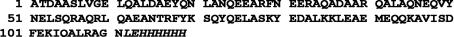The FadA adhesin from F. nucleatum, which is involved in bacterial attachment and invasion of human oral epithelial cells, has been crystallized in space group P61 or P65, and X-ray data have been collected to 1.9 Å resolution.
Keywords: FadA, adhesin, biofilm, periodontal disease, preterm birth
Abstract
Fusobacterium nucleatum is a Gram-negative anaerobe prevalent in the oral cavity that is associated with periodontal disease, preterm birth and infections in other parts of the human body. The bacteria attach to and invade epithelial and endothelial cells in the gum tissue and elsewhere via a 13.7 kDa adhesin protein FadA (Fusobacterium adhesin A). FadA exists in two forms: the intact form (pre-FadA), consisting of 129 amino acids, and the mature form (mFadA), which lacks an 18-residue signal sequence. Both forms have been expressed in Escherichia coli and purified. mFadA has been crystallized. The crystals belong to the hexagonal space group P61 or P65, with unit-cell parameters a = b = 59.3, c = 125.7 Å and one molecule per asymmetric unit. The crystals exhibit an unusually high solvent content of 74%. Synchrotron X-ray data have been collected to 1.9 Å. The crystals are suitable for X-ray structure determination. The crystal structure of FadA may provide a basis for the development of therapeutic agents to combat periodontal disease and other infections associated with F. nucleatum.
1. Introduction
Fusobacteria are involved in a wide range of infections (Roberts, 2000 ▶). Fusobacterium nucleatum is a filamentous Gram-negative anaerobe that colonizes the oral cavity (Moore & Moore, 1994 ▶). It is one of the most abundant Gram-negative anaerobes in the subgingival plaque (Moore & Moore, 1994 ▶). The organism adheres to and invades epithelial and endothelial cells to cause inflammation and infections (Han et al., 2000 ▶, 2005 ▶). It has been suggested that this bacterium migrates from the primary site of infection in the gum tissue to other tissues throughout the body, presumably via the bloodstream (Han et al., 2004 ▶). For example, F. nucleatum has been isolated from the amniotic fluids and placentas of women delivering prematurely (Chaim & Mazor, 1992 ▶). It has been shown in mice that once in the bloodstream, F. nucleatum colonizes specifically in the placenta, causing preterm and term stillbirths as well as unsustained live births (Han et al., 2004 ▶). F. nucleatum binds to a variety of host mammalian cells, including epithelial and endothelial cells, polymorphonuclear leukocytes, monocytes, erythrocytes, fibroblasts and HeLa cells. Binding and invasion of the epithelial and endothelial cells occurs via a recently discovered 13.7 kDa adhesin, Fusobacterium adhesin A (FadA; Han et al., 2005 ▶).
2. Protein expression and purification
The fadA gene from F. nucleatum 12230 was PCR-amplified and cloned into the expression vector pET-21b (Novagen) to generate pYWH417-6 so that the expression of fadA was under the control of the T7 promoter and regulated by IPTG. This construct also created a noncleavable hexahistidine tag at the C-terminus of FadA preceded by a Leu-Glu dipeptide (Fig. 1 ▶). Both the intact (pre-FadA) and the secreted (mFadA) forms were expressed from pYWH417-6. mFadA was purified by incubating IPTG-induced Escherichia coli in PBS at 333 K for 30 min, followed by cobalt ion-affinity chromatography (Talon). The eluted protein was dialyzed against 10 mM Tris–HCl pH 7.4 at 277 K. The purity of mFadA was >99% as determined by SDS–PAGE and Coomassie Blue staining.
Figure 1.
The amino-acid sequence of mature FadA (mFadA) in single-letter code. A hexahistidine tag was inserted at the C-terminal end. The sequence of this tag is shown in italics.
3. Crystallization and X-ray data collection
Crystallization trials were performed using the hanging-drop or sitting-drop method at 295 K using VDX or Cryschem 24-well plates (Hampton Research, Inc.). mFadA was crystallized under a variety of conditions from a stock solution of ∼30 mg ml−1 protein in 10 mM Tris–HCl, 50 mM NaCl, 0.02% NaN3 pH 8.0. The best crystals were obtained by vapour diffusion in sitting drops obtained by combining 2 µl protein stock solution with 2 µl reservoir solution and 0.5 µl 2 M potassium thiocyanate. The reservoir solution consisted of 0.6 ml 0.1 M sodium citrate pH 5.6, 5% dioxane and 0.05% β-octyl glucoside. Hexagonal crystals developed within one week (Fig. 2 ▶).
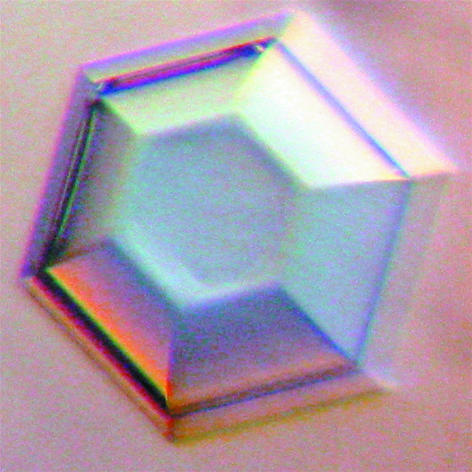
Figure 2.
A hexagonal crystal of mFadA of dimensions 0.7 × 0.7 × 0.4 mm.
The crystals were soaked for 30–60 s in reservoir solution containing 25% glycerol, mounted in a cryo-loop (Hampton Research, Inc.) and immediately flash-cooled in liquid nitrogen for X-ray data collection on an in-house X-ray diffraction system (Bruker Proteum-R) or for transport to a synchrotron-radiation source. Synchrotron data were collected at beamline 4.2.2 at the Advanced Light Source (ALS), Berkeley, California, USA and at beamline BM-19 of the Structural Biology Center at the Advanced Photon Source (APS), Argonne, Illinois, USA. Diffraction images were processed using either DENZO and SCALEPACK from the HKL-2000 suite (Otwinowski & Minor, 1997 ▶) or using the d*TREK package (Pflugrath, 1999 ▶). The crystals belong to the hexagonal space group P61 or P65, with unit-cell parameters a = b = 59.3, c = 125.7 Å. Crystal parameters and data-collection statistics for the best diffracting crystal are given in Table 1 ▶.
Table 1. X-ray data-collection statistics.
Values in parentheses are for the highest resolution shell.
| Crystal system | Hexagonal |
| Space group | P61 or P65 |
| Unit-cell parameters (Å) | a = b = 59.28, c = 125.73 |
| Resolution (Å) | 1.9 (1.97–1.90) |
| Unit-cell volume (Å3) | 388631 |
| No. of molecules per ASU | 1 |
| VM (Å3 Da−1) | 4.74 |
| Rmerge† | 0.073 (0.54) |
| Completeness (%) | 98.2 (86.4) |
| Redundancy | 10.1 (6.1) |
| 〈I/σ(I)〉 | 16.6 (2.6) |
| Radiation source | ALS‡ beamline 4.2.2 |
| Wavelength used (Å) | 1.1271 |
R
merge = 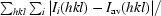
 , where I is the reflection intensity and I
av is the average intensity for multiple measurements of reflection i.
, where I is the reflection intensity and I
av is the average intensity for multiple measurements of reflection i.
Advanced Light Source, Berkeley, CA, USA.
Based on the molecular weight of 13.7 kDa, the value of V M (the volume of the unit cell divided by the total molecular weight of the protein in the unit cell) is 4.74/n, where n is the number of molecules per asymmetric unit. For V M to be within the range observed for other protein crystals, the value of n could be 1 or 2 (Matthews, 1968 ▶). To resolve this ambiguity, the density of the crystals was determined by immersing them in mixtures of xylene and bromobenzene of known density. The crystals sank and floated in solutions of 1.07 and 1.15 g cm−3, respectively. If there were two molecules per asymmetric unit, the crystals should have sunk in a solution of 1.15 g cm−3, according to Westbrook (1985 ▶). This calculation is based on an estimated density of the mother liquor of 1.06 g cm−3. Even if the actual value of the density of the mother liquor is as high as 1.10 g cm−3, the crystals should have sunk in a solution of density 1.15 g cm−3 if the crystals contain two molecules per asymmetric unit. Therefore, the conclusion is that there is only one molecule per asymmetric unit. We went to great lengths to establish this finding as it corresponds to a very high solvent content of 74%. This is unusual but not unprecedented. One has to bear in mind that FadA is likely to be a nonglobular protein since it migrates more slowly on SDS–PAGE than anticipated from its molecular weight (Han et al., 2005 ▶). Thus, FadA may not have many crystal contacts. Instead, it is anticipated that these crystals contain many solvent-filled cavities between the FadA molecules. Crystals of high solvent content usually do not diffract well. Thus, it is somewhat surprising that these crystals diffract to a resolution of 1.9 Å (Table 1 ▶).
The crystals of FadA are suitable for crystal structure determination. Molecular replacement is unlikely to work in this case since there is no homologous protein structure in the Protein Data Bank. Selenomethionine labeling or soaking the crystals in a solution of NaBr are being considered in order to solve the structure by MAD phasing.
The structure of FadA may reveal potential binding sites to receptors on the surface of epithelial cells, which could serve as drug targets and eventually lead to the development of therapeutic agents to combat periodontal disease and other infections.
Acknowledgments
This work was supported by NIH grant RO1 DE14924 to YWH. We thank the staff of beamlines 4.2.2 at the Advanced Light Source, Berkeley, California, USA and beamline BM-19 of the Structural Biology Center at the Advanced Photon Source, Argonne, Illinois, USA for beam-time allocation and expert assistance in X-ray data collection.
References
- Chaim, W. & Mazor, M. (1992). Arch. Gynecol. Obstet.251, 1–7. [DOI] [PubMed] [Google Scholar]
- Han, Y. W., Ikegami, A., Rajanna, C., Kawsar, H. I., Zhou, Y., Li, M., Sojar, H. T., Genco, R. J., Kuramitsu, H. K. & Deng, C. X. (2005). J. Bacteriol.187, 5330–5340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han, Y. W., Redline, R. W., Li, M., Yin, L., Hill, G. B. & McCormick, T. S. (2004). Infect. Immun.72, 2272–2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han, Y. W., Shi, W., Huang, T.-J, Kinder Haake, S., Park, N. H., Kuramitsu, H. & Genco, R. (2000). Infect. Immun.68, 3140–3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews, B. W. (1968). J. Mol. Biol.33, 491–497. [DOI] [PubMed] [Google Scholar]
- Moore, W. E. & Moore, L. V. (1994). Periodontol. 2000, 5, 66–77. [DOI] [PubMed] [Google Scholar]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol.276, 307–326. [DOI] [PubMed]
- Pflugrath, J. W. (1999). Acta Cryst. D55, 1718–1725. [DOI] [PubMed] [Google Scholar]
- Roberts, G. L. (2000). Br. J. Biomed. Sci.57, 156–162. [PubMed] [Google Scholar]
- Westbrook, E. M. (1985). Methods Enzymol.114, 187–196. [DOI] [PubMed] [Google Scholar]