The X-ray structure of the tetragonal form of apo acyl-CoA-binding protein (ACBP) from the Harderian gland of the South American armadillo Chaetophractus villosus has been solved.
Keywords: acyl-CoA-binding protein, ACBP, armadillo Harderian gland
Abstract
The X-ray structure of the tetragonal form of apo acyl-CoA-binding protein (ACBP) from the Harderian gland of the South American armadillo Chaetophractus villosus has been solved. ACBP is a carrier for activated long-chain fatty acids and has been associated with many aspects of lipid metabolism. Its secondary structure is highly similar to that of the corresponding form of bovine ACBP and exhibits the unique flattened α-helical bundle (up–down–down–up) motif reported for animal, yeast and insect ACBPs. Conformational differences are located in loops and turns, although these structural differences do not suffice to account for features that could be related to the unusual biochemistry and lipid metabolism of the Harderian gland.
1. Introduction
Acyl-CoA-binding protein (ACBP) binds CoA thioesters of C14–C22 fatty acids (LCFA-CoAs) with nanomolar affinity (Kragelund et al., 1999 ▶) and is involved in the intracellular transport and storage of these compounds. Because LCFA-CoAs modulate the activity of several proteins and genes, ACBP also plays an important role in metabolism, cell signalling, membrane biogenesis and gene expression (Faergeman & Knudsen, 1997 ▶; Faergeman et al., 2004 ▶; Fulceri et al., 1999 ▶; Gaigg et al., 2001 ▶; Helledie et al., 2002 ▶; Knudsen et al., 2000 ▶; Kragelund et al., 1999 ▶).
Sequence analysis demonstrates that the 10 kDa ACBP domain belongs to a highly conserved family of eukaryotic proteins (Burton et al., 2005 ▶) and can be found alone or as part of larger modular proteins (Leung et al., 2006 ▶; Suk et al., 1999 ▶). NMR and X-ray structures of the stand-alone ACBPs from Taurus bovis, Plasmodium falciparum and Saccharomyces cerevisiae are available (Kragelund et al., 1993 ▶; Teilum et al., 2005 ▶; van Aalten et al., 2001 ▶; PDB codes 1aca, 1st7, 1hb6, 1hb8 and 1hbk). In addition, structures of multidomain proteins containing ACBP modules have been solved (PDB codes 2cqu and 2cop). ACBP adopts a unique open helix-bundle fold (up–down–down–up) with a shallow and exposed cavity that serves as a binding site (Kragelund et al., 1993 ▶; Teilum et al., 2005 ▶; van Aalten et al., 2001 ▶). The fatty-acid part of the ligand lines the cavity and is covered by the solvated adenosine-3′-phosphate moiety.
Comparison of bovine, insect and yeast ACBPs shows a nearly identical architecture of helical elements with conformational variation in the connecting loops. This variation might be related to binding specificity, affinity for different ligands (van Aalten et al., 2001 ▶) and crystallographic contacts (see below), as well as to differential interaction with membranes and macromolecules (Vallejo, 2006 ▶). The loop between helix 1 and 2, at the rim of the binding site, is of particular interest because it exhibits significant changes in length and sequence among animal, yeast and insect ACBPs and may be related to species differences in ligand selectivity (van Aalten et al., 2001 ▶).
Mammalian Harderian glands are lipogenic organs that produce an abundant secretion of glycerol ether lipids. These unique compounds are synthesized by peroxisomal enzymes from a separated pool of LCFA-CoAs (Horie & Suga, 1989 ▶). In the Harderian glands of the South American armadillo Chaetophractus villosus, the expression pattern of ACBP is unusual (Cavagnari et al., 2001 ▶). This, along with the peculiarities in lipid composition and metabolism, prompted us to characterize the protein (Cavagnari et al., 1997 ▶, 2001 ▶, 2002 ▶). In this study, we report the X-ray structure of armadillo ACBP. Since only a few ACBP structures have so far been reported, the new data should be useful in comparative studies aimed at understanding the function and evolution of these proteins.
2. Materials and methods
Armadillo ACBP was prepared as previously described (Cavagnari et al., 2002 ▶); the procedure yields ligand-free highly soluble monomeric protein with >98% purity. The purified protein in 20 mM Tris–HCl pH 8.0 was concentrated to 40 mg ml−1 using a Centricon YM-3 (Milipore Corp., Bedford, MA, USA). Crystallization trials were carried out using both sitting-drop and hanging-drop vapour-diffusion methods. Crystals were obtained at 277 K using a 1.0 ml reservoir composed of 24%(w/v) polyethylene glycol 6000, 5% NaCl and 10 mM Tris–HCl buffer pH 8.0. Drops contained 60% reservoir solution and 40% 10 mg ml−1 protein solution. A single crystal was mounted in a thin-walled quartz capillary and used for data collection. Cu Kα X-rays were generated by a Rigaku RU-300 rotating-anode generator operating at 40 kV and 80 mA. Diffraction data were collected at room temperature using a MAR Research image-plate scanner. 100 sequential images were recorded with an oscillation angle of 1.0° per image; data reduction was carried out with DENZO and SCALEPACK (Otwinowski & Minor, 1997 ▶). Molecular replacement, model bulding and refinement were performed with AMoRe (Navaza, 2001 ▶), O (Jones et al., 1991 ▶) and REFMAC5 (Collaborative Computational Project, Number 4, 1994 ▶), respectively. The structure was solved using the program ARP (Perrakis et al., 1999 ▶) and water molecules not clearly defined were removed manually with O (Jones et al., 1991 ▶). The quality of intermediate and final models was monitored using PROCHECK (Laskowski et al., 1993 ▶).
3. Results and discussion
Using the conditions described in §2, long needle-like crystals appeared after 2–3 d. On longer incubation (6–10 d), bipyramidal crystals were observed (0.3 × 0.3 × 0.3 mm; Fig. 1 ▶). The bipyramidal crystals exhibited diffraction consistent with the tetragonal space group P41, with unit-cell parameters a = b = 49.11, c = 130.48 Å and three molecules in the asymmetric unit. A diffraction data set was collected to 3.5 Å, with a completeness of 100% and an R merge of 13.6%. Data-collection statistics are summarized in Table 1 ▶.
Figure 1.
Crystals of armadillo ACBP. The crystals are approximately 0.3 × 0.3 × 0.3 mm in size.
Table 1. Data collection and refinement.
Values in parentheses refer to the outermost shell (3.62–3.50 Å).
| Space group | P41 |
| Unit-cell parameters (Å, °) | a = b = 49.11, c = 130.48, α = β = γ = 90.0 |
| Resolution range (Å) | 25.0–3.50 |
| Unique reflections | 3901 |
| Completeness (%) | 100 (99.5) |
| 〈I/σ(I)〉 | 12.9 (5.29) |
| Rmerge (%) | 13.6 (26.2) |
| No. of molecules in ASU | 3 |
| No. of protein atoms | 2125 |
| No. of solvent molecules | 37 |
| Rcryst† (%) | 19.5 |
| Rfree (10% of data) (%) | 25.7 |
| R.m.s. deviations from ideality | |
| Bond lengths (Å) | 0.008 |
| Angles (°) | 1.5 |
| Ramachandran statistics | |
| Residues in most favoured regions | 180 [77.9%] |
| Residues in additional allowed regions | 48 [20.8%] |
| Residues in generously allowed regions | 1 [0.4%] |
| Residues in disallowed regions | 2 [0.9%] |
R = 
 .
.
The search model was the crystal structure of bovine ACBP (PDB code 1hb8). A solution was found with space group P41 or P43 and a CC of 0.621 (R factor 0.510) within the resolution range 15–30 Å. Examination of the crystal packing revealed that space group P43 produced superpositions between symmetry-related molecules whereas the enantiomorph P41 did not, implying that the correct solution corresponded to P41. By alternating simulated annealing, manual rebuilding and addition of solvent molecules, the model was further refined. The final model has an R factor of 0.195 (R free = 0.257), displays good stereochemistry with only one residue (Met46) in a disallowed region of the Ramachandran statistics and contains 37 water molecules.
Although some disorder can be seen in a few solvent-exposed side chains, the backbone and most side chains are well defined in the electron-density map. The solved structure (PDB code 2fdq) is a bundle of four helices (residues 2–12, 21–35, 49–60 and 66–84) with an up–down–down–up topology (Fig. 2 ▶). The overall structure is nearly identical to that of bovine ACBP (PDB code 1hb8): the pairwise r.m.s.d. after superposition on CA atoms is less than 0.4 Å, with most of the variation concentrated in the connecting turns and loops.
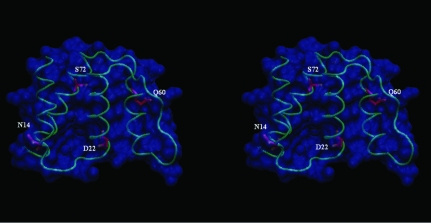
Figure 2.
Stereo image of the crystal structure of armadillo ACBP. Amino acids shown as stick models correspond to sequence differences between armadillo and bovine ACBP. The largest conformational difference between these two proteins is in the loop containing Asn14 (see text).
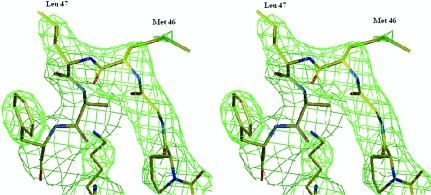
This newest structure reveals local conformational changes to accommodate the four residue differences between bovine and armadillo ACBP variants (Fig. 2 ▶). Of particular interest is the substitution of His for Asn at position 14 in the loop connecting helix 1 and 2, which perturbs the backbone and may have an impact on the fine-tuning of ligand selectivity. The r.m.s.d. between the two structures at position 14 is 0.7 Å, distinctly larger than the mean for the complete structure. Another interesting feature observed in the present structure is the conformational variability of the loop containing Met46 as well as that of the residue itself. In the structure of the orthorhombic crystal form of bovine ACBP this region is a type I turn, whereas it is a type II turn (Venkatachalam, 1968 ▶) in the tetragonal form. This causes the side chain of Met46 to point in opposite directions, suggesting that the Met46 side chain can adopt many possible orientations. The difference between the two crystal forms was ascribed to steric restrictions imposed by the contacts between the three molecules in the asymmetric unit. In the armadillo ACBP the 45–48 section of the backbone adopts a type II β-turn (Fig. 3 ▶), which is consistent with the crystal-packing hypothesis. Nevertheless, this conformational flexibility uncovers the ability of this protein to oscillate between two conformations that may be of functional interest. In fact, it was observed by simulation that this turn (and particularly Met46) is involved in ACBP ligand binding (Vallejo, 2006 ▶). It is worth noting that in the tetragonal form of both bovine (van Aalten et al., 2001 ▶) and armadillo ACBP the Met46 dihedral angles are in a disallowed region of the Ramachandran plot. Similarly, in the NMR structure of apo bovine ACBP Asp48 is in a disallowed region of the plot, building a conformational tension that is absent in ligand-bound ACBP (Kragelund et al., 1993 ▶). Taken together, these results may indicate that the turn 45–48 is involved in the conformational events associated with ligand transactions between lipid bilayers and ACBP.
Figure 3.
Stereo image of the loop containing the surface-exposed residues Met46 and Leu47.
Overall, the structure of the ACBP from armadillo Harderian gland has been solved and shows the same fold as the other known ACBP structures (van Aalten et al., 2001 ▶; Teilum et al., 2005 ▶; Kragelund et al., 1993 ▶). One difference is in the loop that contains residue 14, where a change of His for Asn affects the conformation in a protein region that may be related to ligand selectivity (van Aalten et al., 2001 ▶). Another feature confirmed by the armadillo ACBP structure is the conformational variability of Met46, although the relationship between the above structural features and the peculiar lipid biochemistry in Harderian glands remains to be established. The data reported herein should be valuable in the systematic analysis of the structure–function relationship and conformational variation within the ACBP family of proteins as a whole.
Supplementary Material
PDB reference: armadillo ACBP, 2fdq, r2fdqsf
Acknowledgments
We thank D. Vallejo and J. R. Grigera for helpful discussions and D. Milikowski and B. Cavagnari for technical assistance. MRE is a member of the National Research Council of Argentina (CONICET). This work was partially supported by a SECYT-UNS 24/F035 grant.
References
- van Aalten, D. M., Milne, K. G., Zou, J.-Y., Kleywegt, G. J., Bergfors, T., Ferguson, M. A., Knudsen, J. & Jones, T. A. (2001). J. Mol Biol.309, 181–192. [DOI] [PubMed] [Google Scholar]
- Burton, M., Rose, T. M., Faergeman, N. J. & Knudsen, J. (2005). Biochem. J.392, 299–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavagnari, B. M., Cordoba, O. L., Affanni, J. M. & Santome, J. A. (1997). Comput. Biochem. Physiol. B Biochem. Mol. Biol.118, 173–180. [DOI] [PubMed]
- Cavagnari, B. M., Milikowski, D., Haller, J. F., Zanek, M. C., Santome, J. A. & Ermacora, M. R. (2002). Int. J. Biol. Macromol.31, 19–27. [DOI] [PubMed] [Google Scholar]
- Cavagnari, B. M., Sterin-Speziale, N., Affanni, J. M., Knudsen, J. & Santome, J. A. (2001). Biochim. Biophys. Acta, 1545, 314–325. [DOI] [PubMed] [Google Scholar]
- Collaborative Computational Project, Number 4 (1994). Acta Cryst. D50, 760–763. [Google Scholar]
- Faergeman, N. J., Feddersen, S., Christiansen, J. K., Larsen, M. K., Schneiter, R., Ungermann, C., Mutenda, K., Roepstorff, P. & Knudsen, J. (2004). Biochem. J.380, 907–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faergeman, N. J. & Knudsen, J. (1997). Biochem. J.323, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulceri, R., Giunti, R., Knudsen, J., Leuzzi, R., Kardon, T. & Benedetti, A. (1999). Biochem. Biophys. Res. Commun.264, 409–412. [DOI] [PubMed] [Google Scholar]
- Gaigg, B., Neergaard, T. B., Schneiter, R., Hansen, J. K., Faergeman, N. J., Jensen, N. A., Andersen, J. R., Friis, J., Sandhoff, R., Schroder, H. D. & Knudsen, J. (2001). Mol. Biol. Cell, 12, 1147–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helledie, T., Jorgensen, C., Antonius, M., Krogsdam, A. M., Kratchmarova, I., Kristiansen, K. & Mandrup, S. (2002). Mol. Cell. Biochem.239, 157–164. [PubMed] [Google Scholar]
- Horie, S. & Suga, T. (1989). Biochem. J.262, 677–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, T. A., Zou, J.-Y., Cowan, S. W. & Kjeldgaard, M. (1991). Acta Cryst. A47, 110–119. [DOI] [PubMed] [Google Scholar]
- Knudsen, J., Neergaard, T. B., Gaigg, B., Jensen, M. V. & Hansen, J. K. (2000). J. Nutr.130, 294S–298S. [DOI] [PubMed]
- Kragelund, B. B., Andersen, K. V., Madsen, J. C., Knudsen, J. & Poulsen, F. M. (1993). J. Mol. Biol.230, 1260–1277. [DOI] [PubMed] [Google Scholar]
- Kragelund, B. B., Knudsen, J. & Poulsen, F. M. (1999). Biochim. Biophys. Acta, 1441, 150–161. [DOI] [PubMed] [Google Scholar]
- Laskowski, R. A., MacArthur, M. W., Moss, D. S. & Thornton, J. M. (1993). J. Appl. Cryst.26, 283–291. [Google Scholar]
- Leung, K. C., Li, H. Y., Xiao, S., Tse, M. H. & Chye, M. L. (2006). Planta, 223, 871–881. [DOI] [PubMed] [Google Scholar]
- Navaza, J. (2001). Acta Cryst. D57, 1367–1372. [DOI] [PubMed] [Google Scholar]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol.276, 307–326. [DOI] [PubMed]
- Perrakis, A., Morris, R. & Lamzin, V. S. (1999). Nature Struct. Biol.6, 458–463. [DOI] [PubMed] [Google Scholar]
- Suk, K., Kim, Y. H., Hwang, D. Y., Ihm, S. H., Yoo, H. J. & Lee, M. S. (1999). Biochim. Biophys. Acta, 1454, 126–131. [DOI] [PubMed] [Google Scholar]
- Teilum, K., Thormann, T., Caterer, N. R., Poulsen, H. I., Jensen, P. H., Knudsen, J., Kragelund, B. B. & Poulsen, F. M. (2005). Proteins, 59, 80–90. [DOI] [PubMed] [Google Scholar]
- Vallejo, D. (2006). Personal communication.
- Venkatachalam, C. M. (1968). Biopolymers, 6, 1425–1436. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: armadillo ACBP, 2fdq, r2fdqsf