) | 99.6 (100.0) |
| Redundancy | 3.8 (3.6) | 3.8 (3.9) | 3.6 (3.7) |
| Mean (I)/sd(I) | 16.7 (4.9) | 12.4 (2.6) | 16.3 (4.7) |
| I/σ(I) | 5.1 (3.1) | 5.8 (1.9) | 4.5 (2.6) |
| Rsym (%) | 6.8 (23.8) | 7.0 (37.9) | 6.4 (24.9) |
| Rano (%) | 7.6 (26.3) | 6.1 (15.0) |
3. Results and discussion
Native FliI tends to form insoluble aggregates when concentrated. Therefore, we prepared FliI(Δ1–18), which is a highly soluble variant lacking the first 18 residues. Although the N-terminal residues of FliI are required for oligomerization and interaction with FliH (Lane et al., 2006 ▶; Minamino et al., 2001 ▶), N-terminally truncated variants still retain a significant level of ATP-hydrolyzing activity, although not at the wild-type level (Minamino et al., 2006 ▶).
FliI(Δ1–18) crystals diffracted to 2.4 Å resolution. The native crystals belong to the monoclinic space group P21, with unit-cell parameters a = 48, b = 73, c = 126 Å, β = 94°. The Matthews coefficient (V M; Matthews, 1968 ▶) suggests the presence of two molecules in an asymmetric unit, with a solvent content of 48%. The self-rotation function map calculated using the program POLARRFN (Collaborative Computational Project, Number 4, 1994 ▶) indicated the presence of a twofold non-crystallographic symmetry axis in the crystal ac plane.
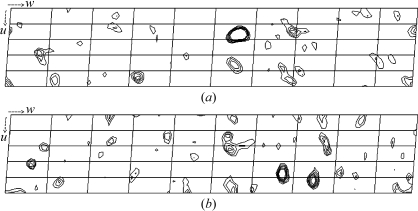
Initially, MAD data collection was attempted for both Os-derivative and Pt-derivative crystals. However, these derivative crystals were highly sensitive to X-ray irradiation. Therefore, we used a helium-cryocooling device to reduce radiation damage and the background of the diffraction image. We were able to collect data at the peak wavelength even though the crystals were still seriously damaged by the strong X-ray beam. Bijvoet difference Patterson maps of the peak-wavelength data from both derivatives showed significant peaks on the Harker section (Fig. 2 ▶) and so did the isomorphous difference Patterson maps, suggesting the usefulness of these data for phasing by the SAD, SIRAS or MIRAS methods.
Figure 2.
Bijvoet difference Patterson maps (v = 0.5 Harker section) at 3.5 Å resolution calculated using the data from the Os derivative (a) and the Pt derivative (b). The contour lines are drawn from 1.5σ to 4.0σ, with an increment of 0.5σ. The height of the highest peak of Os is 9.6σ and that of Pt is 5.2σ.
Acknowledgments
We acknowledge N. Shimizu, M. Kawamoto and K. Hasegawa at SPring-8 for technical help in the use of beamline BL41XU. This work was supported in part by Grants-in-Aid for Scientific Research to KI (16310088) and KN (16087207), and the National Project on Protein Structural and Functional Analyses (to KI) from the Ministry of Education, Science and Culture of Japan, and USPHS grant AI12202 (to RMM).
References
- Akeda, Y. & Galán, J. E. (2005). Nature (London), 473, 911–915. [DOI] [PubMed]
- Auvray, F., Ozin, A. J., Claret, L. & Hughes, C. (2002). J. Mol. Biol.318, 941–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claret, L., Susannah, C. R., Higgins, M. & Hughes, C. (2003). Mol. Microbiol.48, 1349–1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collaborative Computational Project, Number 4 (1994). Acta Cryst. D50, 760–763. [Google Scholar]
- Dreyfus, G., Williams, A. W., Kawagishi, I. & Macnab, R. M. (1993). J. Bacteriol.175, 3131–3138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emerson, S. U., Tokuyasu, K. & Simon, M. I. (1970). Science, 169, 190–192. [DOI] [PubMed] [Google Scholar]
- Fan, F. & Macnab, R. M. (1996). J. Biol. Chem.271, 31981–31988. [DOI] [PubMed] [Google Scholar]
- González-Pedrajo, B., Fraser, G. M., Minamino, T. & Macnab, R. M. (2002). Mol. Microbiol.45, 967–982. [DOI] [PubMed] [Google Scholar]
- Iino, T. (1969). J. Gen. Microbiol.56, 227–239. [DOI] [PubMed] [Google Scholar]
- Lane, M. C., O’Toole, P. W. & Moore, S. A. (2006). J. Biol. Chem.281, 508–517. [DOI] [PubMed] [Google Scholar]
- Leslie, A. G. W. (1992). Jnt CCP4/ESF–EACBM Newsl. Protein Crystallogr.26
- Macnab, R. M. (2003). Annu. Rev. Microbiol.57, 77–100. [DOI] [PubMed] [Google Scholar]
- Macnab, R. M. (2004). Biochim. Biophys. Acta, 1694, 207–217. [DOI] [PubMed] [Google Scholar]
- Matthews, B. W. (1968). J. Mol. Biol.33, 491–497. [DOI] [PubMed] [Google Scholar]
- Minamino, T., Chu, R., Yamaguchi, S. & Macnab, R. M. (2000). J. Bacteriol.182, 4207–4215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minamino, T., Gonzalez-Pedrajo, B., Kihara, M., Namba, K. & Macnab, R. M. (2003). J. Bacteriol.185, 3510–519983-3988. [DOI] [PMC free article] [PubMed]
- Minamino, T., Kazetani, K., Tahara, I., Suzuki, H., Furukawa, Y., Kihara, M. & Namba, K. (2006). J. Mol. Biol.360, 510–519. [DOI] [PubMed] [Google Scholar]
- Minamino, T. & Macnab, R. M. (1999). J. Bacteriol.181, 1388–1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minamino, T. & Macnab, R. M. (2000a). Mol. Microbiol.35, 1052–1064. [DOI] [PubMed] [Google Scholar]
- Minamino, T. & Macnab, R. M. (2000b). Mol. Microbiol.37, 1494–1503. [DOI] [PubMed] [Google Scholar]
- Minamino, T. & Namba, K. (2004). J. Mol. Microbiol. Biotechnol.7, 5–17. [DOI] [PubMed] [Google Scholar]
- Minamino, T., Tame, J. R. H., Namba, K. & Macnab, R. M. (2001). J. Mol. Biol.312, 1027–1036. [DOI] [PubMed] [Google Scholar]
- Thomas, J., Stafford, G. P. & Hughes, C. (2004). Proc. Natl Acad. Sci. USA, 101, 3945–3950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogler, A. P., Homma, M., Irikura, V. M. & Macnab, R. M. (1991). J. Bacteriol.173, 3564–3572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonekura, K., Maki, S., Morgan, D. G., DeRosier, D. J., Vonderviszt, F., Imada, K. & Namba, K. (2000). Science, 290, 2148–2152. [DOI] [PubMed] [Google Scholar]