Abstract
The hyperthermophilic archaeon Acidianus ambivalens expresses a membrane-bound aa3-type quinol oxidase, when grown aerobically, that we have studied by resonance Raman spectroscopy. The purified aa3 oxidase, which does not contain bound quinol, undergoes a reversible slow conformational change at heme a3 upon reduction, as indicated by a change in the frequency of its heme formyl stretching mode, from 1,660 cm−1 to 1,667 cm−1. In contrast, upon reduction of the integral membrane enzyme or the purified enzyme preincubated with decylubiquinol, this mode appears at 1,667 cm−1 much more rapidly, suggesting a role of the bound quinol in controlling the redox-linked conformational changes. The shift of the formyl mode to higher frequency is attributed to a loss of hydrogen bonding that is associated with a group having a pKa of ≈3.8. Based on these observations, a crucial element for proton translocation involving a redox-linked conformational change near the heme a3 formyl group is postulated.
The hyperthermophilic archaeon Acidianus ambivalens contains one of the simplest known respiratory chains (1, 2). This microorganism is a strict chemolitoautotroph, growing optimally at 80°C and at pH 2.5 (3, 4). Membranes from aerobically grown cells contain almost exclusively a-type hemes; only traces of b-type hemes are detected (C.M.G. and M.T., unpublished data). The a-type hemes belong to a single aa3-type quinol oxidase (1, 5), which is the only heme protein present in the membranes, and contain a hydroxyethylgeranylgeranyl side chain instead of a hydroxyethylfarnesyl side chain as seen in most bacterial and eukaryotic aa3 oxidases (6). The major quinone in aerobically grown A. ambivalens is caldariella quinone, which is a thiophenobenzoquinone present in the Sulfolobales (7). The detergent-solubilized enzyme oxidizes caldariella quinol at a high rate (150 s−1 at 40°C, well below its physiological temperature where the rate is expected to be much higher; C.M.G. and M.T., unpublished data), and with high specificity, and hence it is a strict quinol oxidase (1). The oxidase from A. ambivalens is composed of five different subunits, the largest of which (65 kDa) contains the ligands to heme a and to the heme a3-CuB binuclear center (8). Accordingly, it contains these three redox metal cofactors and lacks a CuA center, just as all other quinol oxidases (9) and is therefore a member of the heme-copper oxidase superfamily, although its proton pumping properties have yet to be determined. The overall homology to subunit I of the other members of the heme-copper oxidase superfamily is about 18%, but all of the ligands that coordinate the redox centers are conserved. The remaining subunits are completely distinct from those of other oxidases.
In both cytochrome c and quinol oxidases, oxygen reduction occurs at a heme-CuB binuclear center. Closely associated with the center is a low-spin heme that is part of the electron transfer pathway. In cytochrome c oxidases, CuA serves as the electron entry point in the enzyme (10–13), whereas in bo3 quinol oxidases an enzyme-bound ubiquinol is believed to be the electron entry point that passes electrons to the heme o3-CuB center via the low-spin heme b (9). Similarly, in the aa3 quinol oxidase from Bacillus subtilis the low-spin heme a is thought to be the electron acceptor from membranous quinol (14). The location of a quinol-binding site in quinol oxidases has been debated extensively. In particular, for bo3 from Escherichia coli, several studies indicate that there could be two binding sites, one of which binds ubiquinol-8 tightly (15–20). Recent mass spectrometric analysis suggests that a binding site is located at the interface of subunits I and II (19). Given that this aa3 quinol oxidase of A. ambivalens is a member of the large heme-copper oxidase superfamily, it is essential to learn its structure and function to understand alternative strategies to reduce dioxygen and translocate protons. This may lead to new insights concerning the functional mechanisms of mammalian oxidases as well as how the properties of this enzyme and the role of the quinol are related to the unusual physiological properties of its parent extremophile, A. ambivalens.
The absence of alternative oxidases and the presence of only a-type hemes in A. ambivalens grown under aerobic conditions are unique features that allow spectroscopic measurements to be done on whole membranes and intact cells in addition to those on the purified enzyme. Recently, the kinetics of the electron transfer steps in this enzyme were studied, and a physiological role for the membrane-bound quinol serving as the fourth redox center was suggested (5). In the present study, the aa3 quinol oxidase from A. ambivalens has been characterized by resonance Raman scattering, a technique that has been shown to be very powerful in studying heme-copper oxidases (13, 21–23). It is shown that the heme a3 formyl group undergoes a redox-induced change in its hydrogen bonding, at a much faster rate in the membrane-integrated than in the purified enzyme. It is proposed that a functional role of the bound quinone, in addition to being involved in the electron transfer chain, is to facilitate the conformational changes in the enzyme required for its proper function. In addition, the nature of the redox-linked changes is studied in relation to the physiological conditions of A. ambivalens, and a redox/proton translocation coupling site near the heme a3 formyl group is postulated.
EXPERIMENTAL PROCEDURES
A. ambivalens cells were grown as described (24). Membranes and purified oxidase were prepared as in Giuffre et al. (5), by using lauryl maltoside as the detergent. In the Raman measurements, the membrane fraction (containing exclusively the aa3 oxidase) was studied in the intact state and in a lauryl maltoside-solubilized state. The purified enzyme lacks bound quinone, as determined from quinone extraction and HPLC analysis, which consistently showed the absence of caldariella quinone (C.M.G. and M.T., unpublished data), and it showed a very good signal-to-noise level in the Raman measurements. Decylubiquinone and lauryl maltoside were purchased from Sigma.
Resonance Raman measurements were carried out with an excitation wavelength at 413.1 nm from a Krypton-Ion laser (Spectra-Physics). Details of the instrumentation have been described (25). The sample cell was spun at 6,000 rpm to avoid local heating. The Raman scattered light was dispersed through a polychromator (Spex Industries, Metuchen, NJ) equipped with a 1,200 grooves/mm grating and detected by a liquid nitrogen-cooled charge-coupled device camera (Princeton Instruments, Trenton, NJ). A holographic notch filter (Kaiser Optical Systems, Ann Arbor, MI) was used to remove the laser scattering. Frequency shifts in the Raman spectra were calibrated by using acetone-CCl4 or indene as a reference.
The concentration of the protein samples used for the Raman measurements was ≈60 μM in different buffers for the desired pH (100 mM sodium phosphate, pH 7.4 and pH 2.6; 100 mM sodium acetate, pH 3.5–5.0; 100 mM Mes, pH 6.0). To obtain the reduced samples, the oxidized enzyme was reduced by the addition of an aliquot of a freshly prepared anaerobic solution of dithionite to the degassed protein solution. Absorption spectra were recorded before and after the Raman measurements to check the stability of the species studied. All measurements were carried out at ≈22°C.
RESULTS
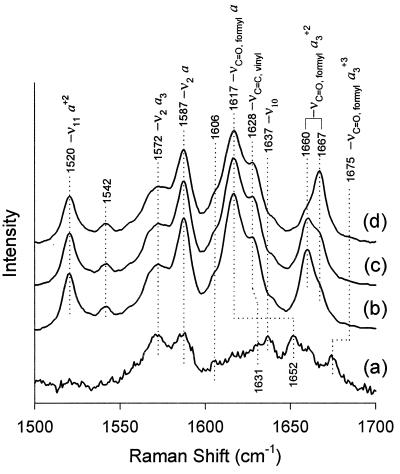
The high-frequency region (1,300–1,700 cm−1) of the resonance Raman spectra of hemeproteins is comprised of porphyrin in-plane vibrational modes that are sensitive to the electron density in the porphyrin macrocycle and also to the oxidation, coordination, and spin state of the central iron atom (25). In aa3 oxidases, the carbonyl stretching modes from the heme formyl groups (in the 1,600–1,700 cm−1 region) are resonance enhanced and can be used as useful probes of the conformation near the hemes. Fig. 1 shows the resonance Raman spectra, with the mode assignments (unpublished results), of the purified A. ambivalens oxidase in the dithionite-reduced form as a function of elapsed time after anaerobic reduction. For comparison, the spectrum of the oxidized enzyme also is shown. Upon reduction, the reduced spectrum obtained is characteristic of that of all oxidases. However, a time-dependent change subsequently occurs in the Raman spectrum, involving a slow shift in frequency of the formyl stretching mode of heme a3, from 1,660 to 1,667 cm−1. Immediately after reduction, the frequency of the a3 formyl stretching mode (1,660 cm−1) observed in this enzyme is the lowest among the heme a3-containing oxidases, where this mode occurs in the 1,662 to 1,669 cm−1 range (26–38). The absence of spectral changes in the other modes, including the oxidation state marker, ν4, at 1,355 cm−1 (data not shown), indicates that the oxidation, coordination, and spin states are not changing as a function of time after reduction in either of the hemes. However, the formyl group of heme a3 undergoes a very slow (t1/2 ≈18 min) redox-induced conformational change. When the 1,667-cm−1 conformer of the reduced enzyme is reoxidized and then rereduced, similar results are obtained, i.e., the 1,660-cm−1 conformer appears first, and then slowly switches to the 1,667-cm−1 conformer.
Figure 1.
Resonance Raman spectra of reduced aa3 in the high-frequency region as a function of time. The excitation frequency was 413.1 nm, and the laser power at the sample was ≈10 mW. The purified enzyme (60 μM) was reduced anaerobically by dithionite, in 100 mM Mes, pH 6.0, 1 mg/ml lauryl maltoside. The spectra shown are (b) immediately (≈1 min) after reduction, (c) after 15 min, and (d) after 50 min. Also shown for comparison is the spectrum of the oxidized enzyme (spectrum a, laser power ≈0.1 mW).
A redox-coupled conformational change in heme a3 would have to be much faster to play a physiological role in the enzymatic function. The purified enzyme lacks bound quinone. On the contrary, the membrane-integrated enzyme has a bound quinol (5). When the reduction of the membrane fraction or the detergent-solubilized membranes was carried out under similar conditions as used for the purified enzyme, the 1,667-cm−1 line appeared within our 1-min limit of reducing the enzyme and measuring a spectrum (spectra not shown, data listed in Table 1). The other vibrational modes occurred at the same frequencies as in the purified enzyme. These data suggest that the conformational changes associated with heme a3 occur at a much faster rate than in the purified enzyme, thus indicating a role of the bound quinol in the redox-coupled structural change.
Table 1.
Selected resonance Raman frequencies (Ex. 413.1 nm) of various preparations of A. ambivalens aa3 quinol oxidase in the reduced state
| Enzyme | ν4 | ν11(a) | ν2 (a) | νC=O (formyl, a3) |
|---|---|---|---|---|
| Purified, t = 1 min* | 1,355 | 1,520 | 1,587 | 1,660 |
| Purified, t ∼ 30 min | 1,355 | 1,520 | 1,587 | 1,667 |
| Purified + UQ, t ∼ 10 min† | 1,355 | 1,520 | 1,587 | 1,667 |
| Intact membrane | 1,355 | 1,519 | 1,587 | 1,667 |
| Membrane extract‡ | 1,356 | 1,519 | 1,588 | 1,667.5 |
| Bovine§ | 1,354 | 1,517 | 1,586 | 1,664 |
The enzyme samples were reduced by addition of a degassed dithionite solution under anaerobic conditions.
Purified aa3in 100 mM phosphate buffer, pH 7.4, 0.3 mg/ml lauryl maltoside, spectrum recorded ∼1 min after reduction by dithionite.
The purified oxidized aa3 was preincubated with 1.6 mg/ml decylubiquinone (UQ) for 1 hr and then was reduced by dithionite. Spectrum was recorded after 10 min.
Membranes dissolved in lauryl maltoside (2g LM/1 g protein).
Data from Argade et al. (30) on bovine cytochrome c oxidase.
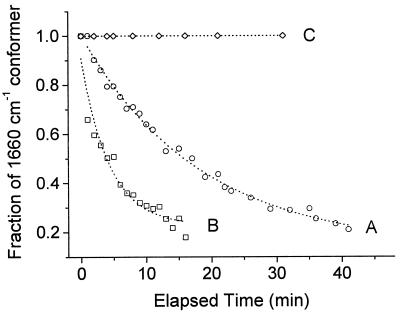
To test the above hypothesis, reduction was carried out on the purified enzyme that had been preincubated with a synthetic quinone, decylubiquinone, which is capable of donating electrons to the enzyme (8) and also is easier to handle experimentally than the highly insoluble caldariella quinol. Fig. 2 presents the time evolution of the a3 formyl stretching frequency upon reduction in the presence (trace B) and in the absence (trace A) of decylubiquinol, at pH 6. The purified enzyme that was preincubated with quinone showed a much faster rate (t1/2 ≈ 4 min) of conversion of the 1,660-cm−1 line relative to that (t1/2 ≈18 min) in the absence of quinone. Thus, the presence of quinol plays a role in modulating the structure of the enzyme that facilitates the redox-coupled conformational changes in heme a3.
Figure 2.
Effect of quinol on the rate of conformational change in heme a3. The decrease in the heme a3 formyl line at 1,660 cm−1 was monitored as a function of time after reduction, in the presence and in the absence of quinol, at two different pH values. The enzyme was reduced anaerobically by dithionite at pH 6, 100 mM Mes, in the absence (A) and in the presence (B) of 1.6 mg/ml decylubiquinol. At pH 2.6 (C), the population of the 1,660-cm−1 conformer remained invariant in both the presence and the absence of decylubiquinol. The relative population of the 1,660-cm−1 (to that of 1,667 cm−1) conformer was calculated by curve fitting using two Lorentzians.
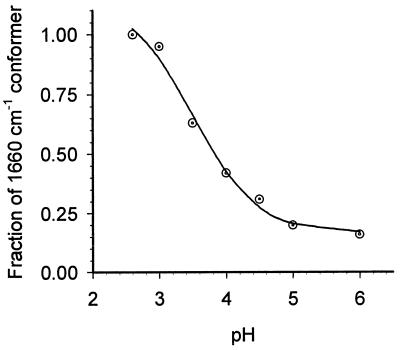
The oxidase from A. ambivalens is stable over a wide pH range. When the purified enzyme was reduced at pH 2.6, the formyl line appeared at 1,660 cm−1 just as seen at pH 6, but did not convert to 1,667 cm−1 even after a prolonged period of incubation (Fig. 2, trace C). Moreover, this band did not shift in the presence of decylubiquinol (data not shown, but trace superimposable on trace C in Fig. 2). At higher pHs, in the range of 3.5 to 6.0, however, the extent of conversion of the 1,660-cm−1 conformer to that at 1,667 cm−1 varied as a function of pH, as shown in Fig. 3. At each pH, the fractional population of the 1,660-cm−1 conformer was measured at 100 min after reduction, where the absence of any additional changes indicates that equilibrium has been reached. An examination of the populations of the two conformers as a function of pH yields a pKa ≈3.8 ± 0.4, associated with the conformational equilibrium.
Figure 3.
Variation in the population of the 1,660-cm−1 conformer as a function of pH (in the absence of decylubiquinol). The relative population of the 1,660-cm−1 conformer was calculated as in Fig. 2. Immediately after reduction, the 1,660-cm−1 conformer is the only species present at all pH values. The population of the 1,660-cm−1 conformer observed 100 min after reduction is shown (in ⊙). The sigmoidal curve (solid line) indicates the best fit that yielded an apparent pKa of 3.8 ± 0.4.
Although A. ambivalens functions in a surrounding medium at acid pH (≈2.5), it does not necessarily imply that the pH would be that low inside the cell. For example, for the closely related archaeon Thermoplasma acidophilum, also growing at pH 2, an internal pH of 6.6 has been measured (39). The fact that the aa3 oxidase is the only oxidase expressed when the archaeon is grown under aerobic conditions indicates that this enzyme participates in cell respiration. To accomplish the dioxygen chemistry correctly, the working pH range of this oxidase is expected to be much higher than ≈2.5. This is consistent with the observation of near-zero enzyme activity below pH ≈3 (data not shown).
DISCUSSION
Conformational Control of Hydrogen Bonding of the Heme a3 Formyl Group.
The assignment of the vibrational modes in aa3 oxidases is well established (30). With the excitation frequency used in this study (413.1 nm), modes from both the low-spin heme a (S = 0) and the high-spin heme a3 (S = 2) in the reduced enzyme contribute to the Raman spectrum. A crucial feature that makes the Raman spectrum of aa3 very useful is a substantial separation between the various lines arising from a and a3. The heme modes of the reduced aa3 oxidase of A. ambivalens in the high-frequency region, which will be discussed in detail elsewhere, match well with those in the bovine enzyme (30). Of particular importance in the present study are the C-O stretching modes of the formyl group (-CHO) of heme a3. In analogy with other oxidases, the formyl mode from heme a3 in the oxidized enzyme is assigned at 1,675 cm−1 and that in the reduced enzyme is located in the 1,660–1,667-cm−1 region (Fig. 1).
As a function of time after the initial reduction, no changes were detected in any of the oxidation-sensitive, coordination-sensitive, or spin-sensitive lines. In addition, the frequency of the formyl stretching mode in heme a (1,628 cm−1) remained unchanged. After the reduction of the enzyme, only the formyl group of heme a3 displayed a slow change in its frequency. The shift of the formyl stretching mode (νC=O) of reduced a3 is attributed to a change in its hydrogen-bonding interactions (28, 40, 41). The weaker the hydrogen bonding to the formyl group, the higher the stretching frequency, given that there is no change in the oxidation, coordination, or spin state of heme a3. Thus, at neutral pH, the conversion of the heme a3 formyl frequency from 1,660 cm−1 to 1,667 cm−1 results from a loss of hydrogen bonding associated with a group or groups having a pKa of ≈3.8. The observation of a very high frequency of the a3 formyl mode (1,675 cm−1) in the oxidized enzyme indicates that the formyl group is not hydrogen-bonded in ferric heme a3. The fact that the conformational changes were observed only in ferrous heme a3 and not in heme a, demonstrates that they are coupled exclusively to proton-dependent conformational changes at the binuclear center.
The data and conclusions reported here are in contrast to the change in hydrogen bonding to the heme a formyl group upon reduction reported previously (40). In that study, upon reduction an increase of the hydrogen-bond strength of the formyl group of heme a of ≈2–2.5 kcal/mol was estimated (40). Such an estimation is not straightforward as the frequency of the formyl mode changes as a function of the oxidation state of the heme iron. It was argued that the differing extents of the frequency shift of the heme a formyl upon reduction between cytochrome c oxidase and model heme a complexes reflect a change in the redox-dependent hydrogen-bond strength in the enzyme. In the present study, the contributions from the change in oxidation state and the change in the H bonding can be distinguished as separate steps. Hence, we are able to conclude that the frequency shift of the formyl is caused by a change in hydrogen bonding after reduction and is not associated with the change in the electronic structure of the formyl group because of the change in the oxidation state of the heme.
To account for the pH and substrate dependence of the formyl stretching mode, several questions must be addressed. First, which groups at the binuclear center have pKa values that could give rise to the observed titration behavior? Second, with which residue is the proton associated that hydrogen-bonds to the formyl group? Third, what is the nature of the conformational change that occurs subsequent to the reduction of the enzyme? Fourth, how does the addition of quinol accelerate the conformational change? Fifth, are the observed changes related to functionally important processes? The last two questions will be addressed in the next section; the first three are considered below.
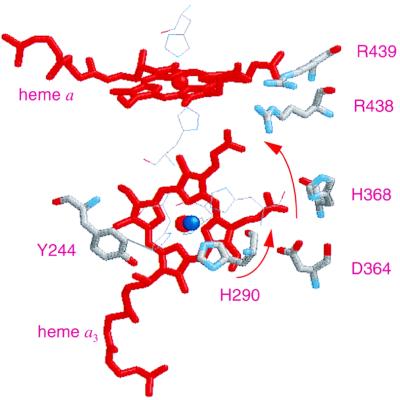
Several groups with labile protons are located near the heme a3 periphery and are candidates for the observed titration. The low pKa suggests the involvement of a carboxylate group or possibly an imidazole residue. Asp-364 (Asp-367 in A. ambivalens) in the structure of bovine cytochrome c oxidase is located near heme a3 (see Fig. 4), and, interestingly, is highly conserved in the heme-copper oxidase superfamily although its functional role is controversial. In some mutagenesis studies, Asp-364 has been shown to have no effect on structural integrity, catalytic activity, and proton translocation (42, 43), whereas in other studies it has been shown to have very significant effects (44, 45). The propionic acid side chain (at position 7 in the A ring of the porphyrin) of heme a3 which is located next to the formyl group (at position 8) is only 3.1 Å away from Asp-364 in the Paracoccus structure (Protein Data Bank code 1AR1) (46, 47), although the oxygen atoms of both the propionate and Asp-364 are about 6 Å away from the oxygen atom of the formyl group. In addition, Asp-364 has been postulated to be closely coupled with the propionate such that the two groups share only one proton in the pH 4 to 11.5 range (48). Thus, a pKa of 3.8 is reasonable for this coupled pair. The other heme a3 propionate (position 6) is connected to a hydrogen-bonded network, in both bovine (49, 50) and Paracoccus (46, 47) cytochrome c oxidases, that involves two arginines, Arg-438 and Arg-439 (bovine numbering, see Fig. 4), which also are conserved in the aa3 oxidase of A. ambivalens (Arg-442 and Arg-443, respectively). This network recently has been proposed as a part of the exit pathway for the pumped protons (51). Noticeably, two of the four heme propionates in P. denitrificans cytochrome c oxidase show redox-dependent changes detected by Fourier transform IR, which were attributed to either protonation-deprotonation or environmental changes (52).
Figure 4.
Structural model of the redox centers in A. ambivalens aa3 oxidase. The model was created based on the structural coordinates of Paracoccus cytochrome c oxidase, taking only the first 491 residues of the amino acid sequence into consideration. The residues are numbered as in bovine sequence. Shown in the picture are heme a, heme a3 (red), CuB (blue), and some of the neighboring residues (in thick lines), His-290 (one of the three histidine ligands to CuB), Tyr-244, Asp-364, His-368, Arg-438, and Arg-439. The residues shown in thin lines but not labeled are the histidine ligands to hemes a and a3 and CuB. The arrows denote the proposed path of proton translocation from His-290 to the Asp-364–propionate pair to the Arg-438–propionate region.
The closest residue with a labile proton to the formyl oxygen atom is His-290 (His-293 in A. ambivalens), which is one of the CuB histidine ligands. The oxygen atom of the heme a3 formyl group is located at a hydrogen-bonding distance (3.1 Å) from the Nδ of this residue, H325, in the Paracoccus structure (Protein Data Bank code 1AR1) and is 4.0 Å away in the bovine structure (Protein Data Bank code 2OCC). This variability of the formyl oxygen to the histidine Nδ distance suggests a degree of conformational flexibility in the structure that is consistent with data from recent x-ray absorption spectroscopy measurements, from which the loss of one of the CuB histidine ligands upon reduction was postulated (53). Thus, a reorganization of the CuB geometry, caused by deprotonation of His-290, is a plausible explanation for the changes we detect. Indeed, the pKa of the residue was calculated to be 4.4 (48) in reasonable agreement with our observed value of 3.8, although a distinction between this and the propionate–Asp-364 pair cannot be made at present.
A Model for Redox-Linked Proton Translocation at the Binuclear Site.
The data obtained in the present work may be discussed in the framework of mechanisms for proton translocation. In A. ambivalens there is a conformational change at the binuclear site that results in a loss of hydrogen bonding to the heme a3 formyl group. We postulate that this region near the formyl group is the center for redox coupling in the enzyme and describe a mechanism in the scheme illustrated in Fig. 5. This model is based on the central postulate that the hydrogen bond to the formyl group comes from His-290 and that the state of protonation of His-290 is controlled by the redox events at the binuclear center. Therefore, in this model, His-290 is the central element that couples the oxygen reduction chemistry to proton translocation.
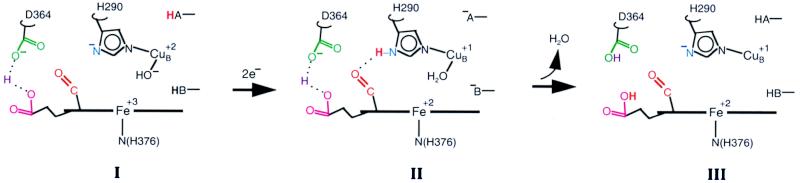
Figure 5.
Model of proton pumping. The residues are numbered following the sequence of bovine aa3. The sequence of structures shown is I, the oxidized form; II, the transient reduced form; and III, the final reduced form. Only selective residues around the heme a3-CuB site are shown. Two unidentified groups capable of donating protons are represented as −BH and −AH. The red proton shown in bold denotes the translocable proton. Asp-364 and the propionate share the proton placed in between them. In the oxidized form (structure I), the formyl of a3 is not hydrogen-bonded as the H290 side chain is an imidazolate. Upon reduction, the formyl group forms hydrogen bonding transiently with the −NδH of imidazole (structure II). In structure III, the hydrogen bond is broken again and the proton is transferred to the propionate–Asp-364 pair.
In the scheme, when the enzyme is fully oxidized, CuB is coordinated by a hydroxide group and three histidine residues (H240, H290, and H291). Because the formyl group is not hydrogen-bonded in the oxidized state, His-290 is taken as being deprotonated in the oxidized enzyme, as postulated by Wikström et al. (54, 55), and serves as the second charge to balance Cu2+. We also assume that the heme propionates are ionized in the oxidized state with the provision that one proton is shared by Asp-364 and the propionate on ring A of heme a3, as concluded from the electrostatic calculations reported recently by Kannt et al. (48).
Upon reduction, when two electrons enter the binuclear site, two protons also must enter to maintain the charge neutrality of the binuclear center, a principle established earlier (56). One of the protons combines with the hydroxide and the other protonates His-290. The protonated form of His-290 is a kinetic intermediate and the imidazole Nδ proton is stabilized by a hydrogen bond to the formyl group. This structure is stabilized for tens of minutes in the purified enzyme in the absence of quinol, but is transient when the enzyme is in the intact membrane. Upon reduction the protons can enter the binuclear site via one or more proton channels. The analysis of the primary sequence of A. ambivalens and a three-dimensional model indicate that although neither the K nor the D channel is strictly conserved, equivalent pathways can be proposed (C.M.G. and M.T., unpublished data). A similar situation is found for the ba3 oxidase of T. thermophilis, which nevertheless was proved recently to pump protons (57). The binding of the initial proton to the hydroxide is consistent with the electrostatic calculations on Paracoccus, and it also has been pointed out that His-290 could form a hydrogen bond to the heme a3 formyl group (48). Whether or not this hydrogen bond occurs without breaking the bond between the copper and the histidine remains to be determined.
In this model the protonated form of His-290 is metastable; its lifetime depends on the properties of the exit channel. Recently, Puustinen and Wikström (51), based on site-directed mutagenesis studies of Arg-438 and Arg-439, proposed that these arginines, along with the propionate on the D ring of heme a3, form part of a proton exit channel. Close to these groups is the A ring propionate—Asp-364 pair that has been proposed to share one proton. This pair may accept a proton from His-290, which in turn causes release of a proton to the exit channel. However, we postulate that in A. ambivalens this pathway is blocked unless quinol is present, and thus in its absence the release of the proton from His-290 is very slow. It is the blockage of the exit channel that has allowed for the stabilization of the proton on His-290 and its hydrogen bond to the formyl group. Although the location of the quinol is unknown, it must occupy a position critical for the proton translocation. When present, the proton can shuttle rapidly through the binuclear center during its pumping cycle and out the exit channel.
In the model presented here we invoke a specific role in proton pumping for the formyl group on heme a3. However, there are proton pumping oxidases that lack the formyl group, so its role cannot be essential. In heme b or heme o oxidases that lack the formyl group, an amino acid residue or a water molecule could provide the necessary stabilization. It is also possible that a proton network between the propionate group, D364, and H290 forms in certain oxidases and the heme a3 chromophore evolved to “hard-wire” the hydrogen bond acceptor into the active site.
It is important to note that in the model presented here the protonation of His-290 is transient. For both the oxidized and reduced forms of the binuclear center, the residue is deprotonated at equilibrium. Thus, the change in its protonation state depends directly on the chemistry at the binuclear site and is not restricted by the need to cycle through changes in redox state at each step. Local electrostatic changes during the redox process will change the proton affinity of the group and thereby couple the chemical events to the proton translocation.
CONCLUSIONS
The model discussed above postulates a putative redox coupling element in A. ambivalens quinol oxidase. The observations that led to this model were made possible in this unique oxidase, as the conditions were discovered in which the deprotonation event became very slow. A similar mechanism of redox coupling could be operative in the canonical oxidases as well, but in those cases the deprotonation events have escaped detection because they are too rapid. To determine whether similar processes occur in bovine cytochrome c oxidase, it will be necessary to find ways to slow down the deprotonation event.
The proposal of His-290 as the crucial element in the proton pumping pathway is consistent with many other models for pumping that have been postulated and data that have been reported. The recent x-ray absorption spectroscopy study on cytochrome bo3 revealed coordination of CuB by three histidines and one hydroxide in the cupric state, and two histidines and one water molecule in the cuprous state (53). This study is fully consistent with our conclusions because conformational changes associated with the protonation of His-290 upon reduction could result in either lengthening of the Cu-N bond or breakage of it. If the bond is broken the model described here may be adapted readily into the histidine-cycle model of Wikström et al. (54, 55) as well as the histidine-cycle/shuttle model proposed by Iwata et al. (46). However, it is to be noted that in Wikström et al.’s model, a different histidine residue, His-291, is proposed to be involved in the histidine cycle. In addition, because under physiological conditions the protons must pass rapidly through His-290 and go to the region of the heme propionates, the protonation of His-290 may be an obligatory transient intermediate along the pathway in the mechanism recently proposed by Michel (58).
Acknowledgments
We thank A. Kannt and H. Michel from the Max Planck Institute for Biophysics, Frankfurt/M, for the structural model (shown in Fig. 4) of the A. ambivalens oxidase. We thank one of the reviewers for suggesting the discussion regarding the hard-wired hydrogen bonding by the formyl group. This work was supported in part by National Institutes of Health Grants GM54806 and GM54812 to D.L.R.; Praxis XXI (BIO1075/94 and 37/96) and the European Union G-Project on Biotechnology of Extremophiles (Bio-4-CT96-0488) to M.T.; and Programa Gulbenkian de Doutoramento em Biologia e Medicina and Praxis XXI (BD9793/96) to C.M.G.
References
- 1.Anemuller S, Schmidt C L, Pacheco I, Schafer G, Teixeira M. FEMS Microbiol Lett. 1994;117:275–280. [Google Scholar]
- 2.Schafer G. Biochim Biophys Acta. 1996;1277:163–200. doi: 10.1016/s0005-2728(96)00104-1. [DOI] [PubMed] [Google Scholar]
- 3.Zillig W, Yeats S, Holz I, Bock A, Gropp F, Rettenberger M, Lutz S. Nature (London) 1985;313:789–791. doi: 10.1038/313789a0. [DOI] [PubMed] [Google Scholar]
- 4.Fuchs T, Huber H, Burggraf S, Stetter K O. Syst Appl Microbiol. 1996;19:56–60. [Google Scholar]
- 5.Giuffre A, Gomes C M, Antonini G, D’Itri E, Teixeira M, Brunori M. Eur J Biochem. 1997;250:383–388. doi: 10.1111/j.1432-1033.1997.0383a.x. [DOI] [PubMed] [Google Scholar]
- 6.Lubben M, Morand K. J Biol Chem. 1994;269:21473–21479. [PubMed] [Google Scholar]
- 7.Tricone A, Lanzotti V, Nicolaus B, Zillig W, De Rosa M, Gambacorta A. J Gen Microbiol. 1989;135:2751–2757. [Google Scholar]
- 8.Purschke W G, Schmidt C L, Petersen A, Schafer G. J Bacteriol. 1997;179:1344–1353. doi: 10.1128/jb.179.4.1344-1353.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Trumpower B L, Gennis R B. Annu Rev Biochem. 1994;63:675–716. doi: 10.1146/annurev.bi.63.070194.003331. [DOI] [PubMed] [Google Scholar]
- 10.Malatesta F, Nicoletti F, Zickermann V, Ludwig B, Brunori M. FEBS Lett. 1998;434:322–324. doi: 10.1016/s0014-5793(98)01006-0. [DOI] [PubMed] [Google Scholar]
- 11.Hill B C. J Biol Chem. 1994;269:2419–2425. [PubMed] [Google Scholar]
- 12.Babcock G T, Wikström M. Nature (London) 1992;356:301–309. doi: 10.1038/356301a0. [DOI] [PubMed] [Google Scholar]
- 13.Ferguson-Miller S, Babcock G T. Chem Rev. 1996;96:2889–2907. doi: 10.1021/cr950051s. [DOI] [PubMed] [Google Scholar]
- 14.Lauraeus M, Morgan J E, Wikström M. Biochemistry. 1993;32:2664–2670. doi: 10.1021/bi00061a026. [DOI] [PubMed] [Google Scholar]
- 15.Musser S M, Stowell M H, Lee H K, Rumbley J N, Chan S I. Biochemistry. 1997;36:894–902. doi: 10.1021/bi961723r. [DOI] [PubMed] [Google Scholar]
- 16.Sato-Watanabe M, Mogi T, Miyoshi H, Anraku Y. Biochemistry. 1998;37:5356–5361. doi: 10.1021/bi9727592. [DOI] [PubMed] [Google Scholar]
- 17.Sato-Watanabe M, Mogi T, Ogura T, Kitagawa T, Miyoshi H, Iwamura H, Anraku Y. J Biol Chem. 1994;269:28908–28912. [PubMed] [Google Scholar]
- 18.Puustinen A, Verkhovsky M I, Morgan J E, Belevich N P, Wikström M. Proc Natl Acad Sci USA. 1996;93:1545–1548. doi: 10.1073/pnas.93.4.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tsatsos P H, Reynolds K, Nickels E F, He D Y, Yu C A, Gennis R B. Biochemistry. 1998;37:9884–9888. doi: 10.1021/bi9809270. [DOI] [PubMed] [Google Scholar]
- 20.Welter R, Gu L Q, Yu L, Yu C A, Rumbley J, Gennis R B. J Biol Chem. 1994;269:28834–28838. [PubMed] [Google Scholar]
- 21.Han S, Ching Y C, Rousseau D L. Nature (London) 1990;348:89–90. doi: 10.1038/348089a0. [DOI] [PubMed] [Google Scholar]
- 22.Han S, Ching Y C, Rousseau D L. Proc Natl Acad Sci USA. 1990;87:8408–8412. doi: 10.1073/pnas.87.21.8408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kitagawa T, Ogura T. Prog Inorg Chem. 1997;45:431–479. [Google Scholar]
- 24.Teixeira M, Batista R, Campos A P, Gomes C, Mendes J, Pacheco I, Anemuller S, Hagen W R. Eur J Biochem. 1995;227:322–327. doi: 10.1111/j.1432-1033.1995.tb20392.x. [DOI] [PubMed] [Google Scholar]
- 25.Wang J, Caughey W S, Rousseau D L. In: Methods in Nitric Oxide Research. Feelisch M, Stamler J S, editors. New York: Wiley; 1996. pp. 427–454. [Google Scholar]
- 26.Shapleigh J P, Hosler J P, Tecklenburg M M, Kim Y, Babcock G T, Gennis R B, Ferguson-Miller S. Proc Natl Acad Sci USA. 1992;89:4786–4790. doi: 10.1073/pnas.89.11.4786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Das T K, Pecoraro C, Tomson F L, Gennis R B, Rousseau D L. Biochemistry. 1998;37:14471–14476. doi: 10.1021/bi981500w. [DOI] [PubMed] [Google Scholar]
- 28.Callahan P M, Babcock G T. Biochemistry. 1983;22:452–461. doi: 10.1021/bi00271a031. [DOI] [PubMed] [Google Scholar]
- 29.Ching Y C, Argade P V, Rousseau D L. Biochemistry. 1985;24:4938–4946. doi: 10.1021/bi00339a032. [DOI] [PubMed] [Google Scholar]
- 30.Argade P V, Ching Y C, Rousseau D L. Biophys J. 1986;50:613–620. doi: 10.1016/S0006-3495(86)83500-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Heibel G E, Hildebrandt P, Ludwig B, Steinrucke P, Soulimane T, Buse G. Biochemistry. 1993;32:10866–10877. doi: 10.1021/bi00091a042. [DOI] [PubMed] [Google Scholar]
- 32.Oertling W A, Surerus K K, Einarsdöttir O, Fee J A, Dyer R B, Woodruff W H. Biochemistry. 1994;33:3128–3141. doi: 10.1021/bi00176a048. [DOI] [PubMed] [Google Scholar]
- 33.Tsubaki M, Matsushita K, Adachi O, Hirota S, Kitagawa T, Hori H. Biochemistry. 1997;36:13034–13042. doi: 10.1021/bi971106i. [DOI] [PubMed] [Google Scholar]
- 34.Lauraeus M, Wikström M, Varotsis C, Tecklenburg M M, Babcock G T. Biochemistry. 1992;31:10054–10060. doi: 10.1021/bi00156a027. [DOI] [PubMed] [Google Scholar]
- 35.Ogura T, Sone N, Tagawa K, Kitagawa T. Biochemistry. 1984;23:2826–2831. doi: 10.1021/bi00307a045. [DOI] [PubMed] [Google Scholar]
- 36.de Paula J C, Peiffer W E, Ingle R T, Centeno J A, Ferguson-Miller S, Babcock G T. Biochemistry. 1990;29:8702–8706. doi: 10.1021/bi00489a028. [DOI] [PubMed] [Google Scholar]
- 37.Heibel G E, Anzenbacher P, Hildebrandt P, Schafer G. Biochemistry. 1993;32:10878–10884. doi: 10.1021/bi00091a043. [DOI] [PubMed] [Google Scholar]
- 38.Gerscher S, Dopner S, Hildebrandt P, Gleissner M, Schafer G. Biochemistry. 1996;35:12796–12803. doi: 10.1021/bi960896d. [DOI] [PubMed] [Google Scholar]
- 39.Hsung J C, Haug A. Biochim Biophys Acta. 1975;389:477–482. doi: 10.1016/0005-2736(75)90158-3. [DOI] [PubMed] [Google Scholar]
- 40.Babcock G T, Callahan P M. Biochemistry. 1983;22:2314–2319. doi: 10.1021/bi00279a002. [DOI] [PubMed] [Google Scholar]
- 41.Choi S, Lee J J, Wei Y H, Spiro T G. J Am Chem Soc. 1983;105:3692–3707. [Google Scholar]
- 42.Thomas J W, Puustinen A, Alben J O, Gennis R B, Wikstrom M. Biochemistry. 1993;32:10923–10928. doi: 10.1021/bi00091a048. [DOI] [PubMed] [Google Scholar]
- 43.Qian J, Shi W, Pressler M, Hoganson C, Mills D, Babcock G T, Ferguson-Miller S. Biochemistry. 1997;36:2539–2543. doi: 10.1021/bi962721+. [DOI] [PubMed] [Google Scholar]
- 44.Kawasaki M, Mogi T, Anraku Y. J Biochem. 1997;122:422–429. doi: 10.1093/oxfordjournals.jbchem.a021770. [DOI] [PubMed] [Google Scholar]
- 45.Pfitzner U, Odenwald A, Ostermann T, Weingard L, Ludwig B, Richter O M. J Bioenerg Biomembr. 1998;30:89–97. doi: 10.1023/a:1020515713103. [DOI] [PubMed] [Google Scholar]
- 46.Iwata S, Ostermeier C, Ludwig B, Michel H. Nature (London) 1995;376:660–669. doi: 10.1038/376660a0. [DOI] [PubMed] [Google Scholar]
- 47.Ostermeier C, Harrenga A, Ermler U, Michel H. Proc Natl Acad Sci USA. 1997;94:10547–10553. doi: 10.1073/pnas.94.20.10547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kannt H, Lancaster C R D, Michel H. Biophys J. 1998;74:708–721. doi: 10.1016/S0006-3495(98)73996-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tsukihara T, Aoyama H, Yamashita E, Tomizaki T, Yamaguchi H, Shinzawa-Itoh K, Nakashima R, Yaono R, Yoshikawa S. Science. 1995;269:1069–1074. doi: 10.1126/science.7652554. [DOI] [PubMed] [Google Scholar]
- 50.Yoshikawa S, Shinzawa-Itoh K, Nakashima R, Yaono R, Yamashita E, Inoue N, Yao M, Fei M J, Libeu C P, Mizushima T, et al. Science. 1998;280:1723–1729. doi: 10.1126/science.280.5370.1723. [DOI] [PubMed] [Google Scholar]
- 51.Puustinen A, Wikström M. Proc Natl Acad Sci USA. 1999;96:35–37. doi: 10.1073/pnas.96.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Behr J, Hellwig P, Mantele W, Michel H. Biochemistry. 1998;37:7400–7406. doi: 10.1021/bi9731697. [DOI] [PubMed] [Google Scholar]
- 53.Osborne J P, Cosper N J, Stalhandske C M, Scott R A, Alben J O, Gennis R B. Biochemistry. 1999;38:4526–4532. doi: 10.1021/bi982278y. [DOI] [PubMed] [Google Scholar]
- 54.Wikström M, Morgan J E, Verkhovsky M I. J Bioenerg Biomembr. 1998;30:139–145. doi: 10.1023/a:1020576031758. [DOI] [PubMed] [Google Scholar]
- 55.Morgan J E, Verkhovsky M I, Wikström M. J Bioenerg Biomembr. 1994;26:599–608. doi: 10.1007/BF00831534. [DOI] [PubMed] [Google Scholar]
- 56.Rich P R, Junemann S, Meunier B. J Bioenerg Biomembr. 1998;30:131–138. doi: 10.1023/a:1020524014920. [DOI] [PubMed] [Google Scholar]
- 57.Kannt A, Soulimane T, Buse G, Becker A, Bamberg E, Michel H. FEBS Lett. 1998;434:17–22. doi: 10.1016/s0014-5793(98)00942-9. [DOI] [PubMed] [Google Scholar]
- 58.Michel H. Proc Natl Acad Sci USA. 1998;95:12819–12824. doi: 10.1073/pnas.95.22.12819. [DOI] [PMC free article] [PubMed] [Google Scholar]