The bicupin YwfC from B. subtilis was crystallized in two crystal forms and diffraction data were collected to 2.2 Å resolution.
Keywords: cupin fold, gene duplication, functional diversity
Abstract
A central tenet of evolutionary biology is that proteins with diverse biochemical functions evolved from a single ancestral protein. A variation on this theme is that the functional repertoire of proteins in a living organism is enhanced by the evolution of single-chain multidomain polypeptides by gene-fusion or gene-duplication events. Proteins with a double-stranded β-helix (cupin) scaffold perform a diverse range of functions. Bicupins are proteins with two cupin domains. There are four bicupins in Bacillus subtilis, encoded by the genes yvrK, yoaN, yxaG and ywfC. The extensive phylogenetic information on these four proteins makes them a good model system to study the evolution of function. The proteins YvrK and YoaN are oxalate decarboxylases, whereas YxaG is a quercetin dioxygenase. In an effort to aid the functional annotation of YwfC as well as to obtain a complete structure–function data set of bicupins, it was proposed to determine the crystal structure of YwfC. The bicupin YwfC was crystallized in two crystal forms. Preliminary crystallographic studies were performed on the diamond-shaped crystals, which belonged to the tetragonal space group P422. These crystals were grown using the microbatch method at 298 K. Native X-ray diffraction data from these crystals were collected to 2.2 Å resolution on a home source. These crystals have unit-cell parameters a = b = 68.7, c = 211.5 Å. Assuming the presence of two molecules per asymmetric unit, the V M value was 2.3 Å3 Da−1 and the solvent content was approximately 45%. Although the crystals appeared less frequently than the tetragonal form, YwfC also crystallizes in the monoclinic space group P21, with unit-cell parameters a = 46.7, b = 106.3, c = 48.7 Å, β = 92.7°.
1. Introduction
The double-stranded β-helix (cupin) domain is found in proteins that perform a diverse range of functions. A characteristic feature of the cupin scaffold is a sequence motif that is spread across two β-strands separated by a less conserved region. This motif has a consensus sequence G(X)5HXH(X)3,4E(X)6G and G(X)5PXG(X)2H(X)3N with an intervening sequence that can vary from 11 to 50 amino-acid residues. Another feature often associated with proteins having a cupin scaffold is the presence of a metal cofactor coordinated by three histidines and a glutamate residue. Members of the cupin superfamily of prokaryotic and eukaryotic proteins include several enzymes, factors that bind sugars as well as other compounds and seed storage proteins in higher plants (Dunwell et al., 2000 ▶). This variety in function seen in proteins with the cupin scaffold thus allows one to address general questions regarding the origin and evolution of diverse biochemical activities in related structural contexts (Anantharaman et al., 2003 ▶). For example, sequence-based functional correlations have established that a set of conserved histidines that were employed in sugar binding in the ancestral non-enzymatic domains evolved into a metal-coordinating centre, yielding a superoxide dismutase (Anand et al., 2002 ▶). Another evolutionary variation in this family is demonstrated by the iron-2-oxoglutarate dioxygenases, where the active-site cavity is modified to bind 2-oxoglutarate. The evolutionary strategy of variation in function in the rigid cupin domain is thus likely to be that of exploration of target or substrate space as opposed to evolution of reaction space as seen in the case of proteins with the TIM-barrel scaffold (Anantharaman et al., 2003 ▶).
Gene-fusion or gene-duplication events lead to the fusion of two or more cupin domains to form single-chain multidomain proteins. It has been suggested that this gene-duplication event occurred in a prokaryote, with subsequent evolution leading to two-domain proteins in higher plants and other eukaryotes (Dunwell et al., 2000 ▶). These gene-duplication events also aid in the subsequent diversification of function either by the absence of the cupin-sequence motif in the second cupin domain or by a change in the inter-motif spacing. Bicupin proteins with two cupin domains thus occur either as homo-bicupins (with the same inter-motif spacing between the two cupin domains) or hetero-bicupins (with different intermotif spacing). The limited structure–function information on bicupins, however, does not allow any conclusions to be drawn as to whether these gene-fusion events also provide an evolutionary mechanism to explore reaction space.
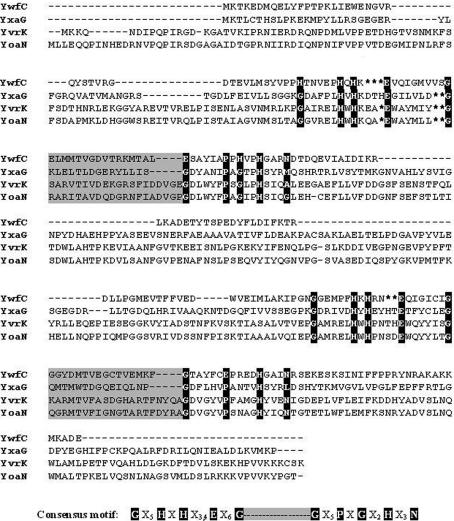
Bacillus subtilis is a good model system for the study of prokaryotic cupin diversity. Sequence analysis revealed that B. subtilis probably has the largest number of plant-related cupins (Dunwell et al., 2001 ▶), ∼20 cupin genes, comprising 0.5% of the total number of proteins coded by the genome. Phylogenetic studies using these cupin sequences provide evidence for two types of gene-duplication events that could have occurred during the evolution of B. subtilis. The first evolutionary event is believed to have resulted in an increase in the number of cupin genes, while the second involved gene fusion, thereby forming bicupin proteins. This analysis sounds plausible owing to the abundance of doublet genes (568 genes or 14% of the genome) and triplets (273 or 7% of the genome). There are four bicupins in B. subtilis: the quercetin 2,3-dioxygenase (YxaG), with an inter-motif spacing of 15 amino acids between the two cupin motifs, the protein YwfC, with an inter-motif spacing of 16 amino acids, and the oxalate decarboxylases YvrK and YoaN, with an inter-motif spacing of 20 residues (Dunwell et al., 2000 ▶). A sequence comparison of these proteins is shown in Fig. 1 ▶.
Figure 1.
Sequence alignment of the four bicupins from B. subtilis. Residues in the so-called cupin motifs that coordinate a metal cofactor are highlighted. The consensus cupin motif is shown below the sequence alignment (adapted from Dunwell et al., 2000 ▶). YxaG is a quercetin dioxygenase, whereas YvrK and YoaN are oxalate decarboxylases. The function of YwfC remains to be determined.
The function(s) of YwfC are as yet unknown. Although there are at least 18 different functional subclasses in the cupin superfamily (Dunwell et al., 2001 ▶), very few of these functional attributes have been confirmed by experimental biochemical evidence. The ambiguity in functional annotation is further compounded by the recent influx of cupin structures, the majority of which are from various structural genomics consortia. The limited primary sequence similarity between these cupin structures suggests that the total number of proteins and possibly functional variants in the cupin superfamily could be much larger than was previously anticipated. In a function-based sequence-clustering analysis, it was noted that cupins could be clustered into various subgroups that include five AraC-type transcription factors, three phosphomannose isomerases and the cysteine dioxygenases. This classification mechanism led to a clear distinction between the transcription factors and cupins with other functions. While transcription factors tend to have pI values between 6.10 and 8.48, the other proteins are generally more acidic, with pI values that range between 4.41 and 5.90 (Dunwell et al., 2000 ▶). YxaG has a pI of 5.56, YwfC of 5.19, YvrK of 5.10 and YoaN of 5.36. Sequence-based features thus suggest that YwfC could either have enzymatic and/or receptor roles. Preliminary biochemical data on this protein suggests a role of a receptor, akin to the auxin-binding protein. The structure of this protein would help in confirming this observation and perhaps also suggest other functional roles for this protein.
2. Materials and methods
2.1. Protein purification and expression
The gene encoding YwfC was cloned between the NheI and XhoI restriction sites of the bacterial expression vector pET-22b (incorporating a C-terminal polyhistidine tag) to simplify protein purification. After transforming the plasmid into BL21 (DE3) cells (Novagen Inc.), the cells were grown to an optical density at 600 nm of 0.5; at this point, the cells were induced with 1 mM IPTG (final concentration). Following this, the temperature for growth was lowered to 298 K and cells were grown for a further 7–8 h before being spun down and stored at 193 K until use. The cells were resuspended in lysis buffer (50 mM Tris–HCl buffer containing 250 mM NaCl pH 7.5). After sonication for 10 min on ice, the cell debris was separated from the crude cell lysate by centrifugation for 30 min at 10 000 rev min−1 in a Sorvall Centrifuge. After equilibration with cobalt-based Talon (Clontech Inc.) resin (approximately 4 ml resin suspension for the cell-free lysate from 10 g cell paste) and a washing step with buffer B (50 mM Tris–HCl, 250 mM NaCl, 5 mM imidazole pH 7.5), the C-terminal histidine-tagged YwfC was eluted from the column in elution buffer (50 mM Tris–HCl, 250 mM NaCl, 200 mM imidazole pH 7.5). The partially purified protein was further subjected to size-exclusion chromatography on a Sephacryl HiPrep 16/60 S-200 HR column (Amersham Biosciences Inc.). Based on the elution volume of YwfC in the size-exclusion chromatography experiment, we infer that this protein is a monomer in solution (data not shown).
2.2. Crystallization and data collection
Initial screening for crystallization conditions for this protein was performed using crystallization kits from Hampton Research (Crystal Screens 1 and 2 and PEG/Ion Screen). The conditions were examined using the hanging-drop method at 293 K, where the drop (4 µl) contained 2 µl protein solution (∼15 mg ml−1) and 2 µl reservoir solution. Although needle-shaped crystals could be obtained in more than one crystallization condition, a range of polyethylene glycols had to be examined as precipitants in order to obtain single diffraction-quality crystals. Modification of the crystallization protocol was the key to obtaining crystals of a different morphology to the needle/plate-like crystal forms obtained by the hanging-drop method. In the microbatch method, rod-like crystals belonging to the monoclinic space group were obtained in a condition containing PEG 8000 and NaCl, whereas diamond-like crystals were obtained in conditions where CaCl2 replaced NaCl. The crystals for which diffraction data are reported in this manuscript were obtained from a condition containing 20% PEG 8000 and 0.2 M CaCl2 (tetragonal form) and 20% PEG 8000 and 0.2 M NaCl (monoclinic form).
2.3. Data collection
A single crystal of YwfC was mounted on a cryo-loop using 20% PEG 400 as the cryoprotectant. The diffraction data were collected at 100 K on a MAR imaging-plate system mounted on a Bruker FRE591 rotating-anode X-ray generator. The data were processed using MOSFLM (Leslie, 1992 ▶) and were scaled using the program SCALA (Collaborative Computational Project, Number 4, 1994 ▶). The data-collection statistics are reported in Table 1 ▶.
Table 1. Summary of data-collection and processing statistics.
Values in parentheses are for the outer shell.
| Wavelength (Å) | 1.5418 | 1.5418 |
| Space group | P422 | P21 |
| Unit-cell parameters (Å, °) | a = b = 68.7, c = 211.5 | a = 46.7, b = 106.3, c = 48.7, β = 92.7 |
| Resolution limit | 35.27–2.21 (2.33–2.21) | 35.92–2.21 (2.33–2.21) |
| Total No. of observations | 573250 (76162) | 130612 (13994) |
| Total No. of unique reflections | 25906 (3264) | 22801 (2805) |
| Completeness (%) | 98.1 (87.0) | 96.4 (82.0) |
| Multiplicity | 22.1 (23.3) | 5.7 (5.0) |
| Rmerge† | 10.2 (38.1) | 9.7 (40.0) |
| 〈I〉/σ(I) | 31.3 (10.1) | 15.8 (4.1) |
R
merge = 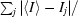
 , where I
j is the intensity of the jth reflection and 〈I〉 is the average intensity.
, where I
j is the intensity of the jth reflection and 〈I〉 is the average intensity.
3. Results and discussion
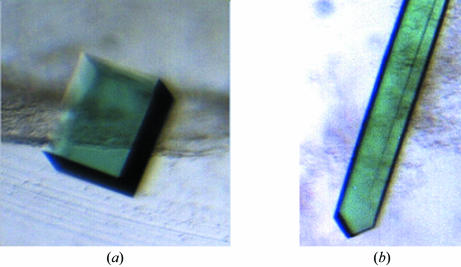
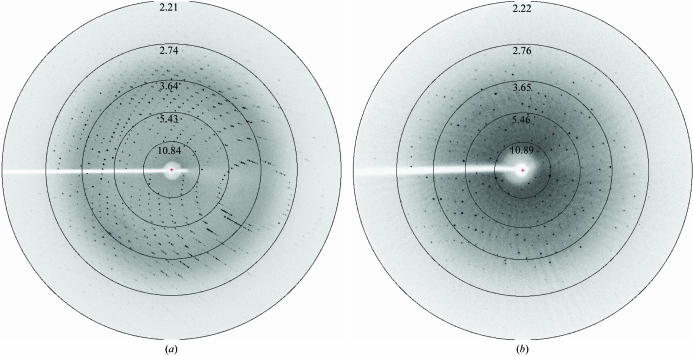
Crystals of YwfC appeared about two weeks after setting up the crystallization experiments in the microbatch method. These crystals had dimensions of ∼0.2 × 0.2 × 0.3 mm and diffracted to better than 2.2 Å resolution. These crystals (Fig. 2 ▶ a) belong to the tetragonal space group P422, with unit-cell parameters a = b = 68.7, c = 211.5 Å. Rod-like crystals (Fig. 2 ▶ b) of approximate dimensions 0.1 × 0.1 × 0.4 mm belonging to the monoclinic space group P21 appear about four weeks after setting up the crystallization condition and have unit-cell parameters a = 46.7, b = 106.3, c = 48.7 Å, β = 92.7°. These rod-like crystals appear less frequently than the tetragonal crystal form. The data-collection statistics for both these crystal forms are reported in Table 1 ▶ and diffraction patterns are shown in Fig. 3 ▶. Based on the molecular weight and the space groups, we infer that there are two molecules of YwfC in the asymmetric unit of the crystal in both crystal forms. In the case of the tetragonal space group, this corresponds to a solvent content of about 45% (Matthews, 1968 ▶). However, the elution profile of YwfC in a size-exclusion experiment suggests that the protein is a monomer in solution. Initial molecular-replacement (MR) trials were performed using Phaser (Read, 2001 ▶; Storoni et al., 2004 ▶) using a cupin protein from Thermotoga maritima (PDB code 1vj2) that had 32% sequence identity over 100 residues. This MR strategy was based on a recent study in which it was noted that phase information from ∼30% of the model is often sufficient to solve the structure of the entire protein using current crystallographic software (Rajavel et al., 2006 ▶). Accordingly, phase improvement using iterated rounds of model building, NCS averaging and maximum-likelihood density modification using RESOLVE (Terwilliger, 2000 ▶) are presently under way. We also propose to supplement the phase information using the single isomorphous replacement technique. Optimization of conditions to obtain isomorphous derivatives of YwfC is in progress.
Figure 2.
Crystals of YwfC. (a) Crystals belonging to the tetragonal space group, with approximate dimensions 0.2 × 0.2 × 0.3 mm. (b) Rod-like crystals belonging to the monoclinic space group, with dimensions ∼0.1 × 0.1 × 0.4 mm.
Figure 3.
Diffraction patterns of the YwfC crystals. (a) Tetragonal form. (b) Monoclinic form. Both crystal forms of YwfC diffract to better than 2.2 Å resolution.
Acknowledgments
The work reported in this manuscript was funded in part by a grant from the Indian Space Research Organization. BG is an International Senior Research Fellow of the Wellcome Trust, UK.
References
- Anand, R., Dorrestein, P. C., Kinsland, C., Begley, T. P. & Ealick, S. E. (2002). Biochemistry, 41, 7659–7669. [DOI] [PubMed] [Google Scholar]
- Anantharaman, V., Aravind, L. & Koonin, E. V. (2003). Curr. Opin. Chem. Biol.7, 12–20. [DOI] [PubMed] [Google Scholar]
- Collaborative Computational Project, Number 4 (1994). Acta Cryst. D50, 760–763. [Google Scholar]
- Dunwell, J. M., Culham, A., Carter, C. E., Sosa-Aguirre, C. R. & Goodenough, P. W. (2001). Trends Biochem. Sci.26, 740–746. [DOI] [PubMed] [Google Scholar]
- Dunwell, J. M., Khuri, S. & Gane, P. J. (2000). Microbiol. Mol. Biol. Rev.64, 153–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leslie, A. G. W. (1992). Jnt CCP4/ESF–EACBM Newsl. Protein Crystallogr.26
- Matthews, B. W. (1968). J. Mol. Biol.33, 491–497. [DOI] [PubMed] [Google Scholar]
- Rajavel, M., Warrier, T. & Gopal, B. (2006). Proteins, 64, 923–930. [DOI] [PubMed] [Google Scholar]
- Read, R. J. (2001). Acta Cryst. D57, 1373–1382. [DOI] [PubMed] [Google Scholar]
- Storoni, L. C., McCoy, A. J. & Read, R. J. (2004). Acta Cryst. D60, 432–438. [DOI] [PubMed] [Google Scholar]
- Terwilliger, T. C. (2000). Acta Cryst. D56, 965–972. [DOI] [PMC free article] [PubMed] [Google Scholar]