Abstract
Many diverse animal species regenerate parts of an organ or tissue after injury. However, the molecules responsible for the regenerative growth remain largely unknown. The screen reported here aimed to identify genes that function in regeneration and the transdetermination events closely associated with imaginal disc regeneration using Drosophila melanogaster. We screened a collection of 97 recessive lethal P-lacZ enhancer trap lines for two primary criteria: first, the ability to dominantly modify wg-induced leg-to-wing transdetermination and second, for the activation or repression of the lacZ reporter gene in the blastema during disc regeneration. Of the 97 P-lacZ lines, we identified six genes (Krüppelhomolog- 1, rpd3, jing, combgap, Aly and S6 kinase) that met both criteria. Five of these genes suppress, while one enhances, leg-to-wing transdetermination and therefore affects disc regeneration. Two of the genes, jing and rpd3, function in concert with chromatin remodeling proteins of the Polycomb Group (PcG) and trithorax Group (trxG) genes during Drosophila development, thus linking chromatin remodeling with the process of regeneration.
Keywords: Drosophila, Imaginal discs, Regeneration, Transdetermination, Wingless, Blastema, Chromatin remodeling, Enhancer trap, P-elements, Genetic screen
1. Introduction
There are three different mechanisms that organisms use to re-grow and replace lost or damaged body parts, and often, more than one mechanism can function within different tissues of the same organism. Muscle and bone, for example, repair themselves by activating a resident stem cell population, while the liver regenerates by compensatory proliferation of normally quiescent differentiated cells (Fausto, 2006; Shi and Garry, 2006). Appendage/fin regeneration in lower vertebrates occurs by a process termed epimorphic regeneration (Akimenko et al., 2003; Morgan, 1901; Poss et al., 2003; Stoick-Cooper et al., 2007), which proceeds in three distinct stages: (1) wound healing and migration of the surrounding epithelial cells to form the wound epidermis, (2) formation of the regeneration blastema – a mass of undifferentiated and proliferating cells of mesenchymal origin and (3) regenerative outgrowth and pattern re-formation. Whether these diverse modes of regeneration share a common molecular and genetic basis is not known.
Regeneration in the Drosophila imaginal discs, the primordia of the adult fly appendages, closely parallels epimorphic limb/fin regeneration in lower vertebrates. Cells in the imaginal discs are rigidly determined to form specific adult structures (e.g., legs and wings) by the third larval instar. If the discs are fragmented at this time and cultured in vivo, they will regenerate (Karlsson, 1980; Schubiger and Hadorn, 1968; Tiong et al., 1977). Disc regeneration begins 12 h after wounding, when transient heterotypic contacts are made between peripodial (squamous epithelium) and columnar cells (disc proper) near the cut edges of the wound (Bosch et al., 2005; Reinhardt and Bryant, 1981; Reinhardt et al., 1977). These initial contacts involve microvilli-like extensions and provide temporary wound closure. Then, approximately 24 h after wounding, homotypic cell contacts (between columnar or between squamous cells) are made involving the close apposition of cell membranes and cellular bridges, which eventually (48 h after wounding) restore the physical continuity of the disc (Reinhardt and Bryant, 1981; Reinhardt et al., 1977). Before and during wound healing, cell division is randomly distributed throughout the disc (Graves and Schubiger, 1982). However, once completed (36–48 h after wounding), division is only observed in cells near the wound site (Kiehle and Schubiger, 1985). These cells are known as the regeneration blastema. Thus, like appendage regeneration in lower vertebrates, disc regeneration involves wound healing followed by blastema formation.
Blastema cells are responsible for the regeneration and repatterning of the entire missing disc fragment (Abbott et al., 1981; Kiehle and Schubiger, 1985). Thus, these cells exhibit remarkable developmental plasticity. For example, in anterior- only leg disc fragments, some blastema cells will switch to posterior identity and establish a novel posterior compartment in the regenerate (Abbott et al., 1981). Gibson and Schubiger (1999) found that this anterior/posterior conversion occurs during heterotypic wound healing, when hedgehog (hh)- expressing peripodial cells induce ectopic engrailed (en) expression in the apposing anterior columnar cells. In addition, the disc blastema, like its vertebrate counterpart, is able to form a normal regenerate (complete leg disc and adult leg) when isolated from the remaining disc fragment(Karpen and Schubiger, 1981; Brockes and Kumar, 2005). Regenerative plasticity is also observed when a few blastema cells switch fate to that of another disc type (e.g., leg-to-wing), in a phenomenon known as transdetermination (Gehring, 1966; Gehring et al., 1968; Hadorn, 1963;Wildermuth, 1968). Transdetermination events are closely associated with regenerative disc growth. Clonal analysis, for example, has shown that blastema cells first regenerate the missing disc structures, and only then, are they competent to transdetermine (Gehring, 1967;Wildermuth, 1968).
Little is known about how the regeneration blastema forms in the fragmented leg disc, although ectopic Wingless (Wg/Wnt1) expression is detected along the cut site, both prior to and during blastema formation (Gibson and Schubiger, 1999) (unpublished observations). Wg is a developmental signal in many different tissues and animals; in flies Wg patterns all of the imaginal discs, functioning as both a morphogen and mitogen to regulate disc cell fate and growth (Johnston and Sanders, 2003; Strigini and Cohen, 1999). In lower vertebrates, Wnt ligands are key regulators of blastema formation during epimorphic regeneration (Kawakami et al., 2006; Stoick-Cooper et al., 2007). Thus, activation of Wg within the disc blastema is potentially important for regeneration. This idea is consistent with the observation that ubiquitous expression of wg during the second or third larval instars, in unfragmented leg discs, is sufficient to induce a regeneration blastema in the proximodorsal region of the disc, known as the weak point (Johnston and Schubiger, 1996; Sustar and Schubiger, 2005). Moreover, ubiquitous expression of wg mimics the pattern deviations associated with leg disc fragmentation and subsequent regeneration, including the duplication of ventral with concomitant loss of dorsal pattern elements and leg-to-wing transdetermination events (Johnston and Schubiger, 1996; Maves and Schubiger, 1995). Thus, leg disc regeneration can be examined using two experimental protocols: fragmentation or ubiquitous wg expression. However, it is important to point out that only fragmentation-induced regeneration involves wound healing.
Precisely which molecules and signaling pathways are required for the process of regeneration remain poorly understood, partly because the organisms historically used to study regeneration (e.g., newts and salamanders) have been refractory to genetics and molecular manipulations. Recently, however, the use of new genetic techniques together with ‘regeneration’ model systems – such as planarians, hydra and zebrafish have given researchers the opportunity to examine the mechanisms of regeneration and to identify the genes, proteins and signaling pathways that regulate different regenerative processes (Broun et al., 2005, 1999; Cebria et al., 2002; Nechiporuk and Keating, 2002; Reddien et al., 2005; Schummer et al., 1992; Technau and Bode, 1999; ten Dijke and Hill, 2004; Thummel et al., 2006). For example, a large scale RNAi-based screen was performed to survey gene function in planarian tissue homeostasis and regeneration (Reddien et al., 2005). Out of ~1000 genes examined, RNAi knock-down of 240 displayed regeneration-related phenotypes, including defects in wound healing, blastema formation and blastema cell differentiation (Reddien et al., 2005). Despite these studies, however, it remains unclear whether regeneration requires only the modulation of genes expressed at the time of injury, the reactivation of earlier developmental genes and/or signaling pathways, or the activation of novel genes specific to the process of regeneration. Thus, a major interest in the field of regenerative biology is the identification of gene products that regulate blastema formation, blastema growth and regenerative cellular plasticity. We have performed a genetic screen aimed at identifying genes that regulate cellular plasticity and regeneration using Drosophila prothoracic leg discs. We screened a collection of 97 recessive lethal P-element lacZ (PZ) insertion lines (Russell et al., 1998) and identified six genes that function in wg-induced leg disc regeneration, including genes with functional ties to Wg signaling as well as chromatin remodeling proteins.
2. Results
2.1. PZ insertions that modify wg-induced regeneration and transdetermination
We have focused on the regeneration of prothoracic leg discs because the process of regeneration in these discs has been well-characterized both at the descriptive and molecular level (Johnston and Schubiger, 1996; Maves and Schubiger, 1995; Maves and Schubiger, 1998; Sustar and Schubiger, 2005). Leg disc regeneration can be induced either by disc fragmentation and in vivo culture or by ubiquitous wg expression in mid-second or early third instar larvae (Johnston and Schubiger, 1996; Sustar and Schubiger, 2005). Regeneration induced by both experimental protocols causes leg-to-wing transdetermination in the proximodorsal region of the leg disc, known as the weak point (Maves and Schubiger 1995; Johnston and Schubiger 1996; Sustar and Schubiger 2005). Transdetermination events, therefore, are closely associated with regenerative disc growth and proliferation (Tobler, 1966; Schubiger, 1973; Sustar and Schubiger, 2005). For example, when regeneration is augmented by additional disc injury both the frequency and area of transdetermination increase (Tobler, 1966). Conversely, when regenerative proliferation and growth are inhibited, transdetermination is not observed (Schubiger, 1973; Sustar and Schubiger, 2005). These observations indicate that we can screen mutations in genes for their function in leg disc regeneration by examining the frequency of transdetermination. Here we focus specifically on leg-to-wing transdetermination because it involves the ectopic activation of the wing selector gene vestigial (vg) in leg disc cells (Maves and Schubiger, 1995). Furthermore, activation of vg in leg-to-wing transdetermination has been molecularly characterized and is known to require the vg-Boundary Enhancer (vgBE) (Kim et al., 1996; Maves and Schubiger, 1998; Williams et al., 1991). Since both fragmentation and ubiquitous wg expression induce regeneration and transdetermination, we conducted our screen by overexpressing wg – a much simpler procedure than fragmentation and in vivo culture.
Previously, 470 recessive lethal P-lacZ enhancer trap lines were screened for ectopic expression after imaginal disc injury and regeneration (Russell et al., 1998). In this study, Russell et al. (1998) induced disc regeneration in vivo by random cell death using two different methods: (1) a temperature sensitive cell–lethal mutation in suppressor of forked [su(f)12] and (2) a short exposure to gamma radiation. Out of the 470 PZ insertions, a total of 118 were identified with ectopic lacZ expression after gamma radiation, and 47 of those were also activated after su(f)-induced cell death (Russell et al., 1998). The design of this screen identified genes that potentially function in radiation repair and/or apoptosis, as well as disc regeneration. Therefore, we re-examined the 118 PZ insertion lines, of which only 97 remain as viable fly stocks, for their role in wg-induced leg disc regeneration.
Ubiquitous expression of wg (Act5C>wg) during the third larval instar induces leg-to-wing transdetermination with a high frequency (>90% of leg discs analyzed) (Maves and Schubiger, 1998; Sustar and Schubiger, 2005). This protocol was modified in order to identify PZ insertions that both enhance and suppress wg-induced leg-to-wing transdetermination and disc regeneration. A protocol of ubiquitously expressing wg (Act5C>wg) during the mid-second larval instar (60 h after egg deposition, AED) was selected because leg-to-wing transdetermination was observed at a moderate frequency of 32% (n = 60 discs) (Fig. 1A, B and Table 1). Hereafter, we refer to this frequency as the wg-induced transdetermination control. Leg-to-wing transdetermination was detected by the ectopic expression of Vg in leg disc cells (Maves and Schubiger, 1995) (Fig. 1B). To identify genes that function in regeneration, we screened for PZ insertions that when heterozygous mutant (PZ/+) would dominantly enhance or suppress the frequency of the wg-induced transdetermination control. Fig. 1 outlines the genetic details of the screen. Out of an initial collection of 97 lethal PZ insertions, we tested 90 for their ability to modify leg disc regeneration. Seven PZ insertions were excluded from the screen because the fly stocks were no longer recessive lethal (see Supplementary Table 1). Each PZ line was examined in three replicate experiments to ensure a consistent interaction (Supplementary Table 1). Of the 90 lines, 17 significantly suppressed and 2 enhanced the wg-induced transdetermination control (Table 1). To rule out possible interactions due to genetic background and to further confirm a functional interaction with wg, we tested additional mutant alleles of 10 genes that were identified as modifiers (taiman, Secβ61, polo, Krüppel homolog-1, ken and barbie, cyclinA, jing, combgap, schnurri and Nup154), and found that 9 similarly modified wg-induced leg-to-wing transdetermination (Supplementary Table 1). schnurri (shn) was excluded from the screen because an amorphic allele of shn (shn1) did not significantly modify the frequency of wg-induced transdetermination (data not shown).
Fig. 1.
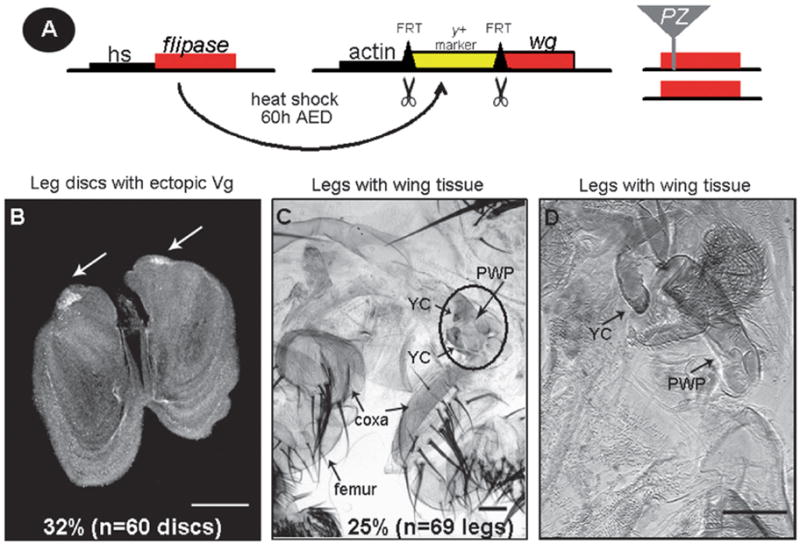
wg-induced regeneration and transdetermination. (A) Leg-to-wing transdetermination was induced in each heterozygous PZ insertion line by ubiquitously expressing wg in mid-second instar larvae (60 h after egg deposition, AED) using the FLP/FRT system(for details see Section 5). (B) Using this protocol, wg overexpression alone (Actin5C>wg, referred to as the wg-induced transdetermination control) induces ectopic Vg expression (white arrows) in leg discs with a moderate frequency of 32% (n = 60 discs).White scale bar, 50 μm. (C and D) Leg cuticle fromActin5C>wg pharate adults (differentiated but not enclosed animals) contain wing tissue with a frequency of 25% (n = 69 legs). (C) Black circle within the leg cuticle highlights ventral wing hinge structures including the yellow club (YC) and pleural wing process (PWP). The proximal leg segments, coxa and femur, are labeled. (D) Leg cuticle with transdetermined wing structures (YC and PWP) at a higher magnification than in (C). Black scale bars, 100 μm.
Table 1.
PZ lines that suppress or enhance wg-induced leg-to-wing transdetermination
| Genotype | LacZ expression | In situ expression | Frequency (%) of Vg expression in leg discs (n = discs) | Frequency (%) of legs with wing tissue (n = discs) |
|---|---|---|---|---|
| Act>wg | 32 (60) | 25 (69) | ||
| Suppressors | ||||
| l(3)01629 | Class 4 | N/A | 4 (113)* | 11(19) |
| taiman (tai01351) | Class 4 | N/A | 5 (97)* | 8 (24) |
| Secβ6107214 | Class 3 | Class 3 | 6 (107)* | 6 (46)** |
| polo01673 | Class 4 | N/A | 7 (53)** | 15 (34) |
| Krüppel-homolog-1 (Kr-h110642) | Class 3 | Class 4# | 8 (128)* | 10 (30) |
| Syx1301470 | Class 3 | N/A | 8 (97)* | 6 (31)** |
| ken and barbie (ken02970) | Class 1 | Class 1 | 9 (114)* | 3 (40)** |
| oho23B03575 | Class 2 | Class 2 | 9 (88)* | 13 (23) |
| cyclin A (cycA03946) | Class 1 | Class 1 | 10 (119)* | 19 (54) |
| l(2)00248 | Class 3 | N/A | 10 (96)* | 12 (30) |
| CG309471483a | Class 1 | Class 1 | 10 (80)** | 15 (20) |
| l(3)05203 | Class 4 | N/A | 11 (74)** | 23 (40) |
| rpd304556 | Class 3 | Class 3/4 | 12 (122)** | 35 (20) |
| jing01094 | Class 4 | Class 4 | 13 (113)** | 3 (32)** |
| combgap (cg07659) | Class 4 | Class 4 | 13 (100)** | 10 (20) |
| Nup15401501 | Class 3 | Class 3 | 13 (68)** | 8 (26) |
| Aly02267 | Class 4 | Class 4 | 15 (117)** | 5 (40)** |
| Enhancers | ||||
| l(3)01344 | Class 4 | Class 2# | 59 (92)** | 25 (24) |
| S6 kinase (S6k07084) | Class 3 | Class 3/4 | 48 (82)** | 19 (48) |
Data shown are from prothoracic leg discs and adult legs. 90 PZ insertion lines (see Supplementary Table 1) were examined for their ability when heterozygous mutant to dominantly modify wg-induced regeneration and transdetermination. Transdetermination frequency in leg discs and adult cuticle were generated from animals heat shocked at 60 h AED (37 °C) to induce ubiquitous wg expression.
Asterisk(s) indicate p-values <0.001;
indicate p-values within 0.01–0.05. The lacZ and in situ expression patterns of each PZ insertion falls into 4 different classes after ubiquitous wg expression (see Fig. 2, for details). The number sign (#) indicates that the lacZ expression pattern differed significantly from the in situ expression pattern (Fig. 5). N/A, in situ expression is not available.
To verify that ectopic Vg expression in the leg discs reflects actual leg-to-wing transdetermination events, we allowed animals overexpressing wg and heterozygous for the PZ insertion (Act>wg/PZ, Act>wg; PZ/+), to develop through metamorphosis and examined the presence of wing cuticle in the adult legs (Fig. 1C and D). As shown in Table 1, nearly all of the PZ insertions (15 out of 17) that suppressed Vg expression in the leg discs, showed a similar and corresponding reduction of leg-to-wing transdetermination in the adult leg cuticle. In four PZ insertions [l(3)05203, rpd3, S6k and l(3)01344], however, the frequency of ectopic Vg expression in the leg discs did not follow the frequency of leg-to-wing transdetermination in the cuticle. For example, the rpd3 insertion decreased the frequency of Vg expression in the leg discs, yet the fraction of adult legs with wing cuticle increased, although not significantly (Table 1). This difference may be caused by the development of wing cuticular structures which do not originate from vg-expressing leg disc cells; ventral wing hinge structures, such as the yellow club, for example, do not arise from vg-positive cells in the wing disc (Fig. 1C and D). Additionally, the two PZ insertions that enhanced Vg frequency in the leg discs, l(3)01344 and S6k, did not similarly increase the frequency of leg-to-wing transdetermination in the adult cuticle (Table 1). The reasons for this discrepancy could be related to the differences in resolution between the two analyses: we can detect as few as 2–3 Vg-expressing cells in the leg disc through antibody staining, yet identifying small wing cuticular structures, such as a single wing bristle, within the adult leg cuticle is much more difficult to recognize. Nevertheless, we find that the expression of Vg in leg disc cells is a reliable indicator of transdetermination to wing identity and subsequent terminal differentiation of adult wing structures.
2.2. Expression of PZ insertions during leg disc regeneration
Having identified 19 genes that dominantly modify wg-induced leg-to-wing transdetermination, we defined the lacZ expression of these genes to establish possible correlations with disc regeneration. Specifically, we tested for changes of candidate gene expression in the wg-induced regeneration blastema (proximodorsal cells of the leg disc). For this, first thoracic leg discs were harvested from wandering third instar PZ/+ control larvae and PZ/Act>wg or Act>wg/+; PZ/+ experimental larvae 2.5 days (120 h AED) after the induction of wg during the mid-second instar (60 h after egg deposition, AED). Both control and experimental leg discs were stained for β-gal. Among the 19 PZ insertion lines, we observed four different classes of lacZ expression changes: (1) no change from the endogenous lacZ expression pattern, (2) complete or partial loss of lacZ expression, (3) ubiquitous expression throughout the leg disc and (4) blastema-specific expression. Representative examples of the four classes are shown in Fig. 2. Among the entire PZ collection (90 insertions), the most common expression pattern following ubiquitous wg expression was that of no change from the endogenous lacZ expression pattern (Fig. 2A). In contrast, among the 19 PZ insertions that modified wg-induced leg-to-wing transdetermination eight displayed expression limited to the regeneration blastema (Fig. 2B). These findings indicate that the screen found an enrichment for genes activated in the regeneration blastema following ubiquitous wg expression (Fig. 2A). Three of the 19 PZ insertions, ken and barbie (ken), cyclin A (cycA) and CG30947, showed minor or undetectable expression changes upon wg-induced regeneration (data not shown). ken and cycA were ubiquitously expressed, while CG30947 displayed low levels of expression during regeneration (Fig. 2C, Class 1; data not shown). One of the 19 PZ insertions, oho23B, showed complete loss of the endogenous lacZ expression pattern upon wg-induced regeneration, including loss of expression in the presumptive tarsal segments and chordotonal organ (Fig. 2C, Class 2). In seven of the 19 PZ insertions, Krüppel-homolog-1 (Kr-h1), Nup154, rpd3, S6 kinase (S6k), Secβ61, l(2)00248, Syx13, expression was low or absent in prothoracic leg discs of third instar larvae, but increased significantly and ubiquitously in the discs during leg disc regeneration (Fig. 2C, Class 3 and Fig. S1). Finally, a total of eight lines, jing, taiman (tai), combgap (cg), polo, l(3)05203, l(3)01629, l(3)01344, Aly, exhibited lacZ expression limited to the wg-induced regeneration blastema (Fig. 2C, Class 4 and Fig. S2). The blastema-specific expression of these genes during wg-induced regeneration, together with their ability to modify leg-to-wing transdetermination, suggests that they function in disc regeneration as well as regulate disc cell plasticity.
Fig. 2.
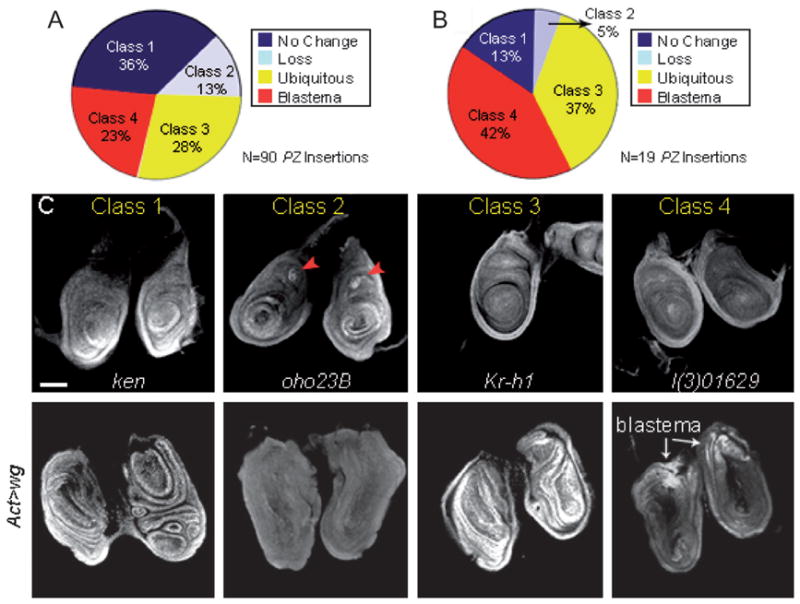
LacZ expression of enhancer trap lines that modify leg disc regeneration. PZ insertions lines were examined for lacZ expression in the regeneration blastema (proximodorsal region) following ubiquitous wg expression. (A) Pie-chart shows the distribution of the 90 PZ insertions into four different classes of lacZ expression: Class 1 lines show no expression change upon ubiquitous wg expression (36%, blue), Class 2 lines show partial or complete loss of expression (13%, light blue), Class 3 lines exhibit ubiquitous expression (28%; yellow) and Class 4 lines display expression limited to the regeneration blastema (23%; red). (B) Pie-chart shows the lacZ expression patterns of the 19 PZ insertions identified to modify wg-induced leg-to-wing transdetermination. Note that there is an enrichment for genes that exhibit blastema-specific expression following ubiquitous wg expression (Class 4, 42%; red). (C) Representative examples of the four classes of lacZ expression patterns. Upper panels show endogenous expression in prothoracic leg disc pairs from four PZ lines [ken and barbie (ken), oho23B, Krüppel-homolog-1 (Kr-h1), and l(3)01629]. The leg discs were dissected from wandering third instar larvae. Lower panels show lacZ expression of ken, oho23B, Kr-h1, and l(3)01629 after ubiquitously expressing wg (Act > wg) during the mid-second instar. Leg disc pairs were dissected 2.5 days (120 h AED) after wg induction. Red arrowheads mark lacZ expression in the chordotonal organ. Scale bar, 50 μm.
2.3. mRNA expression of PZ lines that modify wg-induced regeneration
A major criterion of our screen was to identify genes activated specifically in the blastema during leg disc regeneration. Because P-elements can insert into chromosomes in either the 5′ or 3′ direction, or detect only a subset of enhancers and possibly even disrupt regulation by insertion, it is necessary to establish that the lacZ reporter expression represents the actual expression of the adjacent gene. For many of the PZ insertions identified, both the insertion site and adjacent gene were known. To verify the lacZ expression from the enhancer trap lines, we made cDNA probes to the genes that modified transdetermination, and performed in situ hybridization in wild-type and wg-overexpressing leg discs. Five PZ lines [l(3)01629, tai, Syx13, l(2)00248 and l(3)05203] were not examined, either because the P-element insertions were near uncharacterized genes or full length cDNAs were not available (Table 1). Of the 14 genes examined by in situ hybridization, 11 (ken, cycA, CG30947, oho23B, Secβ61, Nup154, rpd3, S6k, jing, Aly and cg) reliably reproduced the enhancer trap expression pattern from the PZ insertion lines (Fig. 3A–K, Table 1). Expression of the ken, cycA and CG30947 genes did not alter significantly after ubiquitous wg expression (Fig. 3A–C, Class 1), while expression of oho23B in the distal primordia of the leg disc (data not shown) was lost after wg-induced regeneration (Fig. 3D, Class 2). Transcripts from Secβ61, Nup154, rpd3, and S6k were ubiquitously expressed following wg overexpression (Fig. 3E–H, Class 3). In most leg discs (>50%), rpd3 and S6k expression were found at significantly higher levels in the regeneration blastema (n = 26 and 32 discs, respectively; Fig. 3G and H, black arrowheads). Consistent with these observations, we found that the enhancer trap expression of these two genes, following ubiquitous wg expression, was noticeably higher in vg-expressing leg cells that had transdetermined to wing identity (Fig. 4A–A‴ and data not shown). Thus, upon wg-induced regeneration these two genes are expressed throughout the entire leg disc, but at noticeably higher levels in the blastema (Fig. 3G, H and Table 1). In contrast, in situ hybridization of the jing, Aly and cg genes detected blastema-specific expression in the majority of leg discs analyzed (>60%, n = 19, 24 and 27 discs, respectively; (Fig. 3I–K, Class 4). jing-lacZ expression was consistently adjacent to, but never overlapping with, leg disc cells expressing ectopic Vg (Fig. 4B–B‴; data not shown), while the enhancer trap expression of cg and Aly were coincident with vg-expressing leg disc cells (Fig. 4C–C‴; data not shown). Of the remaining three genes, we identified two, l(3)01344 and Kr-h1, whose expression by in situ hybridization differed from the enhancer trap expression pattern (Fig. 5A–B). In situ hybridization of l(3)01344 revealed uniform and low levels of expression in prothoracic leg discs from third instar larvae (data not shown) which was reduced to undetectable levels upon wg-induced regeneration (Fig. 5A, Class 2). This differed significantly from the l(3)01344-lacZ expression pattern which was found in the wg-induced regeneration blastema (Fig. 5A′). Additionally, Kr-h1-lacZ expression was ubiquitous after wg induction (Fig. 5B′), yet by in situ hybridization Kr-h1 transcripts were detected specifically in the regeneration blastema (70%, n = 23; Fig. 5B, Class 4). Finally, we could not reliably detect expression of the polo gene by in situ hybridization. Thus, the enhancer trap expression analysis together with in situ hybridization experiments, has allowed us to identify six genes (jing, Aly, cg, Kr-h1, rpd3 and S6k) that are activated in the regeneration blastema upon ubiquitous wg expression. Heterozygous mutations in the jing, Aly, cg, Kr-h1 and rpd3 genes dominantly suppressed, while S6k enhanced wg-induced leg-to-wing transdetermination (Table 1), supporting the conclusion that they function in regeneration.
Fig. 3.
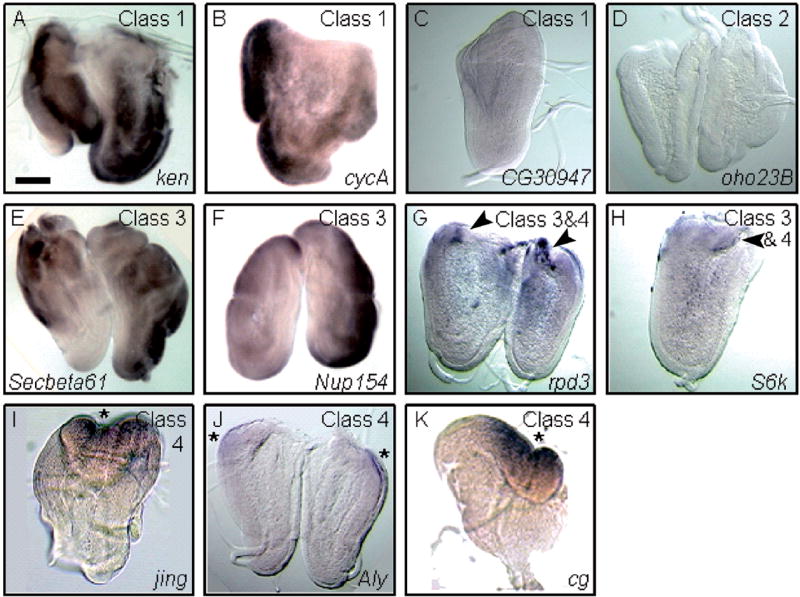
Whole mount in situ hybridization to wg-overexpressing leg discs. (A–K) wg-overexpressing leg discs were hybridized with probes from genes that modify wg-induced leg-to-wing transdetermination. Expression of the ken (A), cycA (B), CG30947 (C), oho23B (D), Secβ61 (E), Nup154 (F), rpd3 (G), S6k (H), jing (I), Aly (J) and cg (K) genes by in situ hybridization reproduced the enhancer trap expression pattern. The ubiquitous expression of the ken and cycA genes and low level expression of CG30947 did not significantly alter upon wg misexpression (Class 1). Expression of oho23B was not detected upon wg-induced leg disc regeneration (Class 2), while the Nup154, S6k, rpd3 and Secβ61 genes were ubiquitous or broadly expressed in the dorsal region of wg-overexpressing leg discs (Class 3). Note that rpd3 and S6k expression is considerably higher in the wg-induced regeneration blastema (black arrowheads in G and H; Class 3 and 4). The jing, Aly, and cg genes exhibit blastema-specific expression following ubiquitous wg expression (Class 4). Asterisks mark the proximodorsal regeneration blastema. Scale bar, 100 μm.
Fig. 4.
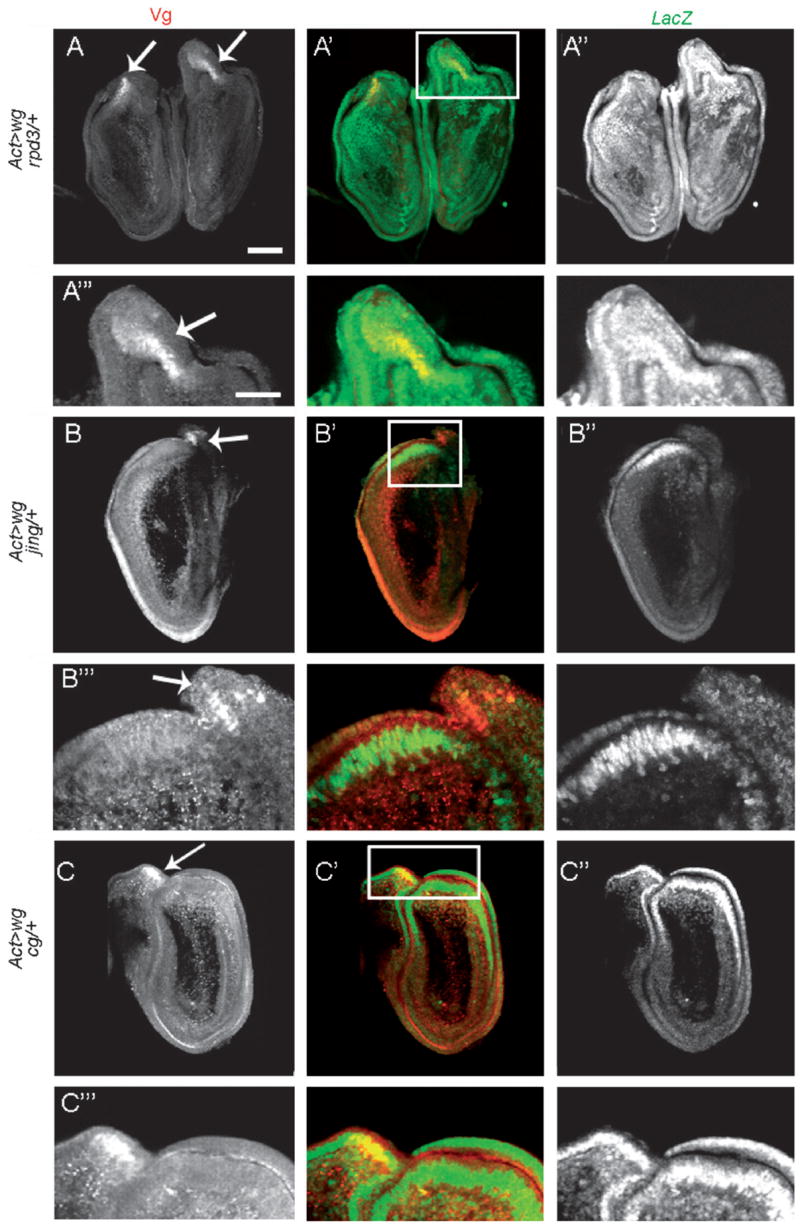
Expression of PZ lines in transdetermined leg disc cells. (A–A″) Actin5C>wg; rpd3/+ prothoracic leg disc pair; (B–B″) Actin5C>wg, jing/+ prothoracic leg disc; (C–C″) Actin5C>wg, cg/+ prothoracic leg disc. (A‴–C‴) are enlargements of the boxed regions in A′–C′, respectively. The single channels are shown in first (Vg expression) and last (LacZ expression) columns. In the merged images Vg expression is in red and LacZ expression is in green. White arrows mark Vg-expressing leg disc cells. (A′–A‴) LacZ expression from PZ insertion rpd3 is ubiquitous after wg overexpression, yet higher expression levels are detected in transdetermined leg disc cells. (B′–B‴) After wg overexpression, jing-lacZ expression is localized to the regeneration blastema and adjacent to, but not overlapping with, vg-expressing leg disc cells. (C′–C‴) LacZ expression from PZ insertion cg is restricted to the regeneration blastema after wg induction, and generally co-localizes with vg-expressing leg disc cells (4/4 discs). White scale bars, 50 μm.
Fig. 5.
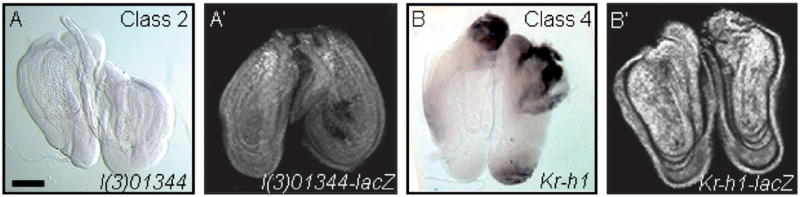
In situ hybridization of the l(3)01344 and Kr-h1 genes reveals different expression patterns from the enhancer trap lines. (A and B) wg-overexpressing leg discs were hybridized with probes to the l(3)01344 and Kr-h1 genes. In situ hybridization of l(3)01344 (A) and Kr-h1 (B) does not reproduce the lacZ expression pattern from the enhancer trap lines which are shown in (A′) and (B′), respectively. (A) The low level of l(3)01344 expression in wild-type leg discs (data not shown) is down-regulated upon wg-induced leg disc regeneration (Class 2). (B) In situ hybridization of the Kr-h1 gene detects blastema-specific expression following ubiquitous wg expression (Class 4). Scale bar, 100 μm.
2.4. Modifications of transdetermination in other discs
Ubiquitous wg expression induces not only leg-to-wing transdetermination, but other transdetermination events to wing in the eye, antenna, maxillary palp and genital discs (Table 2) (Johnston and Schubiger, 1996). To determine whether transdetermination, in general, is altered by our modifier genes we examined the frequencies of wg-induced transdeterminations in these other discs. Of the six blastema-specific genes that modified wg-induced leg-to-wing transdetermination, heterozygous mutations in two (jing and cg) significantly suppressed all wg-induced transdeterminations in the other discs (Table 2). These findings reveal that these genes generally promote regeneration and cell fate plasticity. Interestingly, the remaining four genes (Kr-h1, Aly, rpd3 and S6k) modified wg-induced transdeterminations in a context-dependent manner. For example, animals that were heterozygous mutant for Kr-h1 suppressed transdetermination events in all of the discs except the genital disc (Table 2). This suggests that some genes can modify cell fate plasticity, in general, while others act in a more disc-specific manner.
Table 2.
Effects of modifier genes on transdetermination to wing in eye-antennal and genital discs
| Genotype | Eye-to-wing (%) (n) | Antenna-to-wing (%) (n) | Maxillary palp-to-wing (%) (n) | Genital-to-wing (%) (n) |
|---|---|---|---|---|
| Act>wg (control) | 25 (53) | 28 (71) | 20 (71) | 29 (24) |
| rpd304556 | 0 (28)** | 0 (28)* | 11 (28) | 14 (28) |
| jing01094 | 0 (27)** | 0 (27)* | 4 (27)** | 0 (15)** |
| cg07659 | 0 (29)** | 0 (29)* | 0 (29)** | 5 (22)** |
| Kr-h110642 | 0 (20)** | 0 (20)** | 5 (20)** | 46 (22) |
| Aly02267 | 4 (24)** | 4 (24) | 15 (20) | 20 (20) |
| S6k07084 | 22 (32) | 0 (32)* | 22 (32) | 13 (23) |
Data shown are from eye-antennal (which includes the maxillary palp) and genital discs. The number of discs with ectopic Vg expression per total number of discs examined is given in percentages. Transdetermination frequency in eye-antennal, maxillary palp and genital discs were generated from animals heat shocked at 60 h AED (37 C) to induce ubiquitous wg expression. Asterisk(s) (*) indicate p-values <0.001; (**) indicate p-values within 0.001–0.05.
2.5. Blastema formation is altered in genes that modify leg-to-wing transdetermination
To investigate how these genes function in regeneration we examined blastema formation in animals heterozygous mutant for the PZ insertions. Ubiquitous expression of wg during the third larval instar (72 h AED) induces a regeneration blastema within 48 h (82%, n = 22 discs; Fig. 6A) (Sustar and Schubiger, 2005). As the blastema is formed, the surrounding cells exit the cell cycle (Fig. 6A′) (Sustar and Schubiger, 2005). In four of the six PZ insertions, we observed significant changes in blastema formation and thus regeneration. Leg discs from jing/+ animals rarely formed a blastema, instead, cell replication was randomly distributed throughout the leg discs both 48 and 72 h after wg induction (62%, n = 21 discs; 84.6% n = 26 discs, respectively) (Fig. 6B and B′). Aly/+ and cg/+ discs, in contrast, were able to form a blastema, yet oneday later compared to the wg-expressing controls (Fig. 6A, A′, C, C′, D and D′). Instead of cell replication limited to the blastema at 48 h, Aly/+ and cg/+ leg discs showed a random distribution of cell replication (54.5%, n = 22; 53%, n = 18, respectively) (Fig. 6C and D). However, after 72 h of wg expression a blastema formed in Aly/+ (81%, n = 22) and cg/+ (84%, n = 19) leg discs with a frequency similar to the wg-expressing controls (74%, n = 23) (Fig. 6C′ and D′). Like Aly/+ and cg/+ animals, rpd3/+ animals were able to form a blastema, but at a reduced frequency when compared to the wg-expressing controls (55%, n = 21). Leg discs that failed to form a blastema showed random cell replication (30%) or no replication (15%). Similar patterns of replication were observed in rpd3/ + leg discs after 72 h of wg expression (data not shown). Blastema formation in S6k/+ and Kr-h1/+ animals was not significantly different from the wg-expressing control animals (data not shown). Taken together, these results provide evidence that two wild-type copies of jing, Aly, cg and rpd3 are required for the initiation of regeneration and/or the timing of regenerative proliferation, growth and cellular plasticity.
Fig. 6.
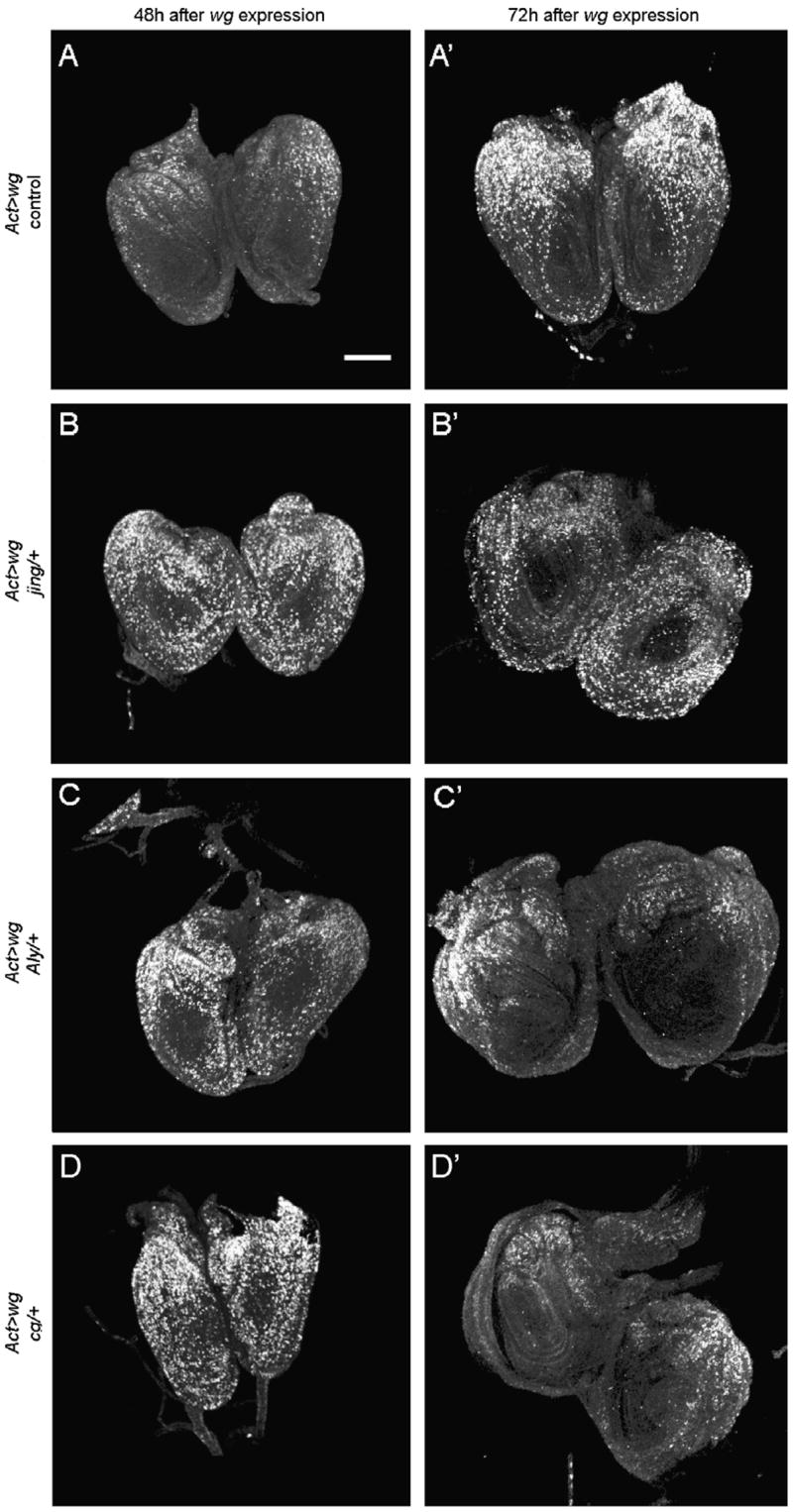
Blastema formation is altered by heterozygousmutations of jing, Alyand cg. BrdUlabeling showscell division in leg discs 48 and 72 h after ubiquitous wg expression (induced at 72 h AED) in control animals expressing wg alone (A and A′), jing/+ animals (B and B′), Aly/+ animals (C and C′) and cg/+ animals (D and D′). (A) At 48 h, cell replication is maintained in the proximodorsal leg cells, the regeneration blastema, and arrested in ventral non-blastema cells. This phenotype is observed in 82% of leg discs analyzed (n = 22). (A′) At 72 h, the blastema grows in size and expands ventrally. (B and B′) In jing/+ leg discs a blastema rarely forms, instead, random cell replication is observed at 48 h and again at 72 h after wg induction. (C, C′ and D, D′) Aly/+ and cg/+ leg discs form a blastema, yet with a one-day delay compared to the wg-expressing control animals. (C and D) At 48 h, the majority of Aly/+ and cg/+ leg discs exhibit randomcell replication, yet one day later, after 72 h of wg expression, (C′ and D′) a regeneration blastema is observed. Note that in all cases the cell cycle is arrested in the center of the discs, the primordia of tarsal segments 2–5. Scale bar, 50 μm.
3. Discussion
3.1. wg-inducible genes involved in leg disc regeneration
Extensive studies have focused on the proliferation and pattern regulation of regenerating imaginal discs (Abbott et al., 1981; Adler, 1981; Bryant et al., 1977; Dale and Bownes, 1985; French et al., 1976; Gibson and Schubiger, 1999; Johnston and Schubiger, 1996; Kiehle and Schubiger, 1985; Mattila et al., 2004; Maves and Schubiger, 1998; Sustar and Schubiger, 2005). In contrast, relatively few studies have investigated the molecular basis for regenerative growth in the discs, including the question of how the regeneration blastema forms and what gene products contribute to its proliferation and remarkable cellular plasticity (Mattila et al., 2005). Here we describe an enhancer trap screen designed to identify genes with changed gene expression during leg disc regeneration as well as required for regenerative proliferation and growth.
This screen identified 19 genes that when heterozygous mutant (PZ/+), dominantly modify wg-induced leg-to-wing transdetermination, which serves as a functional assay for disc regeneration (Table 1). Of the 19 genes, 37% are transcription factors or involved in transcriptional regulation (tai, Krh1, ken, jing, cg, rpd3 and Aly), 21% function in cell cycle regulation and growth (oho23B, S6k, polo and cycA), 10.5% play a role in protein secretion (Secβ61 and Syx13), and 31% are of other or unknown function [l(3)01629, CG30947, l(2)00248, l(3)05203, l(3)01344, Nup154]. The identification of transcription factors as the most frequent class of genes that modify wg-induced leg disc regeneration was similarly observed in a DNA microarray screen designed to identify genes enriched in leg disc cells that transdetermine to wing (Klebes et al., 2005). Together, these findings strongly suggest that transcription factors and their downstream targets play a prominent role in disc cell plasticity.
Using lacZ expression analyses, together with whole mount in situ hybridization experiments, we verified the expression patterns of the 19 genes that modified wg-induced leg-to-wing transdetermination (Figs. 2–5). This analysis identified several different expression patterns upon wg-induced regeneration, including a loss of gene expression, ubiquitous expression and genes with expression limited to the regeneration blastema (Figs. 2–5). Such observations indicate that a complex change of gene expression, both negative and positive, mediates the process of epimorphic regeneration. Six (jing, Alyi cg, rpd3, Kr-h1 and S6k) of the 19 modifiers displayed expression limited to the regeneration blastema (Table 3), indicating that we have identified novel markers of regeneration and transdetermination. The blastema-specific expression patterns of jing, Aly, cg, Kr-h1, rpd3 and S6k raised the intriguing possibility that these genes may be functionally involved in the formation, cell proliferation or maintenance of the blastema during disc regeneration (Table 3). Indeed, upon ubiquitous wg expression jing/+ animals rarely formed a regeneration blastema, indicating that two wild-type copies of jing are required for the initiation of the regenerative process. In contrast, Aly/+ and cg/+ animals formed a normal blastema, but only after a one-day delay (Fig. 6). Therefore, two wild-type copies of the Aly and cg genes are required for the proper timing of regeneration. In addition, we found that the frequency of blastema formation was reduced in rpd3/+ animals, implicating this gene in the process of regeneration. Interestingly, heterozygous mutations in all four of these genes (jing, Aly, cg and rpd3) strongly suppress wg-induced leg-to-wing transdetermination (Table 3). We speculate that the transdetermination frequency declines in these mutant animals because the initiation and/or timing of blastema formation is delayed. This idea is consistent with all previous work which has shown that blastema cells are only competent to transdetermine after they have regenerated the missing disc structures (Gehring, 1967; Wildermuth, 1968). Heterozygous mutations in Kr-h1 and S6k did not significantly alter the formation of the wg-induced regeneration blastema, however, these genes did affect regeneration-induced transdetermination (Table 3). Such results suggest that Kr-h1 and S6k specifically function to modulate the cell fate changes that occur as a consequence of regeneration.
Table 3.
Summary of genes that function in leg disc regeneration
| Gene | Expression during regeneration | Blastema formation |
wg-induced transdeterminations
|
||||
|---|---|---|---|---|---|---|---|
| Leg-to-wing | Eye-to-wing | Antenna-to-wing | Maxillary palp-to-wing | Genital-to-wing | |||
| Kr-h1^ | Blastema-specific | NC | S | S | S | S | NC |
| rpd3 | Ubiquitous/blastema | Reduced | S | S | S | NC | NC |
| jing^ | Blastema-specific | Reduced | S | S | S | S | S |
| cg^ | Blastema-specific | Delayed | S | S | S | S | S |
| Aly | Blastema-specific | Delayed | S | S | S | NC | NC |
| S6k | Ubiquitous/blastema | NC | E | NC | S | NC | NC |
Six genes were identified that met both criteria of our screen, that is, they displayed expression in the regeneration blastema during leg disc regeneration and significantly modified regeneration-induced leg-to-wing transdetermination. Expression patterns are based on whole-mount in situ hybridization in the leg disc. S, suppresses; E, enhances; NC, no significant change.
Indicates that more than one allele was tested and found to modify wg-induced leg-to-wing transdetermination. All of the genes when heterozygous mutant dominantly modify at least one other (besides leg-to-wing) wg-induced transdetermination event.
3.2. Chromatin remodeling genes and regenerative cellular plasticity
We call attention to two genes identified in our screen: jing and rpd3. Our analysis showed that upon regeneration, the expression of jing and rpd3 is blastema-specific and functional assays revealed that both genes function in blastema formation and disc cell plasticity during regeneration. Jing is a Zn-finger transcriptional repressor required for Drosophila wing development and proximodistal (P/D) axis formation in the developing leg (Culi et al., 2006). Drosophila Rpd3 is a histone deacetylase (HDAC) that alters local chromatin architecture by deacetylating histone tails, leading to chromatin compaction and transcriptional silencing (De Rubertis et al., 1996). Both jing and rpd3 function in concert with the Polycomb Group (PcG) and trithorax Group (trxG) proteins during Drosophila development (Collins and Treisman, 2000; Culi et al., 2006). This is significant because two previous studies have reported the differential regulation of several PcG and trxG proteins in regenerating leg disc cells (Klebes et al., 2005; Lee et al., 2005). These studies have also shown that the wg-induced transdetermination frequency is altered in PcG and trxG heterozygous mutant flies. PcG and trxG proteins sustain lineage-specific transcriptional programs, and thus cell identities, over multiple cell divisions through epigenetic modification of chromatin structure, notably through covalent histone modifications (Ringrose and Paro, 2004). Our findings of jing and rpd3 activation in the blastema during disc regeneration and their mutations altering blastema formation supports the idea that regenerative proliferation, plasticity and transdetermination events are regulated or driven by changes in chromatin remodeling (Klebes et al., 2005; Lee et al., 2005).
3.3. Regeneration genes and the process of transdetermination
Investigations into the molecular basis of transdetermination have shown that inputs from the Wg, Decapentapelagic (Dpp) and Hedgehog (Hh) signaling pathways activate key selector genes out of their normal developmental context, such as ectopic Vg activation in the leg disc, which then drives cell-fate switches (Maves and Schubiger, 2003). Several of the genes identified in our screen have functional ties to Wg, Dpp and Hh signaling pathways. For example, Cg is a zinc-finger transcription factor that is required for proper transcriptional regulation of the Hh signaling effector gene Cubitus interruptus (Ci) (Campbell and Tomlinson, 2000). In cg mutant wing and leg discs, Ci expression is lowered in the anterior compartment, resulting in the ectopic activation of wg and dpp and significant disc overgrowth (Campbell and Tomlinson, 2000). Another gene identified in our screen–ken, functions in concert with Dpp to direct the development of the Drosophila terminalia (Lukacsovich et al., 2003). Further characterizations of whether these genes and other modifiers of transdetermination and regeneration affect Wg, Dpp and Hh expression and/or signaling may shed light on the regulation of regeneration and regeneration-induced proliferation and cell fate plasticity.
4. Conclusion
So far, only one previous study has examined the molecular basis of blastema formation in the Drosophila imaginal discs after injury. Mattila et al. (2005) reported that the Jun-N terminal Kinase (JNK) signaling pathway recruits cells into the regenerative cell cycles. Thus, our screen contributes significantly to the identification of genes functionally involved in imaginal disc regeneration. Further characterization of the genes identified in this screen will provide important insights into the mechanism of epimorphic regeneration, not only in the Drosophila imaginal discs, but also in appendage regeneration in lower vertebrates, and ultimately aid in the development of therapies to replace or re-grow tissues lost to disease or injury.
5. Experimental procedures
5.1. Fly stocks and genetics
Flies were reared at 25 °C and on standard Drosophila media containing cornmeal and molasses. The lethal P-lacZ insertion lines used in the screen (see Supplementary Table 1), and the different mutant alleles of the PZ insertions (taiK15101, Secβ61K02307, poloKG03033, Kr-h1K04411, ken1, cycAEY11746, jing3404, cg1, Nup154EY13350, shn1) tested for function in wg-induced regeneration, were received through the Bloomington Stock Center. Each second chromosome PZ line was balanced over a CyO-GFP chromosome and each third chromosome PZ line was balanced over a TM6B, Tb chromosome in a y w hs-flp122 background. Females of the balanced PZ lines were then crossed to y w; Actin5c>y+>wg males (K. Basler) (Fig. 1). Egg collections were taken for 1–2 h after a 1 h pre-collection. To overexpress wg and induce regeneration, the progeny were heat-shocked 60 h after egg deposition (AED) for 75 min at 37 °C (Fig. 1). Using this protocol wg is overexpressed in >90% of the disc cells after heat shock (Maves and Schubiger 1995). Animals heterozygous for the PZ insertion and Actin5c>y+>wg were identified by the absence of GFP (2nd chromosome PZ lines) or Tb marker (3rd chromosome PZ lines), and either dissected 2.5 days (120 h AED) after wg induction or allowed to develop through metamorphosis. For all experiments, the y w hs-flp122; Actin5c>y+>wg animals were used as the control. To examine the formation of the regeneration blastema, ubiquitous wg expression was induced at 72h AED in animals heterozygous mutant for the following PZ insertions: jing01094, Aly02267, cg07659, rpd304556, Krh110642 and S6k07084.
5.2. Immunocytochemistry and in situ hybridization
Discs were fixed in 4% formaldehyde in PBS. The following primary antibodies were used in overnight incubations at 4 °C in PBNT: rabbit anti-Vestigial (1:200, S Carroll), rabbit anti-β-galactosidase (1:1000, Cappel). Confocal images were collected with Bio-Rad MRC 600.
Imaginal discs were hybridized with digoxigenin-labeled anti-sense and sense RNA probes as previously described (O’Neill and Bier, 1994). Probes were made from full-length cDNA containing plasmids (provided by BDGP).
For blastema analysis, BrdU (10 μg/ml) incorporation was performed at both 48 and 72 h after wg expression (induced at 72 h AED) for 20 min before a 30 min fixation.
5.3. Cuticle analysis
Transdetermination is only observed in pharate adults (adult flies unable to eclose after differentiation). Such animals were boiled in 5 N KOH for 15 min, washed two to three times with H2O and mounted in Faure’s mounting media (Ashburner, 1989).
Supplementary Material
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.mod.2007.10.003.
Acknowledgments
We thank all members of our lab for helpful discussions. We are very grateful to Margrit Schubiger, Lynn Riddiford and Celeste Berg for critically reading the manuscript. We thank Sean Carroll for supplying us with antibodies against Vestigial. This work was supported by Grant ROI GM058282 from the National Institutes of Health to G.S and K.M. was supported by Grant Number 5T32 HD07183 from NIH.
Contributor Information
Kimberly D. McClure, Email: kimberly.mcclure@ucsf.edu.
Gerold Schubiger, Email: gerold@u.washington.edu.
References
- Abbott LC, Karpen GH, Schubiger G. Compartmental restrictions and blastema formation during pattern regulation in Drosophila imaginal leg discs. Dev Biol. 1981;87:64–75. doi: 10.1016/0012-1606(81)90061-0. [DOI] [PubMed] [Google Scholar]
- Adler PN. Growth during pattern regulation in imaginal discs. Dev Biol. 1981;87:356–373. doi: 10.1016/0012-1606(81)90159-7. [DOI] [PubMed] [Google Scholar]
- Akimenko MA, Mari-Beffa M, Becerra J, Geraudie J. Old questions, new tools, and some answers to the mystery of fin regeneration. Dev Dyn. 2003;226:190–201. doi: 10.1002/dvdy.10248. [DOI] [PubMed] [Google Scholar]
- Ashburner M. Drosophila: A Laboratory Handbook and Manual. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1989. [Google Scholar]
- Bosch M, Serras F, Martin-Blanco E, Baguna J. JNK signaling pathway required for wound healing in regenerating Drosophila wing imaginal discs. Dev Biol. 2005;280:73–86. doi: 10.1016/j.ydbio.2005.01.002. [DOI] [PubMed] [Google Scholar]
- Brockes JP, Kumar A. Appendage regeneration in adult vertebrates and implications for regenerative medicine. Science. 2005;310:1919–1923. doi: 10.1126/science.1115200. [DOI] [PubMed] [Google Scholar]
- Broun M, Gee L, Reinhardt B, Bode HR. Formation of the head organizer in hydra involves the canonical Wnt pathway. Development. 2005;132:2907–2916. doi: 10.1242/dev.01848. [DOI] [PubMed] [Google Scholar]
- Broun M, Sokol S, Bode HR. Cngsc, a homologue of goosecoid, participates in the patterning of the head, and is expressed in the organizer region of Hydra. Development. 1999;126:5245–5254. doi: 10.1242/dev.126.23.5245. [DOI] [PubMed] [Google Scholar]
- Bryant PJ, Bryant SV, French V. Biological regeneration and pattern formation. Sci Am. 1977;237:66–76. doi: 10.1038/scientificamerican0777-66. [DOI] [PubMed] [Google Scholar]
- Campbell GL, Tomlinson A. Transcriptional regulation of the Hedgehog effector CI by the zinc-finger gene combgap. Development. 2000;127:4095–4103. doi: 10.1242/dev.127.19.4095. [DOI] [PubMed] [Google Scholar]
- Cebria F, Kobayashi C, Umesono Y, Nakazawa M, Mineta K, Ikeo K, Gojobori T, Itoh M, Taira M, Sanchez Alvarado A, Agata K. FGFR-related gene nou-darake restricts brain tissues to the head region of planarians. Nature. 2002;419:620–624. doi: 10.1038/nature01042. [DOI] [PubMed] [Google Scholar]
- Collins RT, Treisman JE. Osa-containing Brahma chromatin remodeling complexes are required for the repression of wingless target genes. Genes Dev. 2000;14:3140–3152. doi: 10.1101/gad.854300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culi J, Aroca P, Modolell J, Mann RS. Jing is required for wing development and to establish the proximodistal axis of the leg in Drosophila melanogaster. Genetics. 2006;173:255–266. doi: 10.1534/genetics.106.056341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dale L, Bownes M. Pattern regulation in fragments of Drosophila wing discs which show variable wound healing. J Embryol Exp Morphol. 1985;85:95–109. [PubMed] [Google Scholar]
- De Rubertis F, Kadosh D, Henchoz S, Pauli D, Reuter G, Struhl K, Spierer P. The histone deacetylase RPD3 counteracts genomic silencing in Drosophila and yeast. Nature. 1996;384:589–591. doi: 10.1038/384589a0. [DOI] [PubMed] [Google Scholar]
- Fausto N. Involvement of the innate immune system in liver regeneration and injury. J Hepatol. 2006;45:347–349. doi: 10.1016/j.jhep.2006.06.009. [DOI] [PubMed] [Google Scholar]
- French V, Bryant PJ, Bryant SV. Pattern regulation in epimorphic fields. Science. 1976;193:969–981. doi: 10.1126/science.948762. [DOI] [PubMed] [Google Scholar]
- Gehring W. Cell heredity and changes of determination in cultures of imaginal discs in Drosophila melanogaster. J Embryol Exp Morphol. 1966;15:77–111. [PubMed] [Google Scholar]
- Gehring W. Clonal analysis of determination dynamics in cultures of imaginal disks in Drosophila melanogaster. Dev Biol. 1967;16:438–456. doi: 10.1016/0012-1606(67)90058-9. [DOI] [PubMed] [Google Scholar]
- Gehring W, Mindek G, Hadorn E. Auto- and allotypical differentiation of the haltere disc blastemata of Drosophila melanogaster after culture in vivo. J Embryol Exp Morphol. 1968;20:307–318. [PubMed] [Google Scholar]
- Gibson MC, Schubiger G. Hedgehog is required for activation of engrailed during regeneration of fragmented Drosophila imaginal discs. Development. 1999;126:1591–1599. doi: 10.1242/dev.126.8.1591. [DOI] [PubMed] [Google Scholar]
- Graves BJ, Schubiger G. Cell cycle changes during growth and differentiation of imaginal leg discs in Drosophila melanogaster. Dev Biol. 1982;93:104–110. doi: 10.1016/0012-1606(82)90243-3. [DOI] [PubMed] [Google Scholar]
- Hadorn E. Differenzierungsleistungen wiederholt fragmentierter Teilstucke mannlicher Genitalscheiben von Drosophila melanogaster nach Kultur in vivo. Dev Biol. 1963;6:617–629. [Google Scholar]
- Johnston LA, Sanders AL. Wingless promotes cell survival but constrains growth during Drosophila wing development. Nat Cell Biol. 2003;5:827–833. doi: 10.1038/ncb1041. [DOI] [PubMed] [Google Scholar]
- Johnston LA, Schubiger G. Ectopic expression of wingless in imaginal discs interferes with decapentaplegic expression and alters cell determination. Development. 1996;122:3519–3529. doi: 10.1242/dev.122.11.3519. [DOI] [PubMed] [Google Scholar]
- Karlsson J. Distal regeneration in proximal fragments of the wing disc of Drosophila. J Embryol Exp Morphol. 1980;59:315–323. [PubMed] [Google Scholar]
- Karpen GH, Schubiger G. Extensive regulatory capabilities of a Drosophila imaginal disk blastema. Nature. 1981;294:744–747. doi: 10.1038/294744a0. [DOI] [PubMed] [Google Scholar]
- Kawakami Y, Rodriguez Esteban C, Raya M, Kawakami H, Marti M, Dubova I, Izpisua Belmonte JC. Wnt/beta-catenin signaling regulates vertebrate limb regeneration. Genes Dev. 2006;20:3232–3237. doi: 10.1101/gad.1475106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiehle CP, Schubiger G. Cell proliferation changes during pattern regulation in imaginal leg discs of Drosophila melanogaster. Dev Biol. 1985;109:336–346. doi: 10.1016/0012-1606(85)90460-9. [DOI] [PubMed] [Google Scholar]
- Kim J, Sebring A, Esch JJ, Kraus ME, Vorwerk K, Magee J, Carroll SB. Integration of positional signals and regulation of wing formation and identity by Drosophila vestigial gene. Nature. 1996;382:133–138. doi: 10.1038/382133a0. [DOI] [PubMed] [Google Scholar]
- Klebes A, Sustar A, Kechris K, Li H, Schubiger G, Kornberg TB. Regulation of cellular plasticity in Drosophila imaginal disc cells by the Polycomb group, trithorax group and lama genes. Development. 2005;132:3753–3765. doi: 10.1242/dev.01927. [DOI] [PubMed] [Google Scholar]
- Lee N, Maurange C, Ringrose L, Paro R. Suppression of Polycomb group proteins by JNK signalling induces transdetermination in Drosophila imaginal discs. Nature. 2005;438:234–237. doi: 10.1038/nature04120. [DOI] [PubMed] [Google Scholar]
- Lukacsovich T, Yuge K, Awano W, Asztalos Z, Kondo S, Juni N, Yamamoto D. The ken and barbie gene encoding a putative transcription factor with a BTB domain and three zinc finger motifs functions in terminalia development of Drosophila. Arch Insect Biochem Physiol. 2003;54:77–94. doi: 10.1002/arch.10105. [DOI] [PubMed] [Google Scholar]
- Mattila J, Omelyanchuk L, Kyttala S, Turunen H, Nokkala S. Role of Jun N-terminal Kinase (JNK) signaling in the wound healing and regeneration of a Drosophila melanogaster wing imaginal disc. Int J Dev Biol. 2005;49:391–399. doi: 10.1387/ijdb.052006jm. [DOI] [PubMed] [Google Scholar]
- Mattila J, Omelyanchuk L, Nokkala S. Dynamics of decapentaplegic expression during regeneration of the Drosophila melanogaster wing imaginal disc. Int J Dev Biol. 2004;48:343–347. doi: 10.1387/ijdb.041847jm. [DOI] [PubMed] [Google Scholar]
- Maves L, Schubiger G. Wingless induces transdetermination in developing Drosophila imaginal discs. Development. 1995;121:1263–1272. doi: 10.1242/dev.121.5.1263. [DOI] [PubMed] [Google Scholar]
- Maves L, Schubiger G. A molecular basis for transdetermination in Drosophila imaginal discs: interactions between wingless and decapentaplegic signaling. Development. 1998;125:115–124. doi: 10.1242/dev.125.1.115. [DOI] [PubMed] [Google Scholar]
- Maves L, Schubiger G. Transdetermination in Drosophila imaginal discs: a model for understanding pluripotency and selector gene maintenance. Curr Opin Genet Dev. 2003;13:472–479. doi: 10.1016/j.gde.2003.08.006. [DOI] [PubMed] [Google Scholar]
- Morgan TH. Regeneration. The Macmillan Company; New York: 1901. [Google Scholar]
- Nechiporuk A, Keating MT. A proliferation gradient between proximal and msxb-expressing distal blastema directs zebrafish fin regeneration. Development. 2002;129:2607–2617. doi: 10.1242/dev.129.11.2607. [DOI] [PubMed] [Google Scholar]
- O’Neill JW, Bier E. Double-label in situ hybridization using biotin and digoxigenin-tagged RNA probes. Biotechniques. 1994;17:870–875. [PubMed] [Google Scholar]
- Poss KD, Keating MT, Nechiporuk A. Tales of regeneration in zebrafish. Dev Dyn. 2003;226:202–210. doi: 10.1002/dvdy.10220. [DOI] [PubMed] [Google Scholar]
- Reddien PW, Bermange AL, Murfitt KJ, Jennings JR, Sanchez Alvarado A. Identification of genes needed for regeneration, stem cell function, and tissue homeostasis by systematic gene perturbation in planaria. Dev Cell. 2005;8:635–649. doi: 10.1016/j.devcel.2005.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhardt CA, Bryant PJ. Wound healing in the imaginal discs of Drosophila. II Transmission electron microscopy of normal and healing wing discs. J Exp Zool. 1981;216:45–61. doi: 10.1002/jez.1402160107. [DOI] [PubMed] [Google Scholar]
- Reinhardt CA, Hodgkin NM, Bryant PJ. Wound healing in the imaginal discs of Drosophila. I Scanning electron microscopy of normal and healing wing discs. Dev Biol. 1977;60:238–257. doi: 10.1016/0012-1606(77)90122-1. [DOI] [PubMed] [Google Scholar]
- Ringrose L, Paro R. Epigenetic regulation of cellular memory by the Polycomb and Trithorax group proteins. Annu Rev Genet. 2004;38:413–443. doi: 10.1146/annurev.genet.38.072902.091907. [DOI] [PubMed] [Google Scholar]
- Russell MA, Ostafichuk L, Scanga S. Lethal P-lacZ insertion lines expressed during pattern respecification in the imaginal discs of Drosophila. Genome. 1998;41:7–13. doi: 10.1139/g97-099. [DOI] [PubMed] [Google Scholar]
- Schubiger G. Regeneration of Drosophila melanogaster male leg disc fragments in sugar fed female hosts. Experientia. 1973;29:631–632. doi: 10.1007/BF01926712. [DOI] [PubMed] [Google Scholar]
- Schubiger G, Hadorn E. Auto- and allotypic differentiation in vivo cultivated foreleg blastemas of Drosophila melanogaster. Dev Biol. 1968;17:584–602. doi: 10.1016/0012-1606(68)90007-9. [DOI] [PubMed] [Google Scholar]
- Schummer M, Scheurlen I, Schaller C, Galliot B. HOM/HOX homeobox genes are present in hydra (Chlorohydra viridissima) and are differentially expressed during regeneration. Embo J. 1992;11:1815–1823. doi: 10.1002/j.1460-2075.1992.tb05233.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi X, Garry DJ. Muscle stem cells in development, regeneration, and disease. Genes Dev. 2006;20:1692–1708. doi: 10.1101/gad.1419406. [DOI] [PubMed] [Google Scholar]
- Stoick-Cooper CL, Weidinger G, Riehle KJ, Hubbert C, Major MB, Fausto N, Moon RT. Distinct Wnt signaling pathways have opposing roles in appendage regeneration. Development. 2007;134:479–489. doi: 10.1242/dev.001123. [DOI] [PubMed] [Google Scholar]
- Strigini M, Cohen SM. Formation of morphogen gradients in the Drosophila wing. Semin Cell Dev Biol. 1999;10:335–344. doi: 10.1006/scdb.1999.0293. [DOI] [PubMed] [Google Scholar]
- Sustar A, Schubiger G. A transient cell cycle shift in Drosophila imaginal disc cells precedes multipotency. Cell. 2005;120:383–393. doi: 10.1016/j.cell.2004.12.008. [DOI] [PubMed] [Google Scholar]
- Technau U, Bode HR. HyBra1, a Brachyury homologue, acts during head formation in Hydra. Development. 1999;126:999–1010. doi: 10.1242/dev.126.5.999. [DOI] [PubMed] [Google Scholar]
- ten Dijke P, Hill CS. New insights into TGF-beta-Smad signalling. Trends Biochem Sci. 2004;29:265–273. doi: 10.1016/j.tibs.2004.03.008. [DOI] [PubMed] [Google Scholar]
- Thummel R, Burket CT, Hyde DR. Two different transgenes to study gene silencing and re-expression during zebrafish caudal fin and retinal regeneration. Sci World J. 2006;6:65–81. doi: 10.1100/tsw.2006.328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiong SY, Girton JR, Hayes PH, Russell MA. Effect of regeneration on compartment specificity of the bithorax mutant of Drosophila melanogaster. Nature. 1977;268:435–436. doi: 10.1038/268435a0. [DOI] [PubMed] [Google Scholar]
- Tobler H. Cell specific determination and the relationship between proliferation and transdetermination in leg and wing primordia in Drosophila melanogaster. J Embryol Exp Morphol. 1966;16:609–633. [PubMed] [Google Scholar]
- Wildermuth H. Differenzierungsleistungen, Mustergliederungund Transdeterminationsmechanismen in hetero- und homoplastischen Transplantaten der Russelprimordien von Drosophila. Wilhelm Roux’s Arch EntwMech Org. 1968;160:41–75. doi: 10.1007/BF00573646. [DOI] [PubMed] [Google Scholar]
- Williams JA, Bell JB, Carroll SB. Control of Drosophila wing and haltere development by the nuclear vestigial gene product. Genes Dev. 1991;5:2481–2495. doi: 10.1101/gad.5.12b.2481. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.mod.2007.10.003.


