A probable copper-ion tolerance protein from the plant pathogen X. campestris has been overexpressed in E. coli, purified and crystallized.
Keywords: CutA1, copper homeostasis, Xanthomonas campestris, structural genomics
Abstract
Divalent metal ions play key roles in all living organisms, serving as cofactors for many proteins involved in a variety of electron-transfer activities. However, copper ions are highly toxic when an excessive amount is accumulated in a cell. CutA1 is a protein found in all kingdoms of life that is believed to participate in copper-ion tolerance in Escherichia coli, although its specific function remains unknown. Several crystal structures of multimeric CutA1 with different rotation angles and degrees of interaction between trimer interfaces have been reported. Here, the cloning, expression, crystallization and preliminary X-ray analysis of XC2981, a possible CutA1 protein present in the plant pathogen Xanthomonas campestris, are reported. The XC2981 crystals diffracted to a resolution of 2.6 Å. They are cubic and belong to space group I23, with unit-cell parameters a = b = c = 130.73 Å.
1. Introduction
XC2981 (gi|21111511) from the plant pathogen Xanthomonas campestris pv. campestris strain 17 (Xcc) is classified as belonging to the CutA1 divalent ion-tolerance protein superfamily in the Pfam database (Bateman et al., 2000 ▶). It contains 110 amino acids and shares 34.8, 31.8, 30.0 and 37.3% identity with similar proteins from the bacteria Escherichia coli, Thermotoga maritima, Aquifex aeolicus and Deinococcus radiodurans and 36.4, 30.6 and 28.2% identity with similar proteins from the archaea Pyrococcus horikoshii, Archaeoglobus fulgidus and Thermoplasma volcanium, respectively. Molecular-genetics studies on the E. coli homologue suggested that some mutations in the cutA locus can lead to copper sensitivity caused by its increased uptake (Fong et al., 1995 ▶). Although copper is an essential metal ion in all organisms, it is also highly toxic when an excessive amount is accumulated in the cell. This is best demonstrated by the finding that several mammalian neurological pathologies, such as Menke’s and Wilson’s diseases, Alzheimer’s disease, prion diseases and Creutzfeld–Jacob syndrome, can be correlated with the malfunction of copper-binding proteins (Bush, 2000 ▶). The homeostasis mechanism of regulated copper intake is thus an important issue that deserves comprehensive studies (Rosenzweig, 2001 ▶; Banci & Rosato, 2003 ▶).
Recently, several quaternary structures of the CutA1 monomer have been reported. That from T. maritima (Savchenko et al., 2004 ▶) was found to adopt a trimeric structure, while those from E. coli and rat formed a dimer of trimers (Arnesano et al., 2003 ▶) and that from P. horikoshii consisted of a multimer of trimers (Tanaka et al., 2004 ▶). Interestingly, differing rotation angles were found between the two trimer subunits when the CutA1 proteins assembled into a hexamer; in the E. coli protein one trimer is rotated by 60° with respect to the other around the axis perpendicular to the trimer planes, while in rat protein only a 25° rotation was observed (Arnesano et al., 2003 ▶). Additionally, significantly different hydrophobic and electrostatic interactions were observed between these trimer interfaces (Arnesano et al., 2003 ▶). The CutA1 copper-binding proteins may thus adopt a variety of quaternary structures depending on differences in the primary sequences or in environmental conditions. In this manuscript, we describe the cloning, expression, crystallization and preliminary X-ray analysis of XC2981 from Xcc. While a major trimeric species was found in a denatured SDS–PAGE analysis, we also detected significant amounts of tetramer and some hexamer (Fig. 1 ▶), suggesting that XC2981 may possibly form a hexamer in solution.
Figure 1.
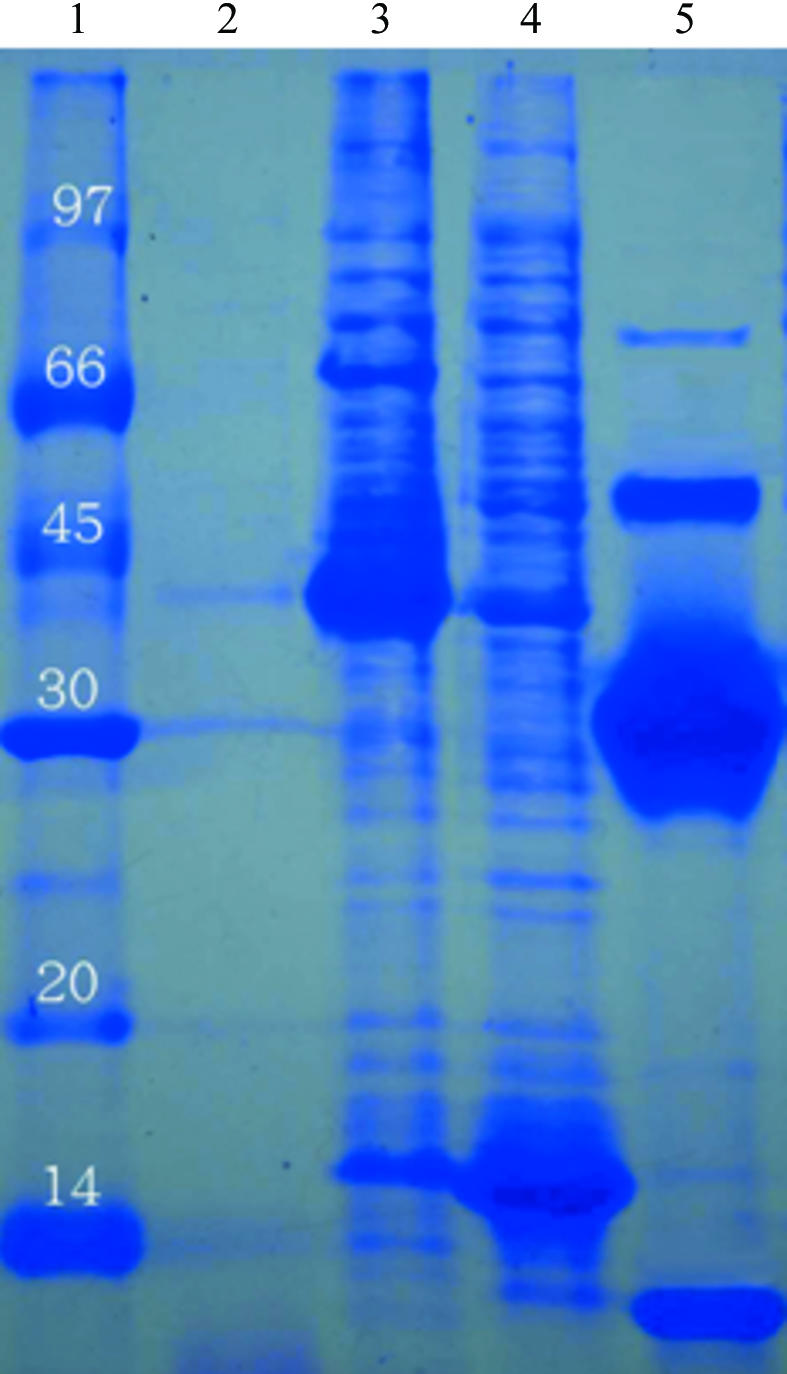
SDS–PAGE monitoring of the overexpression and purification of XC2981. Lane 1, molecular-weight markers in kDa; lane 2, whole cell lysate before IPTG induction; lane 3, whole cell lysate after IPTG induction; lane 4, whole cell lysate after IPTG induction with optional extra SDS; lane 5, purified XC2981 after TEV cleavage without optional extra SDS. It is clear that bands corresponding to monomers (12 kDa), trimers (36 kDa), tetramers (48 kDa) and even hexamers (72 kDa) are present in this gel in the absence of optional extra SDS (lane 5), indicating that XC2981 forms a stable trimer and possibly a hexamer in solution.
2. Materials and methods
2.1. Cloning, expression and purification
The XC2981 gene fragment was PCR-amplified directly from a local Xcc genome (X. campestris pv. campestris strain 17) with a forward 5′-TACTTCCAATCCAATGCTATGAGCGCCTTCTCTCTC primer and a backward 5′-TTATCCACTTCCAATGTCAGGATTCCTTGCGGGTTT primer. A ligation-independent cloning (LIC) approach (Aslanidis & de Jong, 1990 ▶) was carried out to obtain the desired construct according to a previously published protocol (Wu et al., 2005 ▶). The final construct codes for an N-terminal His6 tag, a 17-amino-acid linker and the XC2981 target protein (110 amino acids) under the control of a T7 promoter. Transformed E. coli BL21 (DE3) host cells were grown in LB medium at 310 K until an OD600 of 0.8 was attained. Overexpression of the His6-tagged target protein was induced by the addition of 0.5 mM IPTG at 310 K for 4.5 h. The cells were harvested, resuspended in equilibration buffer (20 mM Tris–HCl, 80 mM NaCl pH 8.0) and lysed using a microfluidizer (Microfluidics). Most tagged target proteins were present in the soluble fraction (Fig. 1 ▶). After centrifugation, the target protein was purified by immobilized metal-affinity chromatography (IMAC) on a nickel column (Sigma), which was eluted with 20 mM Tris pH 8.0, 80 mM NaCl and a gradient of 50–300 mM imidazole. The fractions containing XC2981 were monitored by SDS–PAGE, recombined and dialyzed repeatedly against 50 mM Na2HPO4 pH 8.0, 10% glycerol and 500 mM NaCl. After buffer exchange, the His6 tag and linker were cleaved from XC2981 by TEV (tobacco etch virus) protease at 288 K for 24 h. The purified protein was dialyzed against 20 mM Tris pH 8.0 and 80 mM NaCl several times. For crystallization, XC2981 was further purified on an anion-exchange column (AKTA, Pharmacia Inc.). The fractions eluted with 20 mM Tris pH 8.0, 1 M NaCl were combined and dialyzed against 20 mM Tris pH 8.0 and 80 mM NaCl. The final target protein (110 amino acids) has greater than 99% purity (Fig. 1 ▶) and contains only an extra tripeptide (SNA) at the N-terminal end. The overexpression and purification of XC2981 was monitored by SDS–PAGE as shown in Fig. 1 ▶.
2.2. Crystallization
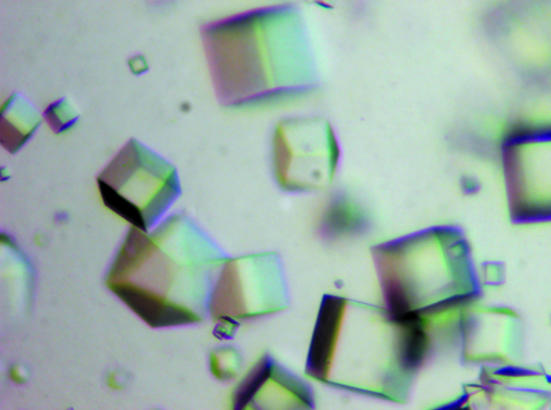
For crystallization, the protein was concentrated to 28 mg ml−1 in 20 mM Tris pH 8.0 and 80 mM NaCl using an Amicon Ultra-10 (Millipore). Screening for crystallization conditions was performed using sitting-drop vapour diffusion in 96-well plates (Hampton Research) at 293 K by mixing 0.5 µl protein solution with 0.5 µl reagent solution. Initial screens included the Hampton sparse-matrix Crystal Screens 1 and 2, a systematic PEG–pH screen and the PEG/Ion Screen and were performed using a Gilson C240 crystallization workstation. Cube-shaped crystals appeared in 3 d from a reservoir solution comprising 0.1 M CAPS buffer pH 10, 2.2 M (NH4)2SO4, 0.2 M Li2SO4.. Crystals suitable for diffraction experiments were grown by mixing 1.5 µl protein solution with 1.5 µl reagent solution at 298 K and reached maximum dimensions of 0.15 × 0.15 × 0.1 mm after 5 d (Fig. 2 ▶).
Figure 2.
Cube-shaped crystals of XC2981 from Xcc grown by the hanging-drop vapour-diffusion method. The crystallization condition used was 0.1 M CAPS buffer pH 10, 2.2 M (NH4)2SO4, 0.2 M Li2SO4. The average dimensions of these crystals were all approximately 0.15 × 0.15 × 0.1 mm.
2.3. Data collection
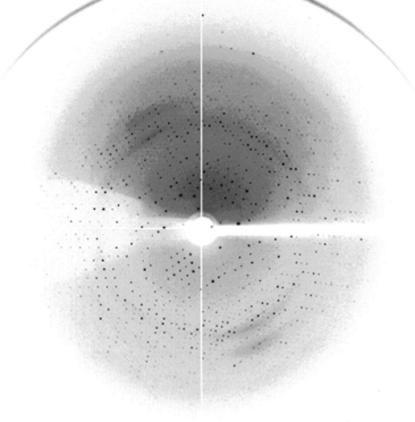
Crystals were soaked in mother liquor and then flash-cooled at 100 K in a stream of cold nitrogen. X-ray diffraction data were collected using the National Synchrotron Radiation Research Center (NSRRC) beamline 13B1, Taiwan. A 2.6 Å resolution native data set was obtained. The data were indexed and integrated using the HKL-2000 software (Otwinowski & Minor, 1997 ▶), giving a data set that had 100% completeness with an overall R merge of 7.0% on intensities. The crystals belong to the cubic space group I23. The data-collection statistics are summarized in Table 1 ▶ and an X-ray diffraction image collected at the NSRRC facility is shown in Fig. 3 ▶.
Table 1. Data-collection statistics for XC2981.
Values in parentheses are for the highest resolution shell.
| Space group | I23 |
| Unit-cell parameters (Å) | a = b = c = 130.70 |
| Temperature (K) | 100 |
| Wavelength (Å) | 0.963927 |
| Resolution range (Å) | 30.0–2.6 (2.69–2.60) |
| Mosaicity (°) | 0.5 |
| Unique reflections | 59537 (1869) |
| Redundancy | 6 (6) |
| Completeness (%) | 100 (100) |
| Rmerge (%) | 7.0 (23.2) |
| Mean I/σ(I) | 8.4 (7.1) |
| Solvent content (%) | 51 |
Figure 3.
Diffraction pattern of XC2981 collected at NSRRC beamline 13B1 from a flash-frozen crystal in reservoir cryoprotectant. The exposure time was 15 s, with an oscillation range of 1° and a crystal-to-detector distance of 200 mm.
3. Results and discussion
The XC2981 gene consists of 333 bp coding for 110 amino-acid residues. The isoelectric point was calculated to be 5.25. Purified XC2981 showed a mixture of monomers (12 kDa), trimers (36 kDa), tetramers (48 kDa) and hexamers (72 kDa) on SDS–PAGE (Fig. 1 ▶), indicating that the protein may possibly adopt a quaternary structure consisting of a hexamer as in E. coli CutA1 or rat CutA1 (Arnesano et al., 2003 ▶). The ‘dimer of trimers’ structure of PhoCutA from P. horikoshii (Tanaka et al., 2004 ▶) is different from the structures of E. coli CutA1 and rat CutA1. The former packs side by side, bridged by copper ions into a multimer (Tanaka et al., 2004 ▶), while in the latter structures monomers stack onto each other to form a hexamer that is mainly stabilized by hydrophobic interactions (Arnesano et al., 2003 ▶). XC2981 contains two Cys residues (Cys13 and Cys34), but not the CXXC motif known to bind copper ions in metallochaperones (Banci & Rosato, 2003 ▶; Tanaka et al., 2004 ▶; Arnesano et al., 2006 ▶). The potential metal-binding features of CutA1 are therefore different from those of metallochaperones. Interestingly, in PhoCutA the copper ion was found to be positioned at the trimer interface and coordinated to Asp48 Oδ, Lys49 C=O and water O atoms from each respective trimer subunit (Tanaka et al., 2004 ▶). It would thus be interesting to see whether or not the quaternary structure of XC2981 is similar to those previously observed.
The high-resolution diffraction data obtained from the native crystals establishes their suitability for X-ray structural analysis (Fig. 3 ▶). Molecular replacement using the AMoRe program (Navaza, 1994 ▶) will first be performed to attempt to obtain protein phases using the deposited coordinates of PDB entries 1naq or 1j2v as the search model. If this approach is unsuccessful, the single-wavelength anomalous diffraction (SAD; Wang, 1985 ▶; Dauter, 2002 ▶) or the multiwavelength anomalous diffraction (MAD) method (Hendrickson & Ogata, 1997 ▶; Terwilliger & Berendzen, 1999 ▶) will be applied using selenomethionine-substituted protein to solve the protein phases. Although XC2981 contains only one methionine, it may still be feasible to solve its structure using SAD or MAD because of its short length (110 amino acids).
Acknowledgments
This work was supported by an Academic Excellence Pursuit grant from the Ministry of Education and the National Science Council, Taiwan to S-HC. We thank the Core Facilities for Protein X-ray Crystallography at the Academia Sinica, Taiwan, at the National Synchrotron Radiation Research Center, Taiwan and at the SPring-8 Synchrotron facility in Japan for assistance during X-ray data collection. The National Synchrotron Radiation Research Center is a user facility supported by the National Science Council, Taiwan and the Protein Crystallography Facility is supported by the National Research Program for Genomic Medicine, Taiwan.
References
- Arnesano, F., Banci, L., Benvenuti, M., Bertini, I., Calderone, V., Mangani, S. & Viezzoli, M. S. (2003). J. Biol. Chem.278, 45999–46006. [DOI] [PubMed] [Google Scholar]
- Arnesano, F., Banci, L., Bertini, I., Ciofi-Baffoni, S., Molteni, E., Huffman, D. L. & O’Halloran, T. V. (2006). Genome Res.12, 255–271. [DOI] [PubMed]
- Aslanidis, C. & de Jong, P. J. (1990). Nucleic Acids Res.18, 6069–6074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banci, L. & Rosato, A. (2003). Acc. Chem. Res.36, 215–221. [DOI] [PubMed] [Google Scholar]
- Bateman, A., Birney, E., Durbin, R., Eddy, S. R., Howe, K. L. & Sonnhammer, E. L. L. (2000). Nucleic Acids Res.28, 263–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bush, A. L. (2000). Curr. Opin. Chem. Biol.4, 184–191. [Google Scholar]
- Dauter, Z. (2002). Acta Cryst. D58, 1958–1967. [DOI] [PubMed] [Google Scholar]
- Fong, S. T., Camakaris, J. & Lee, B. T. (1995). Mol. Microbiol.15, 1127–1137. [DOI] [PubMed] [Google Scholar]
- Hendrickson, W. A. & Ogata, C. M. (1997). Methods Enzymol.276, 494–523. [DOI] [PubMed]
- Navaza, G. (1994). Acta Cryst. A50, 157–163. [Google Scholar]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol.276, 307–326. [DOI] [PubMed]
- Rosenzweig, A. C. (2001). Acc. Chem. Res.34, 119–128. [DOI] [PubMed] [Google Scholar]
- Savchenko, A., Skarina, T., Evdokimova, E., Watson, J. D., Laskowski, R., Arrowsmith, C. H., Edwards, A. M., Joachimiak, A. & Zhang, R. (2004). Proteins, 54, 162–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka, Y., Tsumoto, K., Nakanishi, T., Yasutake, Y., Sakai, N., Yao, M., Tanaka, I. & Kumagai, I. (2004). FEBS Lett.556, 167–174. [DOI] [PubMed] [Google Scholar]
- Terwilliger, T. C. & Berendzen, J. (1999). Acta Cryst. D55, 849–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, B.-C. (1985). Methods Enzymol.115, 90–117. [DOI] [PubMed] [Google Scholar]
- Wu, Y.-Y., Chin, K.-H., Chou, C.-C., Lee, C.-C., Shr, H.-L., Lyu, P.-C., Wang, A. H.-J. & Chou, S.-H. (2005). Acta Cryst. F61, 902–905. [DOI] [PMC free article] [PubMed]