Crystals of the recombinant native and selenomethionine PilS protein belong to the orthorhombic space group P21212, with unit-cell parameters a = 77.88, b = 114.53, c = 31.75 Å. The structure will be solved using the MAD method.
Keywords: PilS, Salmonella typhi, type IV pilin
Abstract
The structure determination of PilS, a type IV pilin, by X-ray crystallography is reported. The recombinant protein from Salmonella typhi was overexpressed, purified and crystallized. The crystals belong to space group P21212, with unit-cell parameters a = 77.88, b = 114.53, c = 31.75 Å. The selenomethionine derivative of the PilS protein was overexpressed, purified and crystallized in the same space group. Data sets have been collected to 2.1 Å resolution from the selenomethionine-derivative crystal using synchrotron radiation for multiwavelength anomalous dispersion (MAD) phasing.
1. Introduction
Type IV pili are found on many bacterial species and play a crucial role in pathogenesis, as they are required for the attachment of bacteria to target host cells. They may also mediate transformation, modulate target-cell specificity and play a role in twitching motility (Forest & Tainer, 1997 ▶). The type IV pili are homopolymers composed of thousands of copies of the single-chain pilin protein. Pilin assembly into type IV pili is required for virulence by bacterial pathogens that cause diseases such as cholera, typhoid, pneumonia, gonorrhoea and meningitis. All type IV pilins share a conserved hydrophobic N-terminal sequence (∼25 residues), an unusual N-methylated N-terminus, a disulfide bond in the C-terminus and a conserved pilus-assembly machinery. The type IV pilins are divided into two subclasses, type IVa and IVb pilins, based on sequence homology, the length of the leader sequence and the identity of the N-methylated N-terminal residue. While type IVa pili are found on a variety of Gram-negative bacteria having a broad host range that includes humans and other mammals, plants, fungi and even bacteria, type IVb pili have exclusively been identified on bacteria that colonize the human intestine and include Vibrio cholerae, Salmonella enterica serovar Typhi (S. typhi) and the enteropathogenic and enterotoxigenic bacterium Escherichia coli. The type IV pilus of the enteropathogenic bacteria S. typhi is believed to be a major adhesion factor during the entry of this pathogen into gastrointestinal epithelial cells.
Type IV pili are polymeric assemblies of the protein pilin and are involved in a number of cellular functions, including surface motility, microcolony and biofilm formation, host-cell adhesion, cell signalling, DNA uptake by natural transformation and phage attachment. For many Gram-negative pathogens, disruption of the pilus assembly results in severely reduced virulence, which highlights the crucial importance of pili in pathogenicity (Craig et al., 2004 ▶). The crystal structures of the full-length type IVa pilins gonococcal (GC) pilin from Neisseria gonorrhoeae MS11 (Parge et al., 1995 ▶) and PAK pilin from Pseudomonas aeroginosa strain K (Craig et al., 2003 ▶) reveal a highly conserved N-terminal hydrophobic tail that serves as a oligomerization domain for fibre formation. The truncated P. aeroginosa PAK pilin (Hazes et al., 2000 ▶) and K122-4 pilin (Audette et al., 2004 ▶) structures have shown that they can retain the biological characteristics of the intact pilin monomer. Only one crystal structure of a type IVb pilin, Tcp of V. cholerae (Craig et al., 2003 ▶), has been solved so far. NMR structures of the truncated type IVb pilins are now available for S. typhi PilS (Xu et al., 2004 ▶) and enteropathogenic E. coli BFP pilin (Ramboarina et al., 2005 ▶). Comparison of these structures reveals a conserved N-terminal α-helix and a structural core that function in pilus assembly, a structurally variable protein surface and, surprisingly, different protein folds for the type IVb pilins.
The target receptor for the S. typhi pilus is a stretch of ten residues from the first extracellular domain of cystic fibrosis transmembrane conductance regulator (CFTR; Tsui et al., 2003 ▶). The structure of the 26 N-terminal amino-acid truncated type IVb structural pilin monomer (ΔPilS) from S. typhi was determined by NMR (Xu et al., 2004 ▶). However, this monomeric NMR structure does not very clearly establish the loops of ΔPilS because of the small number of NOE restraints. Also, previous work on PilS has predicted that selected charged residues are involved in CFTR binding and several of these residues are missing in the NMR structure. We have primarily undertaken this project in order to obtain more detailed structures of the loops and the charged residues of PilS involved in CFTR binding. Secondly, a cysteine-containing region in the C-terminus is proposed to play a role via the formation of disulfides in CFTR binding and in the assembly of PilS to form the pilus of the bacterium. Details of this domain as well as of the mode of arrangement of PilS that could promote the formation of disulfides are partly elusive in the NMR structure.
In the present study, the ΔPilS protein has been crystallized by the sitting-drop vapour-diffusion method. The structure of this protein is being determined by the multiwavelength anomalous dispersion (MAD) method. When completed, this structure will provide further insight into pilus assembly as well as the potential residues that are essential for receptor binding. Also, we are attempting to learn more about the influence of the disulfide bridge of the C-terminal region in CFTR binding and pilus assembly.
2. Materials and results
2.1. Expression and purification of recombinant PilS
The S. typhi ΔPilS protein was subcloned into the modified pET-32a (Novagen) vector with the S-tag and thioredoxin gene removed and was expressed with an N-terminal hexahistidine tag in E. coli strain BL21 (DE3) as inclusion bodies. The pellet was suspended in ice-cold buffer A (50 mM Tris–HCl pH 8.0, 500 mM NaCl) and subjected to sonication. The crude lysate was centrifuged at 42 400g for 45 min at 277 K and the pellet was resuspended in ice-cold denaturing lysis buffer containing 6 M GnHCl and centrifuged as previously. The supernatant was applied onto Talon resin (Clontech) and allowed to bind overnight at 277 K. Loosely bound contaminant proteins were removed by a wash with buffer A containing 20 mM imidazole. The ΔPilS protein was eluted using buffer A containing 150 mM imidazole. The affinity-purified protein was then refolded by rapid dilution in 30× ice-cold buffer B (50 mM Tris pH 8, 500 mM NaCl). The protein was concentrated using an Amicon cell concentrator (Millipore). The concentrated protein was then cleaved using thrombin (2 units per milligram of protein) and the cleaved protein containing residues 26–181 was then subjected to gel filtration on a Hiload 16/60 Superdex-75 column (Pharmacia) previously equilibrated with a high-salt wash buffer (20 mM Tris pH 8, 500 mM Na2SO4). The purified protein was analysed by SDS–PAGE, DLS and MALDI–TOF to confirm purity and homogeneity and concentrated to 17 mg ml−1.
Selenomethionine ΔPilS was also produced in E. coli strain BL21 (DE3) cells (Doublié, 1997 ▶). A 5 ml culture was grown overnight at 310 K in LB medium containing 100 µg ml−1 ampicillin. The cells were pelleted by centrifugation, resuspended in M9 minimal media, pelleted and resuspended twice. The cell suspension was used to inoculate 1 l pre-warmed (310 K) M9 minimal media supplemented with 0.4% glucose, 100 mg l−1 lysine, phenylalanine and threonine, 50 mg l−1 isoleucine, leucine and valine and 25 mg l−1 SeMet. The culture was grown to an OD600 of 0.6, protein expression was induced by the addition of isopropyl β-d-thiogalactopyranoside to a final concentration of 0.3 mM and the growth of cells was continued at 310 K for 6 h. SeMe-substituted ΔPilS was purified as described earlier for the native protein, except that 2-mercaptoethanol was used at 1 mM in all buffers to avoid oxidation of Se.
2.2. Crystallization, data collection and processing of X-ray diffraction data
Initial crystallization screenings were performed by the hanging-drop and sitting-drop vapour-diffusion techniques with 1 ml reservoir solutions using commercially available crystallization screens. Native ΔPilS was crystallized with condition No. 33 from Hampton Research Crystal Screen 1 (4 M sodium formate; Jancarik & Kim, 1991 ▶). Crystals of average dimensions 0.1 × 0.1 × 0.2 mm appeared within 2–3 d (Fig. 1 ▶). SeMet ΔPilS crystals also grew under identical conditions. A mixture of equal amounts of mineral oil and Paratone was used as cryoprotectant and the crystals were flash-cooled in liquid nitrogen.
Figure 1.
Native ΔPilS crystals.
Data collection on the native ΔPilS crystals was performed at 103 K using an R-AXIS IV imaging-plate detector (Molecular Structure Corporation) at a wavelength of 1.5418 Å. All data were indexed, integrated and scaled using the HKL-2000 suite of programs (Otwinowski & Minor, 1997 ▶). MAD data for the SeMet ΔPilS crystals were collected at synchrotron beamline X12C (National Synchrotron Light Source, USA) using a Brandeis B4 charge-coupled device detector. Three data sets were collected to 2.1 Å resolution from a single crystal using wavelengths selected from the selenium absorption spectrum. Data were collected as a series of 1° oscillation images covering a crystal rotation range of 360°. The crystal mosaicity was determined to be 0.3°. The data-collection statistics are summarized in Table 1 ▶. Currently, we are determining the structure of ΔPilS.
Table 1. Data collection and analysis.
Values in parentheses are for the highest resolution shell (1.97–1.90 Å for the native and 2.18–2.10 Å for the SetMet derivative).
| SeMet derivative | ||||
|---|---|---|---|---|
| Native | Peak | Inflection | Remote | |
| Unit-cell parameters (Å) | a = 77.88, b = 114.53, c = 31.75 | |||
| Space group | P21212 | |||
| Resolution (Å) | 1.9 | 2.1 | 2.1 | 2.1 |
| Wavelength (Å) | 1.5418 | 0.97939 | 0.97978 | 0.95 |
| Unique reflections | 23297 | 17461 | 17522 | 16481 |
| Completeness (%) | 99.0 (98.7) | 99.8 (98.3) | 99.9 (98.9) | 99.9 (99.7) |
| Redundancy | 3.9 (4.3) | 7.7 (6.5) | 7.6 (5.1) | 6.5 (5.6) |
| Rsym† | 0.021 (0.17) | 0.069 (0.29) | 0.056 (0.36) | 0.054 (0.44) |
| I/σ(I) | 25.3 (7.4) | 49.9 (7.0) | 49.1 (3.9) | 32.8 (2.7) |
R
sym = 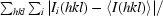
 .
.
3. Discussion
Both the native and selenomethionine ΔPilS proteins crystallize in the orthorhombic form, with unit-cell parameters a = 77.88, b = 114.53, c = 31.75 Å and space group P2 1212. A native data set to 1.9 Å resolution has been collected and crystals of selenomethionine-substituted protein were used to obtain a three-wavelength MAD data set to 2.1 Å. The Matthews coefficient (Matthews, 1968 ▶), self-rotation and native Patterson maps confirm the presence of two molecules in the asymmetric unit, with a V M value of 2.0 Å3 Da−1 and a corresponding solvent content of 37%. Attempts to obtain a molecular-replacement solution using the NMR model (Xu et al., 2004 ▶) did not yield any successful solutions. We are now solving the structure using the MAD method.
PilS forms a dimer in the asymmetric unit (a proposed prerequisite for CFTR binding). Furthermore, several proteins are monomers in solution but in their crystal structures very clearly demonstrate the mechanism of their assembly to form filaments or multimers (Yuan et al., 2003 ▶). As PilS exists as a dimer in the asymmetric unit, we expect that the dimer structure might shed further light on CFTR binding. Also, we expect that the crystal structure will show details of disulfides that might influence the assembly of PilS into the pilus. Understanding of the subunit structure and assembly architecture that produces these extraordinary filaments is crucial for understanding pilus functions and for designing vaccines and therapeutics that are directed towards blocking the S. typhi–CFTR interaction.
References
- Audette, G. F., Irvin, R. T. & Hazes, B. (2004). Biochemistry, 43, 11427–11435. [DOI] [PubMed] [Google Scholar]
- Craig, L., Taylor, R. K., Pique, M. E., Adair, B. D., Arvai, A. S., Singh, M., Lloyd, S. J., Shin, D. S., Getzoff, E. D., Yeager, M., Forrest, K. T. & Tainer, J. A. (2003). Mol. Cell, 11, 1139–1150. [DOI] [PubMed] [Google Scholar]
- Craig, L., Pique, M. E. & Tainer, J. A. (2004). Nature Rev. Microbiol.2, 363–378. [DOI] [PubMed]
- Doublié, S. (1997). Methods Enzymol.276, 523–530. [PubMed] [Google Scholar]
- Forest, K. T. & Tainer, J. A. (1997). Gene, 192, 165–169. [DOI] [PubMed] [Google Scholar]
- Hazes, B., Sastry, P. A., Hayakawa, K., Read, R. J. & Irvin, R. T. (2000). J. Mol. Biol.299, 1005–1017. [DOI] [PubMed] [Google Scholar]
- Jancarik, J. & Kim, S.-H. (1991). J. Appl. Cryst.24, 409–411. [Google Scholar]
- Matthews, B. W. (1968). J. Mol. Biol.33, 491–497. [DOI] [PubMed] [Google Scholar]
- Parge, H. E., Forest, K. T., Hickey, M. J., Christensen, D. A., Getzoff, E. D. & Tainer, J. A. (1995). Nature (London), 378, 32–38. [DOI] [PubMed] [Google Scholar]
- Ramboarina, S., Fernandes, P. J., Daniell, S., Islam, S., Simpson, P., Frankel, G., Booy, F., Donnenberg, M. S. & Matthews, S. (2005). J. Biol. Chem.280, 40252–40260. [DOI] [PubMed] [Google Scholar]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol.276, 307–326. [DOI] [PubMed]
- Tsui, S. M. I., Yip, M. C. C., Hacket, J. & Morris, C. (2003). Infect. Immun.71, 6049–6050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, X. F., Tan, Y. W., Lam, L., Hackett, J., Zhang, M. & Mok, Y. K. (2004). J. Biol. Chem.279, 31599–31605. [DOI] [PubMed] [Google Scholar]
- Yuan, P., Jedd, G., Kumaran, D., Swaminathan, S., Shio, H., Hewitt, D., Chua, N. H. & Swaminathan, K. (2003). Nature Struct. Biol.10, 264–270. [DOI] [PubMed] [Google Scholar]