The crystal structure of human adipocyte lipid-binding protein (aP2) with a bound palmitate is reported at 1.5 Å resolution.
Keywords: aP2, adipocyte lipid-binding protein, palmitate, fatty-acid binding protein
Abstract
Human adipocyte lipid-binding protein (aP2) belongs to a family of intracellular lipid-binding proteins involved in the transport and storage of lipids. Here, the crystal structure of human aP2 with a bound palmitate is described at 1.5 Å resolution. Unlike the known crystal structure of murine aP2 in complex with palmitate, this structure shows that the fatty acid is in a folded conformation and that the loop containing Phe57 acts as a lid to regulate ligand binding by excluding solvent exposure to the central binding cavity.
1. Introduction
Intracellular lipid-binding proteins play important roles in fatty-acid solubilization, transfer and storage in eukaryotic cells. An exemplary intracellular lipid-binding protein is adipocyte lipid-binding protein (aP2; also called FABP4 or A-FABP), which binds a variety of fatty acids and acts as a carrier of fatty acids between the cell membrane and cell organelles (Banaszak et al., 1994 ▶). The aP2 protein is involved in lipolysis and lipogenesis and has been indicated in diseases of lipid and energy metabolism such as diabetes, atherosclerosis and metabolic syndromes (Robl et al., 2000 ▶). Small-molecule inhibitors of human aP2 have been actively pursued as potential therapeutics for treating type 2 diabetes (Lehmann et al., 2004 ▶; Ringom et al., 2004 ▶). The murine aP2 protein consists of a monomeric polypeptide of 131 amino-acid residues. The crystal structure of murine aP2 showed ten antiparallel β-strands arranged in a barrel structure, with an internal ligand-binding cavity capable of receiving medium-chain and long-chain fatty acids and other amphiphilic molecules (Reese-Wagoner et al., 1999 ▶). Here, we present the crystal structure of the human aP2 protein with a bound fatty acid. This new crystal form and structure may assist iterative structure-based drug-design efforts for identifying novel aP2 inhibitors.
2. Materials and methods
2.1. Expression and purification
The cDNA for human aP2 (FABP4, REFSEQ NM_001442) was excised from IMAGE clone 344626 at the BfaI/HincII restriction-endonuclease sites. The 428 bp aP2 cDNA fragment was purified and ligated into the NdeI/EcoRV-digested pET17b vector (Novagen). PCR mutagenesis was performed on the recombinant plasmid at the BfaI–NdeI ligation site (noncleavable) to regenerate the NdeI restriction site. The resulting PCR fragment (540 bp) was cloned into pCR2.1-TOPO (Invitrogen) and sequenced to confirm the mutagenesis. The pCR2.1-TOPO-aP2 clone was digested with the NdeI and XhoI restriction nucleases. The excised 453 bp aP2 cDNA fragment was isolated and ligated into the NdeI/XhoI-digested pET14b vector (Novagen). The resulting plasmid contained the sequence of human aP2 fused at the NdeI site, in frame, to the pET14b vector sequence containing the N-terminal His6 tag. BL21 (DE3) bacterial cells (Novagen) were transformed with the pET14b-aP2 plasmid and single colonies were isolated. 50 ml LB broth (75 µg ml−1 ampicillin) was seeded with a single colony and grown at 310 K to an OD600 of 1.0. 100 mM IPTG was added to a final concentration of 0.4 mM to induce aP2 protein expression. The culture was then allowed to grow for a further 4 h at 310 K.
His-tagged aP2 was isolated by nickel-affinity chromatography with Ni–NTA Superflow resin (Qiagen GmbH, Hilden, Germany). Escherichia coli cell paste containing His-tagged aP2 was resuspended in five volumes of lysis buffer (50 mM NaH2PO4 pH 8.0, 300 mM NaCl, 10 mM imidazole). Complete EDTA-free protease-inhibitor cocktail (Roche Diagnostics GmbH, Basel, Switzerland) was added. The suspension was subjected to two freeze–thaw cycles (193 K for 1 h and thawing at room temperature). The suspension was sonicated until complete lysis was obtained. The lysate was centrifuged at 14 000g for 50 min. The supernatant was applied onto a XK16 column packed with Ni–NTA Superflow resin at 1.0 ml min−1. The column was washed with wash buffer (50 mM NaH2PO4 pH 8.0, 300 mM NaCl, 20 mM imidazole) and step eluted with elution buffer (50 mM NaH2PO4 pH 8.0, 300 mM NaCl, 250 mM imidazole). The peak fractions were pooled and dialyzed against final buffer (50 mM NaH2PO4 pH 7.5, 50 mM NaCl). Attempts to remove the sequestered fatty acid were performed using the delipidating agents Lipidex-1000 and Lipidex-5000 (hydroxyalkoxypropyl dextran types IV and IX, respectively) at 310 and 318 K for up to 4 h. The resulting protein contained ∼50% bound fatty acid as analyzed by NMR (data not shown). Purity was determined to be >95% by SDS–PAGE, LC–MS, Edman sequencing and in-line static light scattering and concentration by UV280. Asymmetric field flow fractionation (A-FFF) combined with in-line dynamic/static light-scattering analysis showed aP2 to be a monodisperse monomer with R h = 2.1 nm (data not shown).
2.2. Crystallization
Sparse-matrix screening using commercially available screens (Hampton Research and Decode Genetics) was performed on the His-tagged aP2 using the hanging-drop vapor-diffusion method. Drops consisting of 2 µl protein plus 2 µl reservoir solution were set up at a protein concentration of 10.9 mg ml−1 in the aforementioned final buffer. Plates were incubated at 295 K. Two hits were identified, containing either 1.6 M trisodium citrate pH 6.5 or 20% PEG 10 000, 0.1 M HEPES pH 7.5. Diffraction-quality plate crystals (Fig. 1 ▶) were obtained by the hanging-drop vapor-diffusion method using 1.5 µl His-tagged aP2 plus 1.5 µl reservoir containing 1.6 M trisodium citrate pH 6.5 after 3 d at 295 K.
Figure 1.
His-tagged aP2 crystals. A single crystal was obtained by breaking apart the cluster.
2.3. Data collection and structure solution
Crystals were briefly soaked in a cryoprotectant consisting of two volumes of ethylene glycol plus eight volumes of the reservoir solution. They were positioned in a 100 K cold nitrogen stream. Data-collection statistics are listed in Table 1 ▶.
Table 1. Data-collection statistics.
Values in parentheses are for the outer shell.
| Beamline | APS 17-ID |
| Wavelength (Å) | 1.0 |
| Space group | P212121 |
| Unit-cell parameters (Å) | a = 32.5, b = 53.9, c = 75.0 |
| Resolution limits | 50–1.50 (1.55–1.50) |
| Observations | 90080 |
| Unique reflections | 20751 |
| Average redundancy | 4.3 (3.2) |
| Completeness | 95.3 (69.3) |
| Average I/σ(I) | 30.1 (3.2) |
| Rsym | 0.047 (0.346) |
The crystal structure of the variant human aP2 was solved by molecular replacement using the murine aP2 structure (murine adipocyte lipid-binding protein; PDB code 1adl) with bound arachidonic acid (20:4) (LaLonde, Levenson et al., 1994 ▶). The structure was first refined with the automated refinement program ARP/wARP and then manually modeled and refined to 1.5 Å resolution with an R factor of 0.175 and an R free of 0.225 (r.m.s. bond lengths 0.010 Å, r.m.s. bond angles 1.3°) using anisotropic B factors and the program REFMAC from the CCP4 program suite (Collaborative Computational Project, Number 4, 1994 ▶). Clear electron density was seen for bound palmitate, citrate and phosphate. The final model includes all native aP2 residues, a 21-amino-acid extension (MGSSHHHHHHSSGLVPRGSHM) from the artificially tagged construct, a bound palmitic acid, a phosphate anion, two partially disordered citrate anions and 153 water molecules. The structure factors and coordinates have been deposited in the Protein Data Bank (code 2hnx).
3. Structure and discussion
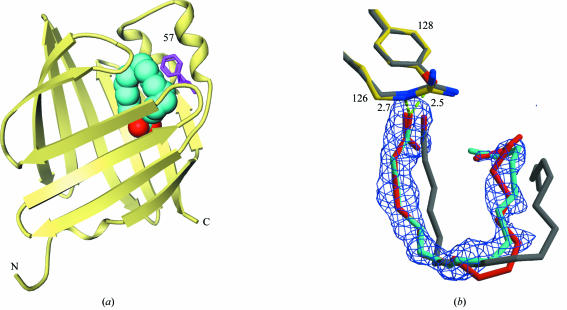
The crystal structure of human aP2 is very similar to that of the murine aP2, with overall r.m.s. differences of 0.3 Å for all Cα atoms. There is clear density for a bound fatty acid in the central cavity of the protein (Fig. 2 ▶). Since no fatty acid was added during the purification, the ligand must have been sequestered in vivo by the aP2 protein during expression and lysis in E. coli. The density fits well with a palmitate, which is a predominant product of E. coli fatty-acid biosynthesis and therefore a reasonable designation for the major component of the bound fatty acid in our crystal. This does not preclude the possibility that a fraction of the bound fatty acid is an oleate. Qualitative data from mass spectroscopy using the purified protein indicated the presence of mostly these two lipid species, but oleic acid does not appear to fit the electron density as well as palmitic acid (Fig. 2 ▶).
Figure 2.
The structure of human aP2 in complex with palmitic acid. (a) Ribbon diagram of human aP2. The bound palmitic acid is shown as a cyan (O atoms in red) CPK model. Phe57 is in magenta. The N- and C-termini are indicated as N and C. (b) The ligand-omitted (F o − F c) electron-density map is contoured at the 2.5σ level. Key hydrogen bonds are depicted in broken green lines with distances labeled. Our structure with palmitate is shown in yellow and cyan. The structural overlay of a folded form of bound oleate observed in the structure of a more distant relative, human brain FABP (PDB code 1hms; 61% identical to human aP2), is shown in gray. Our best attempt to try to fit an oleic acid into the observed electron density is shown in red.
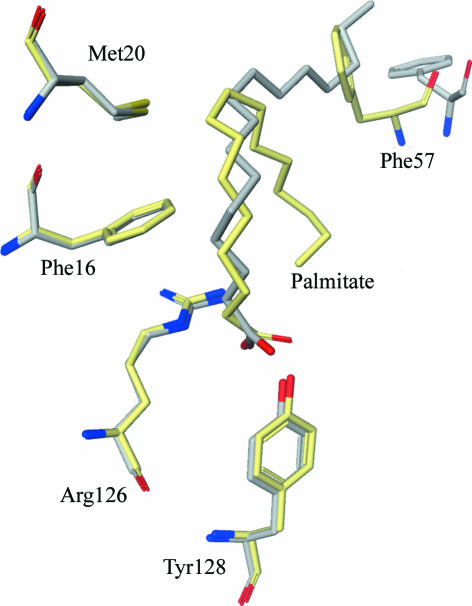
One of the fatty-acid carboxylate O atoms forms hydrogen bonds with the hydroxyl group of Tyr128 (2.5 Å) as well as the guanidium N atom of Arg126 (2.7 Å). These appear to be the key interactions for recognizing fatty-acid ligands (Fig. 2 ▶). The other O atom of the fatty-acid carboxylate interacts with the protein indirectly via water-mediated hydrogen bonds. The hydrophobic chain of the fatty acid binds in a folded conformation in the binding pocket (Fig. 2 ▶), forming numerous van der Waals interactions with protein hydrophobic residues, e.g. Phe16, Met20 and Phe57 (Fig. 3 ▶). This is quite different from the published murine aP2–palmitate and murine aP2–oleate structures, in which the fatty acid is bound in an extended conformation (Fig. 3 ▶) (LaLonde, Bernlohr et al., 1994 ▶). Moreover, the loop containing Phe57 is in a closed conformation in the human structure, as opposed to the open conformation in the murine structures (Fig. 3 ▶). This is surprising to some degree because the mouse and human proteins are >90% identical, without any obvious sequence differences in the area of the observed structural difference. An amino-acid difference in the binding cavity is Gln93 of murine aP2 being a histidine in human aP2, but it is not clear whether this could lead to the aforementioned conformational changes. Interestingly, the closed conformation is similar to the structure of human aP2 in complex with small-molecule inhibitors (Lehmann et al., 2004 ▶). The folded form of the liganded fatty acid is also seen in the epidermal FABP, heart FABP and myelin-type FABP structures (Hohoff et al., 1999 ▶; Zanotti et al., 1992 ▶; Cowan et al., 1993 ▶; Scapin et al., 1993 ▶). While it is possible that these conformational shifts directly correlate with the amino-acid sequence variations or crystal-packing differences between the human and murine aP2, the Phe57 loop clearly acts a lid to regulate the entrance and binding of various fatty-acid ligands.
Figure 3.
Comparison of the structures of mouse (grey) and human (yellow) aP2 in complex with palmitate. Key binding residues are almost perfectly superimposed, except for Phe57 of the lid, which moves according to the bound palmitate conformation.
Supplementary Material
PDB reference: adipocyte lipid-binding protein, 2hnx, r2hnxsf
Acknowledgments
We thank W. Ross Tracey and Jill Sutt for valuable discussions, Boris Chrunyk and Jane Withka for biophysical assay support.
References
- Banaszak, L., Winter, N., Xu, Z., Bernlohr, D. A., Cowan, S. & Jones, T. A. (1994). Adv. Protein Chem. 45, 89–151. [DOI] [PubMed] [Google Scholar]
- Collaborative Computational Project, Number 4 (1994). Acta Cryst. D50, 760–763. [Google Scholar]
- Cowan, S. W., Newcomer, M. E. & Jones, T. A. (1993). J. Mol. Biol.230, 1225–1246. [DOI] [PubMed] [Google Scholar]
- Hohoff, C., Boerchers, T., Ruestow, B., Spener, F. & Van Tilbeurgh, H. (1999). Biochemistry, 38, 12229–12239. [DOI] [PubMed] [Google Scholar]
- LaLonde, J. M., Bernlohr, D. A. & Banaszak, L. J. (1994). Biochemistry, 33, 4885–4895. [DOI] [PubMed] [Google Scholar]
- LaLonde, J., Levenson, M., Roe, J., Bernlohr, D. & Banaszak, L. (1994). J. Biol. Chem.269, 25339–25347. [DOI] [PubMed] [Google Scholar]
- Lehmann, F., Haile, S., Axen, E., Medina, C., Uppenberg, J., Svensson, S., Lundbaeck, T., Rondahl, L. & Barf, T. (2004). Bioorg. Med. Chem. Lett.14, 4445–4448. [DOI] [PubMed] [Google Scholar]
- Reese-Wagoner, A., Thompson, J. & Banaszak, L. (1999). Biochim. Biophys. Acta, 1441, 106–116. [DOI] [PubMed] [Google Scholar]
- Ringom, R., Axen, E., Uppenberg, J., Lundbaeck, T., Rondahl, L. & Barf, T. (2004). Bioorg. Med. Chem. Lett.14, 4449–4452. [DOI] [PubMed] [Google Scholar]
- Robl, J. A., Parker, R. A., Biller, S. A., Jamil, H., Jacobson, B. L. & Kodukula, K. (2000). US Patent 99-US20946-2000015229.
- Scapin, G., Young, A. C., Kromminga, A., Veerkamp, J. H., Gordon, J. I. & Sacchettini, J. C. (1993). Mol. Cell. Biochem.123, 3–13. [DOI] [PubMed] [Google Scholar]
- Zanotti, G., Scapin, G., Spadon, P., Veerkamp, J. H. & Sacchettini, J. C. (1992). J. Biol. Chem.267, 18541–18550. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: adipocyte lipid-binding protein, 2hnx, r2hnxsf