Human S100A13 protein was cloned, expressed, purified and crystallized by the hanging-drop vapour-diffusion method. The crystals obtained belonged to space group P212121 and diffracted to a resolution of 1.8 Å.
Keywords: S100A13, EF-hand calcium-binding proteins
Abstract
S100A13 is a member of the S100 family of EF-hand-containing calcium-binding proteins and plays an important role in the secretion of fibroblast growth factor-1 and interleukin 1α, two pro-angiogenic factors released by the endoplasmic reticulum/Golgi-independent non-classical secretory pathway. Human S100A13 was heterologously expressed in Escherichia coli, purified and crystallized by the hanging-drop vapour-diffusion method using PEG 3350 as the precipitant. The crystals diffracted X-rays from a synchrotron-radiation source to 1.8 Å resolution and the space group was assigned as primitive orthorhombic P212121.
1. Introduction
The S100 family of proteins are small acidic EF-hand calcium-binding proteins implicated in intracellular and extracellular regulatory activities (Donato, 1999 ▶). Within cells, most of the S100 family members exist in the form of homodimers or heterodimers. They have two different types of calcium-binding site. The N-terminal domain contains an S100-specific EF-hand-type low-affinity site, while the C-terminal domain contains a canonical EF-hand-type high-affinity site. S100A13 is a unique member of the S100 family as it is ubiquitously expressed in a broad range of tissues (Wicki et al., 1996 ▶) and does not show the conformational changes upon calcium binding that are thought to be essential for the interaction of other S100 proteins with their target proteins (Ridinger et al., 2000 ▶). S100A13 participates in the release of fibroblast growth factor-1 (FGF1) as a component of the multiprotein complex containing FGF1 and p40-synaptotagmin in response to heat shock (Carreira et al., 1998 ▶; Landriscina, Soldi et al., 2001 ▶). It is also involved in the stress-induced release of interleukin 1α (IL-1α; Mandinova et al., 2003 ▶; Prudovsky et al., 2003 ▶). FGF1 and IL-1α are pro-angiogenic polypeptides which lack the classical signal peptide sequence and are secreted by the nonclassical endoplasmic reticulum/Golgi-independent pathway. The crystal structures of FGF1 and IL-1α demonstrate remarkable structural similarity, despite the absence of sequence similarity (Thomas et al., 1985 ▶; Zhang et al., 1991 ▶). The interaction of S100A13 with these proteins depends on the presence of Cu2+ ions (Landriscina, Bagala et al., 2001 ▶). The Cu2+-binding sites of S100A13 are likely to be formed by Ca2+ binding (Arnesano et al., 2005 ▶), which is mediated by Ca2+ influx through N-type Ca2+-channels (Matsunaga & Ueda, 2006 ▶). Human S100A13 consists of 98 amino-acid residues and has a molecular weight of 11 kDa. The solution structures of the apo- and Ca2+-bound forms of human S100A13 at pH 5.6 (PDB codes 1yus and 1yuu, respectively; Arnesano et al., 2005 ▶) have previously been reported. Here, we report the crystallization of Ca2+-bound human S100A13 at physiological pH (pH 7.5).
2. Materials and results
2.1. Cloning of human S100A13
Human S100A13 cDNA (GenBank accession No. AK097132) cloned from a first-strand cDNA library from human spleen (Origene Technologies) was amplified by polymerase chain reaction (PCR) and subcloned into the NdeI/BamHI site of the pET-16b vector (Novagen).
2.2. Purification of recombinant human S100A13
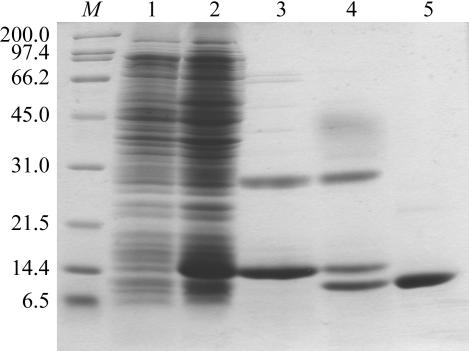
Escherichia coli BL21(DE3) cells harbouring the expression vector pET-16b-human S100A13 were grown at 310 K. The expression of S100A13 with an N-terminal 10×His tag and a Factor Xa protease-cleavage site (MGHHHHHHHHHHSSGHIEGR↓HM…, where the arrow to the C-terminal side of the italicized IEGR sequence indicates the Factor Xa cleavage site and the M in bold is the N-terminal methionine residue of S100A13) was induced at an OD600 of 0.6 with 1 mM isopropyl 1-thio-β-galactopyranoside (IPTG) and the culture continued at 310 K for 4 h. The cells were harvested by centrifugation at 3000g for 10 min at 277 K. The cells were resuspended in 50 mM Tris–HCl pH 8.0, disrupted by sonication and centrifuged at 26 000g for 20 min at 277 K. The supernatant was subjected to His-Bind affinity chromatography precharged with Ni2+ (Novagen). 10×His-tagged human S100A13 was eluted with 1.0 M imidazole, 500 mM NaCl and 20 mM Tris–HCl pH 7.9. The eluted protein was dialyzed against 100 mM NaCl, 5 mM CaCl2 and 50 mM Tris–HCl pH 8.0 at 277 K overnight. The 10×His tag was cleaved off by incubating the His-tagged protein with Factor Xa protease (Novagen, 10 U of enzyme per milligram of protein substrate) at room temperature for 4 h, which resulted in human S100A13 with an additional histidine residue at the N-terminus. The cleavage reaction was stopped by the addition of 1 mM AEBSF (Roche). The cleaved protein was then dialyzed against 25 mM Tris–HCl pH 8.1 and applied onto an Econo-Pac High Q (Bio-Rad) anion-exchange column equilibrated with the same buffer and eluted with a linear gradient of 0–0.1 M NaCl. Fractions containing the purified S100A13 were concentrated to 7.4 mg ml−1 using Vivaspin 20 concentrators (Vivascience). The purity was ascertained by SDS–PAGE and the concentration was determined from the absorbance at 280 nm. The purity of the sample was assessed by SDS–PAGE as shown in Fig. 1 ▶.
Figure 1.
SDS–PAGE (15%) analysis of recombinant human S100A13 protein. Lanes 1 and 2, E. coli cell lysate before and after induction with IPTG, respectively. Lane 3, 10×His-tagged S100A13 eluted from His-Bind Resin. Lane 4, after cleavage of the 10×His tag from S100A13 with Factor Xa protease. Lane 5, S100A13 purified by anion-exchange chromatography. Lane M, protein markers (kDa).
2.3. Crystallization
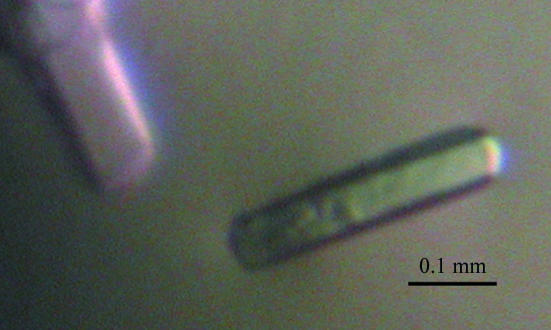
Crystallization experiments were performed at 293 K by the hanging-drop vapour-diffusion method using VDX 24-well crystallization plates (Hampton Research) and the crystallization screening kits Crystal Screens 1 and 2 (Hampton Research) and Wizards I and II (Emerald Biostructures). Calcium chloride was added to the protein solution to a final concentration of 2 mM prior to crystallization in order to obtain a calcium-bound form of S100A13. Crystals appeared in the presence of polyethylene glycol (PEG) 3350 (Hampton Research) as the precipitant. After refinement of the crystallization conditions, crystals suitable for X-ray analysis were obtained in two weeks by mixing 0.5 µl protein solution (7.4 mg ml−1 in 25 mM Tris–HCl pH 8.1, 0.1 M NaCl and 2 mM CaCl2) and 0.5 µl reservoir solution consisting of 22%(w/v) PEG 3350, 0.1 M HEPES–NaOH pH 7.5, 0.2 M NaCl and 1.5%(w/v) 1,2,3-heptanetriol. A drop was equilibrated against 500 µl reservoir solution at 293 K. Fig. 2 ▶ shows typical crystals (approximate dimensions 0.25 × 0.05 × 0.05 mm).
Figure 2.
Crystals of human S100A13 with approximate dimensions of 0.25 × 0.05 × 0.05 mm grown at 293 K using PEG 3350 as the precipitant.
2.4. X-ray data collection and processing
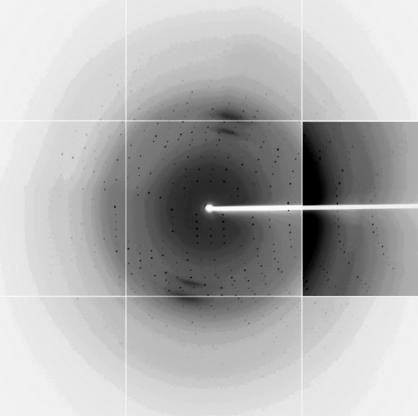
A crystal of S100A13 was picked up in a nylon loop (Hampton Research), transferred to a cryoprotectant solution containing 20%(v/v) glycerol, 17.6%(w/v) PEG 3350, 0.08 M HEPES–NaOH pH 7.5, 0.16 M NaCl and 1.2%(w/v) 1,2,3-heptanetriol and then flash-cooled at 100 K in a nitrogen stream. X-ray diffraction data were collected at beamline BL41XU equipped with an ADSC Quantum 315 detector at SPring-8 (Harima, Japan). An X-ray diffraction data set consisting of 180 images was collected with a wavelength of 1.000 Å, a crystal-to-detector distance of 200 mm, an oscillation angle of 1° and an exposure time of 4.0 s. The crystals diffracted beyond 1.8 Å resolution (Fig. 3 ▶). The diffraction data were indexed, integrated and scaled using HKL-2000 (Otwinowski & Minor, 1997 ▶). The space group of the crystals was determined as P212121 (the data statistics are shown in Table 1 ▶). The crystal contained two S100A13 molecules per asymmetric unit according to the Matthews coefficient (V M = 2.0 Å3 Da−1) and a solvent content of 38%. Structure solution and refinement is in progress.
Figure 3.
An X-ray diffraction image (1° oscillation) of a human S100A13 crystal. The edge of the diffraction image corresponds to a resolution of 1.80 Å. The contrast is enhanced in the right middle panel so that weak diffraction spots can be seen.
Table 1. Crystal parameters of S100A13.
Values in parentheses are for the highest resolution shell.
| Wavelength (Å) | 1.000 |
| Space group | P212121 |
| Unit-cell parameters (Å) | a = 39.7, b = 59.2, c = 77.6 |
| Resolution range (Å) | 50.0–1.80 (1.86–1.80) |
| Observed reflections | 122381 |
| Unique reflections | 17786 |
| Data completeness (%) | 99.1 (93.0) |
| Redundancy | 6.9 (6.3) |
| Rsym† | 0.066 (0.335) |
| 〈I/σ(I)〉 | 30.7 (3.4) |
R
sym = 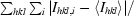
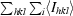 , where I
i is the ith observation of reflection hkl and 〈I
hkl〉 is the weighted average intensity for all observations i of reflection hkl.
, where I
i is the ith observation of reflection hkl and 〈I
hkl〉 is the weighted average intensity for all observations i of reflection hkl.
Acknowledgments
Synchrotron-radiation experiments were performed at SPring-8 (Harima, Japan) with the approval of Japan Synchrotron Radiation Research Institute (Proposal Nos. 2006A2721 and 2006A2728). This work was supported in part by the National Project on Protein Structural and Functional Analyses of Ministry of Education, Culture, Sports, Science and Technology of Japan and by Grants-in-Aid from The Japan Society for the Promotion of Science (JSPS).
References
- Arnesano, F., Banci, L., Bertini, I., Fantoni, A., Tenori, L. & Viezzoli, M. S. (2005). Angew. Chem. Int. Ed.44, 2–5. [DOI] [PubMed]
- Carreira, C. M., La Vallee, T. M., Tarantini, F., Jackson, A., Lathrop, J. T., Hampton, B., Burgess, W. H. & Maciag, T. (1998). J. Biol. Chem.273, 22224–22231. [DOI] [PubMed] [Google Scholar]
- Donato, R. (1999). Biochim. Biophys. Acta, 1450, 191–231. [DOI] [PubMed] [Google Scholar]
- Landriscina, M., Bagala, C., Mandinova, A., Soldi, R., Micucci, I., Bellum, S., Prudovsky, I. & Maciag, T. (2001). J. Biol. Chem.276, 25549–25557. [DOI] [PubMed] [Google Scholar]
- Landriscina, M., Soldi, R., Bagala, C., Micucci, I., Bellum, S., Tarantini, F., Prudovsky, I. & Maciag, T. (2001). J. Biol. Chem.276, 22544–22552. [DOI] [PubMed] [Google Scholar]
- Mandinova, A., Soldi, R., Graziani, I., Bagala, C., Bellum, S., Landriscina, M., Tarantini, F., Prudovsky, I. & Maciag, T. (2003). J. Cell Sci.116, 2687–2696. [DOI] [PubMed] [Google Scholar]
- Matsunaga, H. & Ueda, H. (2006). Cell. Mol. Neurobiol.26, 237–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol.276, 307–326. [DOI] [PubMed]
- Prudovsky, I., Mandinova, A., Soldi, R., Bagala, C., Graziani, I., Landriscina, M., Tarantini, F., Duarte, M., Bellum, S., Doherty, H. & Maciag, T. (2003). J. Cell Sci.116, 4871–4881. [DOI] [PubMed] [Google Scholar]
- Ridinger, K., Schafer, B. W., Durussel, I., Cox, J. A. & Heizmann, C. W. (2000). J. Biol. Chem.275, 8686–8694. [DOI] [PubMed] [Google Scholar]
- Thomas, K. A., Rios-Candelore, M., Gimenez-Gallego, G., DiSalvo, J., Bennett, C., Rodkey, J. & Fitzpatrick, S. (1985). Proc. Natl Acad. Sci. USA, 82, 6409–6413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicki, R., Schafer, B. W., Erne, P. & Heizmann, C. W. (1996). Biochem. Biophys. Res. Commun.227, 594–599. [DOI] [PubMed] [Google Scholar]
- Zhang, J. D., Cousens, L. S., Barr, P. J. & Sprang, S. R. (1991). Proc. Natl Acad. Sci. USA, 88, 3446–3450. [DOI] [PMC free article] [PubMed] [Google Scholar]