Abstract
The region around position 1067 in domain II of 23S rRNA frequently is referred to as the GTPase center of the ribosome. The notion is based on the observation that the binding of the antibiotic thiostrepton to this region inhibited GTP hydrolysis by elongation factor G (EF-G) on the ribosome at the conditions of multiple turnover. In the present work, we have reanalyzed the mechanism of action of thiostrepton. Results obtained by biochemical and fast kinetic techniques show that thiostrepton binding to the ribosome does not interfere with factor binding or with single-round GTP hydrolysis. Rather, the antibiotic inhibits the function of EF-G in subsequent steps, including release of inorganic phosphate from EF-G after GTP hydrolysis, tRNA translocation, and the dissociation of the factor from the ribosome, thereby inhibiting the turnover reaction. Structurally, thiostrepton interferes with EF-G footprints in the α-sarcin stem loop (A2660, A2662) located in domain VI of 23S rRNA. The results indicate that thiostrepton inhibits a structural transition of the 1067 region of 23S rRNA that is important for functions of EF-G after GTP hydrolysis.
Several translational factors of Escherichia coli, including initiation factor 2, elongation factors (EF) Tu and G, and release factor 3, are GTPases that hydrolyze GTP at some point during their function on the ribosome. The same, or similar, contacts of the factors with the large ribosomal subunit appear to be important for factor binding and/or stimulation of GTP hydrolysis. In 23S rRNA, contact regions of elongation factors Tu and G have been identified both in the α-sarcin stem loop around residue G2661 (EF-Tu, EF-G) (1) and in the region around residue A1067 (EF-G) (2), where protein L11 is bound (3). The L7/L12 stalk provides another functionally important contact (4). The 1067 region is the binding site of the peptide antibiotic thiostrepton (5–10) that has been reported to interfere with the function of both elongation factors on the ribosome, notably their GTPase activity (reviewed in ref. 11). Therefore, the 1067 region frequently is referred to as the GTPase center of the ribosome.
Contrary to the generally accepted notion, we report here that GTP hydrolysis by EF-G on the ribosome is not affected by thiostrepton, whereas the steps after GTP hydrolysis, including release of phosphate (Pi), tRNA translocation, and the dissociation of EF-G⋅GDP from the ribosome, are strongly inhibited. We conclude that the 1067 region of 23S rRNA is not necessarily involved in the activation of the factor’s GTPase. Rather, our results suggest that thiostrepton, by binding to the 1067 region, inhibits conformational rearrangements of the EF-G–ribosome complex related to steps after GTP hydrolysis, including phosphate release, translocation, and turnover of EF-G.
MATERIALS AND METHODS
Materials.
70S ribosomes from E. coli MRE 600 and EF-Tu from E. coli K12 were prepared as described (12). EF-G was expressed in E. coli JM109 from plasmid pTZfus (13). Cells were lysed with lysozyme (5 mg/g wet cells) in buffer A (50 mM Tris⋅HCl, pH 7.5/10 mM MgCl2/0.5 mM EDTA/6 mM 2-mercaptoethanol) in the presence of 100 μM PMSF and 30 μM GDP, sodium deoxycholate (12.5 mg/g cells), and DNase I. The first step of EF-G purification was chromatography on DEAE-Sepharose CL6B (Pharmacia) by using a 0- to 0.35-M KCl gradient in buffer A. Fractions containing EF-G were subjected to FPLC on Superdex 75 HiLoad (Pharmacia) by using buffer A containing 10% glycerol and 100 μM PMSF and, subsequently, to chromatography on MonoQ (Pharmacia) by using a 0- to 0.35-M KCl gradient in buffer A with 10% glycerol. EF-G was concentrated and stored in buffer B (50 mM Tris⋅HCl, pH 7.5/70 mM NH4Cl/30 mM KCl/7 mM MgCl2/1 mM DTT) with 50% glycerol.
Initiation factors were isolated from overproducing strains provided by C. Gualerzi (University of Camerino, Italy). Cells were opened by addition of lysozyme (5 mg/g wet cells), sodium deoxycholate (12.5 mg/g), and DNase I in buffer C (20 mM Tris⋅HCl, pH 7.7/60 mM NH4Cl/10 mM MgCl2/5 mM 2-mercaptoethanol/0.1 mM PMSF). To wash initiation factors off the ribosomes, 1 M KCl was added to the S30 fraction and incubated for 10 min, and the ribosomes were removed by centrifugation at 100,000 × g. From the S100 extract, initiation factors were purified to homogeneity by Fast-Flow LC on S-Sepharose by using a gradient of 0.2–0.8 M KCl in 50 mM Mops, pH 6.4/0.25 mM MgCl2/0.05 mM EDTA/5 mM 2-mercaptoethanol/0.1 mM PMSF (14).
Ribosomes were programmed with MFTI-mRNA, which comprised 120 nt and contained a ribosomal-binding site and the coding sequence Met-Phe-Thr-Ile. The plasmid pXR022 coding for MFTI-mRNA (15) was provided by C. Gualerzi. The plasmid was linearized by HindIII, and mRNA was prepared by run-off transcription with T7 RNA polymerase and purified by FPLC on MonoQ.
f[3H]Met-tRNAfMet was prepared and purified as described (16). [14C]Phe-tRNAPhe from E. coli was purified by HPLC on an RP18 column (LiChrospher WP300; Merck) by using a gradient of 0–20% ethanol in 20 mM ammonium acetate, pH 5.5/10 mM magnesium acetate/0.4 M NaCl.
Mutant phosphate-binding protein (PBP) from E. coli containing Cys at position 197 was expressed from a plasmid construct provided by M. Webb (National Institute of Medical Research, London), labeled with N-[2-(1-maleimidyl)ethyl]-7-(diethylamino)coumarin-3-carboxamide (MDCC; Molecular Probes), and purified according to Brune et al. (17).
Biochemical Assays.
[γ-32P]GTP hydrolysis was measured either by extraction (quench–flow samples) or by TLC. For extraction, 50-μl samples were quenched by adding an equal volume of either 40% formic acid, and analyzed by TLC (Fig. 1), or 1 M HClO4/3 mM potassium phosphate, and [32P]phosphate was determined after extraction into isopropyl acetate in the presence of sodium molybdate (12) (Fig. 2). TLC was performed on polyethyleneimine-cellulose in 0.5 M potassium phosphate, pH 3.5. Quench–flow experiments were performed at 37°C as described (18) by using a KinTek quench–flow apparatus.
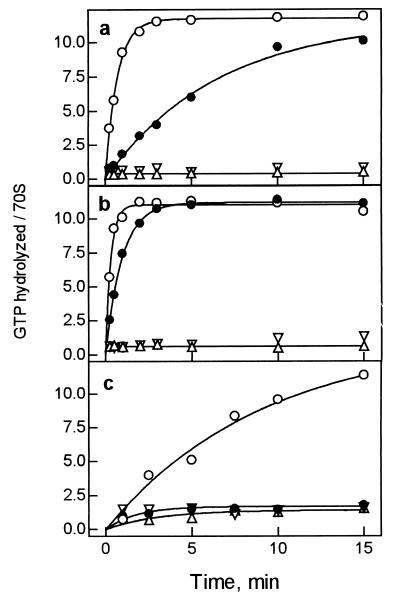
Figure 1.
Effect of thiostrepton on EF-G-dependent GTP hydrolysis on the ribosome. (a) Incubation at 20°C. The reaction was performed with EF-G (0.4 μM), [γ-32P]GTP (5 μM), and vacant ribosomes (0.4 μM) with or without thiostrepton (100 μM). (b) Incubation at 37°C, otherwise as in a. (c) Incubation at 37°C with catalytic amounts of EF-G (0.02 μM) and ribosomes (0.2 μM), otherwise as in a. ○, In the absence of antibiotic; ●, in the presence of thiostrepton; ▵, control without EF-G; ▿, control without ribosomes. Reactions were analyzed by TLC.
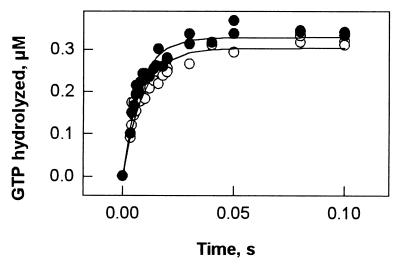
Figure 2.
Rapid kinetics of EF-G-dependent GTP hydrolysis on the ribosome. Quench–flow experiments were performed at 37°C by rapidly mixing solutions (15 μl each) of EF-G preincubated with [γ-32P]GTP and of ribosomes preincubated without (○) or with (●) thiostrepton; final concentrations after mixing were EF-G, 0.8 μM; [γ-32P]GTP, 5 μM; ribosomes, 0.8 μM; thiostrepton, 100 μM. Samples were quenched with 0.8 M HClO4/2.4 mM potassium phosphate, and 32Pi was determined after extraction (Materials and Methods).
Pi release was monitored by the fluorescence change of PBP-MDCC (17) in a stopped-flow apparatus (Applied Photophysics, Surrey, U.K.). The fluorescence of MDCC was excited at 425 nm and monitored after passing a KV450 filter (Schott, Mainz, Germany). To minimize phosphate contaminations, all solutions and the stopped-flow apparatus were preincubated with 200 μM 7-methylguanosine and 0.1 units/ml purine nucleoside phosphorylase (“Pi mop”) (17).
To prepare 70S initiation complexes, ribosomes (0.5 μM) were programmed with 3-fold excess of MFTI-mRNA in the presence of 1.5-fold excess of IF1, IF2, IF3, and f[3H]Met-tRNAfMet and 1 mM GTP in buffer B for 30 min at 37°C. Ternary complex, EF-Tu⋅GTP⋅[14C]Phe-tRNAPhe, was prepared by incubating 1 μM EF-Tu with 1 mM GTP/3 mM phosphoenol pyruvate/0.5 mg/liter pyruvate kinase/0.75 μM [14C]Phe-tRNAPhe for 15 min at 37°C. Ternary complex was added to the initiation complex and incubated for 1 min at 37°C to form pretranslocation complexes. Equal volumes (20 μl each) of 250 μM thiostrepton solution in 5% DMSO, or of 5% DMSO in buffer, and 0.5 μM pretranslocation complex were mixed and incubated for 3 min at 37°C. The amount of [14C]Phe-tRNAPhe or f[3H]Met-tRNAfMet bound to ribosomes was determined by nitrocellulose filtration by directly applying aliquots of the reaction mixture to the filters (Sartorius) and subsequent washing with buffer B. Filters were dissolved and radioactivity was measured in QS361 scintillation cocktail. To induce translocation, EF-G, preincubated with 1 mM GTP for 15 min at 37°C, was added to the pretranslocation complex in 10 μl. The extent of translocation was measured by the formation of fMet-Phe-puromycin (1 mM puromycin, 10 s at 37°C; ref. 18). Control experiments showed that the puromycin reaction was unaffected, that is, complete within 10 s, when EF-G remained bound to the posttranslocation complex because of the presence of thiostrepton (data not shown).
Time courses of fMet-Phe-tRNAPhe dissociation from the pretranslocation complex, measured by nitrocellulose filtration, were evaluated on the basis of a single-step reversible mechanism; fitting was performed by numerical integration by using scientist for Windows software (MicroMath Scientific Software, Salt Lake City). Spontaneous translocation of peptidyl-tRNA (maximum 20%) was measured in parallel and taken into account. The values of koff and kon obtained were used to calculate Kd.
To study the footprint of EF-G on the α-sarcin loop, 0.4 μM ribosomes were incubated with 1.2 μM EF-G and 1 mM GTP in the absence or presence of 100 μM thiostrepton/2% DMSO and/or 200 μM fusidic acid in 20 mM Hepes, pH 7.5/30 mM potassium acetate/70 mM ammonium acetate/7 mM magnesium acetate for 5 min at 37°C, followed by the addition of dimethyl sulfate and further incubation for 10 min at 37°C. Methylated sites in the α-sarcin region were determined by primer extension with avian myeloblastosis virus reverse transcriptase (19) by using an oligodeoxyribonucleotide primer complementary to positions 2721–2740 of 23S rRNA. For quantitative analysis, autoradiographic films were scanned densitometrically.
Because thiostrepton was added together with DMSO (2% final concentration), controls of all assays were performed with DMSO alone; in no case did DMSO have any effect.
RESULTS
Effect of Thiostrepton on the GTPase Activity of EF-G on the Ribosome.
The effect of thiostrepton on ribosome-stimulated GTP hydrolysis by EF-G was studied with vacant ribosomes at 20° and 37°C (Fig. 1 a and b). In the absence of antibiotic, GTP is hydrolyzed rapidly in the presence of stoichiometric amounts of EF-G; because of multiple turnover of the factor, about 90% of all GTP present is hydrolyzed within 3 min. In the presence of thiostrepton, the turnover is strongly inhibited, whereas the full extent of GTP hydrolysis is still observed (Fig. 1b); single-round GTP hydrolysis is not resolved in this time range. At the conditions used in previous reports (20, 21), that is, when catalytic amounts of EF-G were used, the addition of thiostrepton suppressed GTP hydrolysis practically completely (Fig. 1c), hence, the notion that thiostrepton inhibited GTP hydrolysis by EF-G on the ribosome.
Rapid, single-round GTP hydrolysis by EF-G on the ribosome was monitored by the quench–flow technique. As shown in Fig. 2, the addition of thiostrepton affected neither the rate (about 60 s−1, 37°C) nor the amount of GTP hydrolyzed in the first round (more than 0.3 μM, or about 0.8 per ribosome). Thus, the antibiotic does not affect ribosome binding of EF-G⋅GTP and single-round GTP hydrolysis, whereas it strongly inhibits the turnover of EF-G.
Thiostrepton Inhibits Pi Release from EF-G after GTP Hydrolysis.
Pi release was studied according to the method of Brune et al. (17) by using a fluorescent derivative of PBP from E. coli, PBP-MDCC. Binding of Pi to MDCC-labeled PBP is rapid (kon = 108 M−1⋅s−1) and tight (Kd = 0.1 μM), and the formation of the complex strongly increases the fluorescence of MDCC (17).
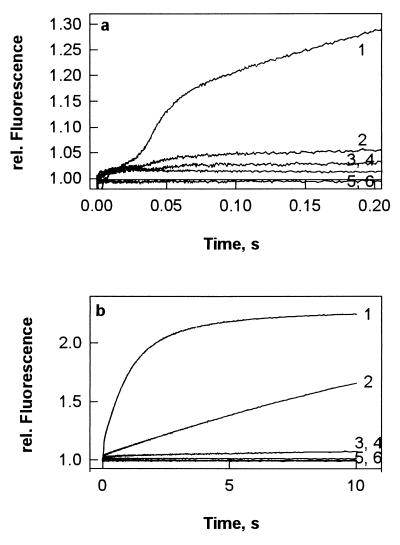
This experimental approach was used to monitor Pi release from EF-G after GTP hydrolysis on vacant ribosomes. After a short lag phase, which probably represents the time required for GTP hydrolysis, a rapid increase of MDCC fluorescence was observed (Fig. 3a), corresponding to Pi dissociation after the first round of GTP hydrolysis, followed by a further slower increase because of multiple rounds of GTP hydrolysis (Fig. 3b). In the presence of thiostrepton, the rapid burst of Pi release was practically completely suppressed (Fig. 3a, trace 2), although, as shown above (cf. Fig. 2), single-round GTP cleavage was not affected. In the presence of thiostrepton, the rate of Pi release in the turnover reaction (Fig. 3b, trace 2) was comparable to the one observed for GTP hydrolysis (cf. Fig. 1a).
Figure 3.
Rapid kinetics of Pi release from EF-G after GTP hydrolysis on the ribosome displayed in short (a) and long (b) time windows. EF-G (0.8 μM) was preincubated with 25 μM GTP for 15 min at 37°C, and then PBP-MDCC (5 μM) was added; ribosome solutions (0.8 μM) were prepared without or with thiostrepton (200 μM). Equal volumes (60 μl) of EF-G/PBP-MDCC (or PBP-MDCC alone) and ribosomes (or buffer) were mixed rapidly in a stopped-flow apparatus in the absence (1) or presence (2) of thiostrepton. Controls were performed without EF-G and ribosomes in the absence (3) or presence (4) of thiostrepton, with EF-G and without ribosomes (5), and with ribosomes and without EF-G (6).
Inhibition of EF-G-Dependent Translocation by Thiostrepton.
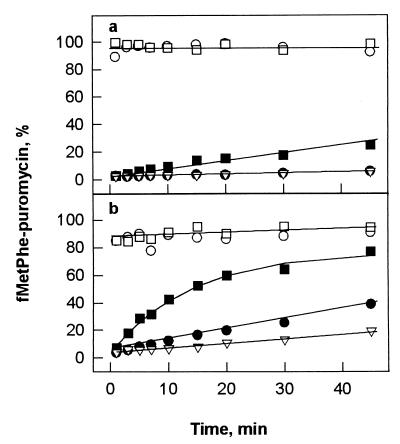
For assaying the effect of thiostrepton on EF-G-dependent translocation, pretranslocation ribosomes were used that were programmed with MFT-mRNA (coding for fMet-Phe-Thr) and carried tRNAfMet and fMet-Phe-tRNAPhe in P and A sites, respectively. Translocation was monitored by the appearance of puromycin-reactive fMet-Phe-tRNAPhe in the P site. Upon addition of EF-G⋅GTP, translocation is unresolvably fast and practically complete (more than 85%) within 10 s both at single-round and multiple-turnover conditions, independently of the temperature (Fig. 4). Thiostrepton inhibits the single-round reaction to different extents at 20° and 37°C, whereas the turnover reaction is blocked practically completely at 20°C and strongly inhibited at 37°C. In the presence of excess EF-G (0.6 μM) at 37°C, the rate of translocation in the presence of thiostrepton (1.2 × 10−3⋅s−1) is about 104 times lower than in the absence of antibiotic (15 s−1, data not shown; measured by stopped flow as in ref. 18). This behavior indicates that thiostrepton strongly inhibits both the translocation of the tRNAs and the subsequent dissociation of EF-G from the ribosome. The latter effect is in keeping with the result of the GTPase experiments with vacant ribosomes (cf. Fig. 1).
Figure 4.
Effect of thiostrepton on EF-G-dependent translocation at 20°C (a) and 37°C (b). Pretranslocation complexes (0.2 μM) were incubated with 0.04 μM (●, ○) or 0.6 μM (■, □) EF-G⋅GTP in the absence (○, □) or presence (●, ■) of 100 μM thiostrepton. ▿, No EF-G added.
Stabilization of Peptidyl-tRNA on the Ribosome by Thiostrepton.
In the pretranslocation complex, peptidyl-tRNA and deacylated tRNA occupy the hybrid A/P* and P/E states, respectively (22, 23). The observed inhibition of translocation by thiostrepton then may, among other factors, be caused by stabilization of the A/P* state of peptidyl-tRNA. To investigate this possibility, we measured the dissociation of fMet-Phe-tRNAPhe from the A site of pretranslocation ribosomes.
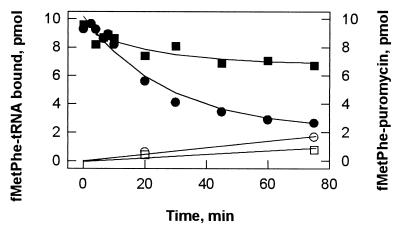
In the absence of thiostrepton, fMet-Phe-tRNAPhe readily dissociates (Fig. 5). The reaction is reversible in that rebinding of released peptidyl-tRNA can be induced by increasing the Mg2+ concentration or by adding polyamines (data not shown); thus, the final level of binding is determined by the equilibrium binding of peptidyl-tRNA to the A site. By evaluating the time course of dissociation using numerical integration (see Materials and Methods), dissociation and association rate constants were estimated. Dissociation constants, Kd, of the complex are 1 μM in the absence and 0.02 μM in the presence of thiostrepton, indicating that the antibiotic stabilizes the binding of fMet-Phe-tRNAPhe to the ribosome about 50-fold (Fig. 5). Hence, the direct effect of thiostrepton in stabilizing peptidyl-tRNA in the pretranslocation A/P* state is small compared with the observed 104-fold inhibition of translocation.
Figure 5.
Dissociation of fMet-Phe-tRNAPhe from the ribosomal A site. Pretranslocation complex (0.2 μM) was incubated without any addition (●) or in the presence of 100 μM thiostrepton (■). ○ and □, Puromycin reaction.
Thiostrepton Inhibits EF-G Footprints on the α-Sarcin Stem Loop.
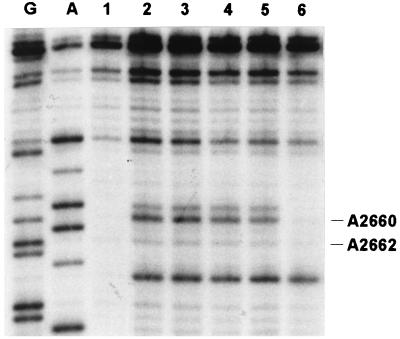
EF-G binding to the ribosome is labile in both GTP- and GDP-bound forms. The complex is stabilized by the antibiotic fusidic acid (24, 25) that binds to EF-G⋅GDP immediately after GTP hydrolysis on the ribosome and inhibits a conformational transition necessary for the dissociation of the factor from the ribosome. In the EF-G–ribosome complex stabilized by fusidic acid, two characteristic sites of presumed EF-G contacts with the ribosome have been identified by footprinting analysis, that is, the 1067 region and the α-sarcin stem loop of 23S rRNA (1). Thiostrepton binding to the 1067 region results in strong DMS footprints; therefore, although it is likely that the antibiotic inhibits the contact of EF-G with the 1067 region, the effect cannot be studied by footprinting. It is possible, however, to study the α-sarcin region that is not affected directly by thiostrepton. To see whether thiostrepton binding to the 1067 region influences the effect of EF-G on the α-sarcin stem loop, we have performed DMS modification experiments with vacant ribosomes (Fig. 6). In the presence of thiostrepton, no footprint of EF-G was found in the α-sarcin stem loop, although, as demonstrated by the inhibition of turnover (compare Figs. 1–3), EF-G was bound to the ribosome. Moreover, no footprint was observed when fusidic acid was present together with thiostrepton. This indicates that the structure of the EF-G–ribosome complex stabilized by thiostrepton differs from the complex stabilized by fusidic acid.
Figure 6.
Effect of thiostrepton on EF-G-dependent protection of α-sarcin stem loop from DMS modification. Lanes: G and A, sequencing lanes; 1, control without DMS modification; 2, ribosomes modified with DMS; 3, ribosomes modified with DMS in the presence of thiostrepton and fusidic acid; 4, ribosomes modified with DMS in the presence of thiostrepton and EF-G⋅GTP; 5, ribosomes modified with DMS in the presence of thiostrepton, fusidic acid, and EF-G⋅GTP; 6, ribosomes modified with DMS in the presence of fusidic acid and EF-G⋅GTP.
DISCUSSION
The 1067 Region of 23S rRNA and GTPase Activation.
The 1067 region of 23S rRNA and the proteins bound to it form an important functional center of ribosome. The binding of thiostrepton to that region has been reported to perturb GTP hydrolysis by both EF-Tu and EF-G (20, 26) as well as to prevent the formation of the fusidic acid-stabilized ribosome complex of EF-G with various nucleotides bound to it (27–29). To reconcile these findings, it was proposed that thiostrepton destabilized the EF-G–ribosome complex such that the dissociation would be too rapid to allow for GTP hydrolysis and the complex would be too labile to be detected by nonequilibrium methods (30).
Our data are inconsistent with that model. Binding of EF-G⋅GTP to the ribosome is not affected by thiostrepton, because a marked change in the fluorescence of peptidyl-tRNA bound to the A site is observed in the presence or absence of the antibiotic (18). Furthermore, as shown here, a single round of EF-G-dependent GTP hydrolysis on the ribosome is not affected by thiostrepton. Previously, the inhibition of EF-G function by thiostrepton was measured at conditions of multiple turnover (20, 21); single-round hydrolysis experiments were not performed. Thus, our present observation of inhibition of turnover is consistent with the previous data, but not with the interpretation put forward at the time. Our results clearly show that thiostrepton, rather than interfering with factor binding and GTP hydrolysis, inhibits the function of EF-G in subsequent steps, i.e., phosphate release, translocation, and the subsequent release of EF-G from the ribosome.
The results indicate that thiostrepton slows down EF-G release, rather than freezing the factor on the ribosome, as fusidic acid does. The extent of inhibition depends on assay conditions, such as temperature (this paper) or ionic conditions (31). This possibly is the reason why the EF-G–ribosome complex containing thiostrepton could not be isolated by conventional nonequilibrium techniques (27, 28, 32, 33).
Thiostrepton and Fusidic Acid Stabilize Different States of EF-G on the Ribosome.
Thiostrepton was shown to interfere with the stabilization of the EF-G⋅GDP–ribosome complex by fusidic acid, and binding of EF-G⋅GDP to the ribosome in the presence of fusidic acid prevented the binding of thiostrepton (28). Likewise, we have found that thiostrepton inhibits the characteristic EF-G-specific protection of bases in the α-sarcin stem loop from DMS modification. Thus, the nucleotide-binding properties and the structural arrangement of EF-G on the ribosome are different in the complexes stabilized by thiostrepton or by fusidic acid, suggesting that different states of the EF-G–ribosome complex are stabilized by the two antibiotics. Because fusidic acid does not affect single-round translocation and is likely to inhibit a structural transition of EF-G immediately before the dissociation of the factor from the ribosome, the respective complex probably represents a late intermediate in the reaction pathway whereas thiostrepton appears to stabilize an earlier intermediate.
Mechanism of Thiostrepton Action on the Ribosome.
Binding of thiostrepton to the 1067 region of 23S rRNA strongly inhibits the functions of the EF-G–ribosome complex after GTP hydrolysis, that is, Pi release, translocation, and subsequent release of EF-G from the ribosome. A possible scenario to explain the inhibitory effect of thiostrepton would be that a conformational change of EF-G is inhibited and, therefore, the subsequent steps cannot take place. Because thiostrepton binds to the ribosome and not to EF-G, the effect on EF-G probably is indirect and caused by an inhibition of a structural change of the ribosome that is coupled to EF-G.
The tertiary structure of the thiostrepton-binding region around position 1067 of 23S rRNA is relatively unstable, suggesting that it may undergo conformational transitions during elongation (34, 35). The region is the binding site of protein L11, the binding of which stabilizes the tertiary structure of the region (10) and increases the affinity of thiostrepton binding (26). The crystal structures of a 46-nt RNA fragment of the 1067 region bound to L11 (36) or the C-terminal domain of L11 (37) show a compact structure of the RNA; a cleft between the RNA and the protein may constitute the binding site of thiostrepton. Mutations in L11 that confer resistance to thiostrepton do not affect the binding of the antibiotic, indicating that the role of L11 is to modulate the structural flexibility of the rRNA region to which it is bound (38). These findings suggest that conformational transitions in that region of 23S rRNA are functionally important and that thiostrepton may interfere with these transitions by stabilizing a conformation, which may the one prevailing in the complex with L11 (39, 40). Other functionally related regions of the ribosome, such as the L10-(L7/12)2 complex forming the stalk of the 50S subunit (9) and the α-sarcin stem loop (41), may be involved as well.
Interestingly, a very similar inhibition pattern, that is, a strong inhibition of both translocation and turnover, was observed with truncated EF-G that lacked domain 4 (18). This parallel supports the view that structural rearrangements involved in both translocation and subsequent dissociation of EF-G are coupled rearrangements of both the factor and the ribosome. The inhibition by thiostrepton of EF-G turnover is consistent with the observation that thiostrepton in vivo seems to inhibit the EF-Tu-dependent A-site binding of aa-tRNA (42). EF-Tu and EF-G bind to overlapping sites on the ribosome, because the binding of the factors is mutually exclusive. Thus, the inhibition of EF-G dissociation from the ribosome after GTP hydrolysis and translocation is likely to interfere with the binding of the ternary complex EF-Tu⋅GTP⋅aa-tRNA to the A site, thereby decreasing the efficiency of translation.
Acknowledgments
We thank Martin Webb for the E. coli strain overproducing mutant PBP, Claudio Gualerzi for strains overproducing initiation factors and for mRNA constructs, and Petra Striebeck for expert technical assistance. The work was supported by the Deutsche Forschungsgemeinschaft (Wi 626/10-2), the Volkswagen-Stiftung, the Alfried Krupp von Bohlen und Halbach-Stiftung, and the Fonds der Chemischen Industrie.
ABBREVIATIONS
- EF-G
elongation factor G
- PBP
phosphate-binding protein
- MDCC
N-[2-(1-maleimidyl)ethyl]-7-(diethylamino)coumarin-3-carboxamide
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
References
- 1.Moazed D, Robertson J M, Noller H F. Nature (London) 1988;334:362–364. doi: 10.1038/334362a0. [DOI] [PubMed] [Google Scholar]
- 2.Sköld S E. Nucleic Acids Res. 1983;11:4923–4932. doi: 10.1093/nar/11.14.4923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Egebjerg J, Douthwaite S R, Liljas A, Garrett R A. J Mol Biol. 1990;213:275–288. doi: 10.1016/S0022-2836(05)80190-1. [DOI] [PubMed] [Google Scholar]
- 4.Traut R R, Dey D, Bochkariov D E, Oleinikov A V, Jokhadze G G, Hamman B, Jameson D. Biochem Cell Biol. 1995;73:949–958. doi: 10.1139/o95-102. [DOI] [PubMed] [Google Scholar]
- 5.Egebjerg J, Douthwaite S, Garrett R A. EMBO J. 1989;8:607–611. doi: 10.1002/j.1460-2075.1989.tb03415.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ryan P C, Lu M, Draper D E. J Mol Biol. 1991;221:1257–1268. doi: 10.1016/0022-2836(91)90932-v. [DOI] [PubMed] [Google Scholar]
- 7.Thompson J, Cundliffe E. Biochimie. 1991;73:1131–1135. doi: 10.1016/0300-9084(91)90156-u. [DOI] [PubMed] [Google Scholar]
- 8.Thompson J, Musters W, Cundliffe E, Dahlberg A E. EMBO J. 1993;12:1499–1504. doi: 10.1002/j.1460-2075.1993.tb05793.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rosendahl G, Douthwaite S. J Mol Biol. 1993;234:1013–1020. doi: 10.1006/jmbi.1993.1655. [DOI] [PubMed] [Google Scholar]
- 10.Xing Y, Draper D E. J Mol Biol. 1995;249:319–331. doi: 10.1006/jmbi.1995.0299. [DOI] [PubMed] [Google Scholar]
- 11.Gale E F, Cundliffe E, Reynolds P E, Richmond M H, Waring M. The Molecular Basis of Antibiotic Action. London: Wiley; 1981. pp. 402–457. [Google Scholar]
- 12.Rodnina M V, Wintermeyer W. Proc Natl Acad Sci USA. 1995;92:1945–1949. doi: 10.1073/pnas.92.6.1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Borowski C, Rodnina M V, Wintermeyer W. Proc Natl Acad Sci USA. 1996;93:4202–4206. doi: 10.1073/pnas.93.9.4202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Soffientini A, Lorenzetti R, Gastaldo L, Parlett J H, Spurio R, La Teana A, Islam K. Protein Expression Purif. 1994;5:118–124. doi: 10.1006/prep.1994.1018. [DOI] [PubMed] [Google Scholar]
- 15.Calogero R A, Pon C L, Canonaco M A, Gualerzi C O. Proc Natl Acad Sci USA. 1988;85:6427–6431. doi: 10.1073/pnas.85.17.6427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rodnina M V, Semenkov Y P, Wintermeyer W. Anal Biochem. 1994;219:380–381. doi: 10.1006/abio.1994.1282. [DOI] [PubMed] [Google Scholar]
- 17.Brune M, Hunter J L, Corrie J E, Webb M R. Biochemistry. 1994;33:8262–8271. doi: 10.1021/bi00193a013. [DOI] [PubMed] [Google Scholar]
- 18.Rodnina M V, Savelsbergh A, Katunin V I, Wintermeyer W. Nature (London) 1997;385:37–41. doi: 10.1038/385037a0. [DOI] [PubMed] [Google Scholar]
- 19.Stern S, Moazed D, Noller H F. Methods Enzymol. 1988;164:481–489. doi: 10.1016/s0076-6879(88)64064-x. [DOI] [PubMed] [Google Scholar]
- 20.Pestka S. Biochem Biophys Res Commun. 1970;40:667–674. doi: 10.1016/0006-291x(70)90956-3. [DOI] [PubMed] [Google Scholar]
- 21.Naaktgeboren N, Roobol K, Gubbens J, Voorma H O. Eur J Biochem. 1976;70:39–47. doi: 10.1111/j.1432-1033.1976.tb10953.x. [DOI] [PubMed] [Google Scholar]
- 22.Moazed D, Noller H F. Nature (London) 1989;342:142–148. doi: 10.1038/342142a0. [DOI] [PubMed] [Google Scholar]
- 23.Wilson K S, Noller H F. Cell. 1998;92:337–349. doi: 10.1016/s0092-8674(00)80927-7. [DOI] [PubMed] [Google Scholar]
- 24.Baca O G, Rohrbach M S, Bodley J W. Biochemistry. 1976;15:4570–4574. doi: 10.1021/bi00666a004. [DOI] [PubMed] [Google Scholar]
- 25.Brot N. In: Molecular Mechanisms of Protein Synthesis. Weissbach H, Pestka S, editors. New York: Academic; 1977. pp. 375–411. [Google Scholar]
- 26.Cundliffe E. In: Structure, Function and Genetics of Ribosomes. Hardesty B, Kramer G, editors. New York: Springer; 1986. pp. 586–604. [Google Scholar]
- 27.Bodley J W, Lin L, Highland J H. Biochem Biophys Res Commun. 1970;41:1406–1411. doi: 10.1016/0006-291x(70)90543-7. [DOI] [PubMed] [Google Scholar]
- 28.Highland J H, Lin L, Bodley J W. Biochemistry. 1971;10:4404–4409. doi: 10.1021/bi00800a009. [DOI] [PubMed] [Google Scholar]
- 29.Cundliffe E, Thompson J. Eur J Biochem. 1981;118:47–52. doi: 10.1111/j.1432-1033.1981.tb05484.x. [DOI] [PubMed] [Google Scholar]
- 30.Thompson J. In: Ribosomal RNA. Structure, Evolution, Processing, and Function in Protein Biosynthesis. Zimmermann R A, Dahlberg A E, editors. Boca Raton, FL: CRC; 1995. pp. 311–325. [Google Scholar]
- 31.Campuzano S, Vazquez D, Modolell J. Biochemistry. 1979;18:1570–1574. doi: 10.1021/bi00575a029. [DOI] [PubMed] [Google Scholar]
- 32.Lin L, Bodley J W. J Biol Chem. 1976;251:1795–1798. [PubMed] [Google Scholar]
- 33.Hausner T P, Geigenmuller U, Nierhaus K H. J Biol Chem. 1988;263:13103–13111. [PubMed] [Google Scholar]
- 34.Ryan P C, Draper D E. Biochemistry. 1989;28:9949–9956. doi: 10.1021/bi00452a012. [DOI] [PubMed] [Google Scholar]
- 35.Wang Y X, Lu M, Draper D E. Biochemistry. 1993;32:12279–12282. doi: 10.1021/bi00097a002. [DOI] [PubMed] [Google Scholar]
- 36.Wimberly B T, Guymon R, McCutcheon J P, White S W, Ramakrishnan V. Cell. 1999;97:491–502. doi: 10.1016/s0092-8674(00)80759-x. [DOI] [PubMed] [Google Scholar]
- 37.Conn G L, Draper D E, Lattman E E, Gittis A G. Science. 1999;284:1171–1174. doi: 10.1126/science.284.5417.1171. [DOI] [PubMed] [Google Scholar]
- 38.Draper D E, Xing Y, Laing L G. J Mol Biol. 1995;249:231–238. doi: 10.1006/jmbi.1995.0291. [DOI] [PubMed] [Google Scholar]
- 39.Draper D E, Xing Y. Nucleic Acids Symp Ser. 1995;33:5–7. [PubMed] [Google Scholar]
- 40.Porse B T, Leviev I, Mankin A S, Garrett R A. J Mol Biol. 1998;276:391–404. doi: 10.1006/jmbi.1997.1541. [DOI] [PubMed] [Google Scholar]
- 41.Miller S P, Bodley J W. Nucleic Acids Res. 1991;19:1657–1660. doi: 10.1093/nar/19.7.1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cundliffe E. Biochem Biophys Res Commun. 1971;44:912–917. doi: 10.1016/0006-291x(71)90798-4. [DOI] [PubMed] [Google Scholar]