Two thermostable DNA nucleases from archaea were crystallized in different space groups; the crystals were suitable for X-ray analysis.
Keywords: nucleases, exonucleases, Archaeoglobus, Methanothermobacter
Abstract
Temperature-tolerant organisms are an important source to enhance the stability of enzymes used in biotechnological processes. The DNA-cleaving enzyme exonuclease III from Escherichia coli is used in several applications in gene technology. A thermostable variant could expand the applicability of the enzyme in these methods. Two homologous nucleases from Archaeoglobus fulgidus (ExoAf) and Methanothermobacter thermoautrophicus (ExoMt) were studied for this purpose. Both enzymes were crystallized in different space groups using (poly)ethylene glycols, 2,4-methyl pentandiol, dioxane, ethanol or 2-propanol as precipitants. The addition of a 10-mer DNA oligonucleotide was important to obtain monoclinic crystals of ExoAf and ExoMt that diffracted to resolutions better than 2 Å using synchrotron radiation. The crystal structures of the homologous proteins can serve as templates for genetic engineering of the E. coli exonuclease III and will aid in understanding the different catalytic properties of the enzymes.
1. Introduction
In living cells, DNA-cleaving enzymes fulfil important roles in DNA-repair and cell-cycle events. Their functions include, for example, various endonucleolytic or exonucleotic activities. A well studied bacterial DNA nuclease is the monomeric 31 kDa exonuclease III (ExoIII) of Escherichia coli. The enzyme possesses various catalytic activities: double-strand 3′-5′ exonuclease activity, exonucleolytic RNase H activity, 3′-phosphomonoesterase activity and endonuclease activity at apurinic/apyrimidic sites, urea-N-glycosides (Kuo et al., 1994 ▶) or O-alkylhydroxylamine-N-glycosides (Kow & Wallace, 1985 ▶). These activities are utilized in various molecular-biology methods, e.g. deletion studies (Trimble & Hozumi, 1987 ▶; DiRita & Gelvin, 1987 ▶), DNA sequencing (Henikoff, 1984 ▶; Puapaiboon et al., 2000 ▶; Brakmann & Lobermann, 2002 ▶) and in vitro recombination of homologous DNA sequences (Koltermann et al., 2002 ▶). A genetically engineered ExoIII of increased thermal stability or a homologous enzyme from a thermophilic organism would expand the applicability of ExoIII, especially in the last-mentioned method.
Two homologous nucleases from the thermophilic archea Archaeoglobus fulgidus and Methanothermobacter thermoautrophicus were chosen for this purpose (Miertzschke & Greiner-Stöffele, 2003 ▶; Pfeifer & Greiner-Stöffele, 2005 ▶). The thermostable Mg2+-dependent DNA-cleaving enzyme from A. fulgidus (ExoAf) has an activity optimum at 353 K but shows nonspecific DNase activity. It is able to degrade supercoiled plasmids and has no preference for blunt or recessed 3′-termini of linear double-stranded DNA. The Mg2+-dependent protein of M. thermoautrophicus is less temperature-tolerant and starts to unfold at 343 K. It shows an AP-site-specific DNA-nicking activity in addition to DNase-like action. However, at salt concentrations higher than 0.5 M KCl nonspecific nicking is inhibited, whilst AP-site nicking remains. Crystallization and subsequent three-dimensional structure determination of ExoAf and ExoMt is expected to provide valuable insights into the structural factors responsible for their thermal stability and for their differing catalytic properties. Based on the structures, a rational enzyme design for variants which possess the catalytic properties of E. coli ExoIII but the thermostability of the enzymes isolated from the thermophiles may be possible.
2. Materials and methods
2.1. Expression and purification
Cloning and expression of the genes xthA (ExoAf) and Mth0212 (ExoMt) in E. coli as well as protein production and purification of ExoAf and ExoMt was performed as described in Miertzschke & Greiner-Stöffele (2003 ▶) and Pfeifer & Greiner-Stöffele (2005 ▶), respectively. ExoAf and ExoMt were used directly after ion-exchange chromatography without a further concentration step. Protein concentration was determined according to Bradford (1976 ▶) using bovine serum albumin fraction V (Lot 3X04340) from Applichem (Darmstadt, Germany) as standard.
2.2. Crystallization
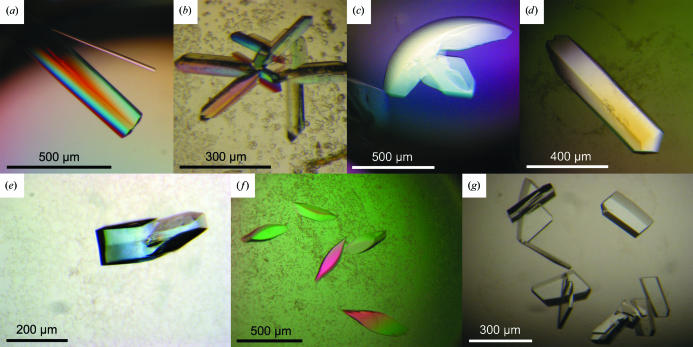
Tailor-made sparse-matrix screens (Jancarik & Kim, 1991 ▶) adapted from commercially available crystallization screens from Hampton Research (Aliso Viejo, CA, USA) and Jena BioScience (Jena, Germany) were performed at 292 K using the sitting-drop vapour-diffusion technique in three-drop 96-well Greiner plates (82 µl reservoir solution) with a hydrophobic surface in order to determine initial crystallization conditions for optimization. 0.2 µl reservoir solution and 0.2 µl protein solution were dispensed using a Cartesian eight-channel dispensing system (Genomic Solutions, Irvine, CA, USA). While we failed to crystallize unliganded ExoMt, a crystalliaztion hit for native ExoAf was optimized by the hanging-drop technique (1 µl reservoir + 1 µl protein solution equilibrated against 500 µl reservoir solution) in 24-well plates (two 12-well PVC trays from Nelipak, Venray, The Netherlands and tray boxes of polystyrol from VWR, Darmstadt, Germany) using self-coated (AquaSil from Hampton Research, Aliso Viejo, CA, USA) cover slides from Roth (Karlsruhe, Germany) to produce large rod-shaped crystals of ExoAf (Fig. 1 ▶ a) grown from polyethylene glycol (PEG) of molecular weight 8000 (Table 1 ▶).
Figure 1.
Crystals of ExoAf (a–d) and ExoMt (e–g). The crystals in (f) and (g) represent the same crystal form, but were grown using different precipitants (ethanol or dioxane, respectively). Detailed crystallization conditions for the various crystal forms are listed in Table 1 ▶.
Table 1. Crystal data and data-collection statistics of crystals grown at 292 K.
Values in parentheses are for the highest resolution shell. dsDNA, double-stranded DNA. r.a., rotating-anode generator. n.d., not determined.
| Crystal form | ExoAf-I | ExoAf-II | ExoAf-III | ExoAf-IV | ExoMt-I | ExoMt-II |
|---|---|---|---|---|---|---|
| Figure | 1(a) | 1(b) | 1(c) | 1(d) | 1(e) | 1(f), 1(g) |
| Reservoir solution | 10–12% PEG 8000, 0.05 M sodium cacodylate pH 5.0, 5 mM CaCl2 | 30% PEG 4000, 0.1 M Tris pH 8.0–8.5 | 30% PEG 4000, 0.2 M Li2SO4, 0.1 M Tris pH 8.5 | 20–26% PEG 3350, 10% 2-propanol, 0.1 M Tris pH 8.0–9.5 | 10% PEG 8000, 5–8% ethylene glycol, 0.1 M NaOAc pH 5.0–5.4 | 5–10% PEG 3350, 10–15% 2-propanol, 0.1 M MES pH 5.0–6.5 |
| Protein solution | 37 mg ml−1 ExoMt, 20 mM MES pH 6.0, 0.6 M NaCl | 17 mg ml−1 (0.57 mM) ExoAf, 0.53 mM dsDNA, 9 mM MES pH 6.0, 0.28 M NaCl | 17 mg ml−1 (0.57 mM) ExoAf, 0.53 mM dsDNA, 9 mM MES pH 6.0, 0.28 M NaCl | 16.5 mg ml−1 (0.55 mM) ExoAf, 0.50 mM dsDNA, 9 mM MES pH 6.0, 0.27 M NaCl | 8 mg ml−1 (0.27 mM) ExoMt, 0.25 mM dsDNA, 15 mM MES pH 5.7, 0.38 M NaCl | 8 mg ml−1 (0.27 mM) ExoMt, 0.25 mM dsDNA, 15 mM MES pH 5.7, 0.38 M NaCl |
| Cryocooling condition | — (298 K) | — (298 K) | Straight from drop | Paraffin oil | 25%(v/v) Ethylene glycol | — (298 K) |
| X-ray source | r.a. | r.a. | r.a./BESSY BL14.2 | r.a. | r.a./BESSY BL14.1 | r.a. |
| Wavelength (Å) | 1.5418 | 1.5418 | 1.5418/0.9537 | 1.5418 | 1.5418/0.9537 | 1.5418 |
| Data-collection temperature (K) | 298 | 298 | 298/100 | 100 | 298/100 | 298 |
| Space group† | P6x | P2x | P21 | P6x | P21 | P4x |
| Resolution limit (Å) | 2.5 | 3.2 | 2.2/1.7‡ | 2.3 | 3.0/1.8‡ | 3.0 |
| Unit-cell parameters | ||||||
| a (Å) | 252.7 | 84.5 | 69.6/67.8 | 92.0 | 45.0/44.4 | 101.3 |
| b (Å) | 252.7 | 77.9 | 61.3/60.8 | 92.0 | 99.3/96.8 | 101.3 |
| c (Å) | 76.2 | 89.0 | 77.5/72.4 | 89.0 | 81.5/80.8 | 80.7 |
| β (°) | 92.3 | 95.0/91.2 | 97.8/97.8 | |||
| VM§ (Å3 Da−1) | 2.0–3.9 | 2.0–3.3 | 2.5/2.3¶ | 3.6 | 2.7/2.6¶ | 2.3 or 3.4 |
| Solvent content§ (%) | 37–68 | 37, 50 or 62 | 51/46¶ | 66 | 54/52¶ | 46 or 64 |
| Molecules per ASU§ | 12–6 | 5–3 | 2¶ | 1 | 2¶ | 3–2 |
| Total reflections | n.d. | n.d. | 507681 | n.d. | 306722 | n.d. |
| Unique reflections | n.d. | n.d. | 64871 | n.d. | 61516 | n.d. |
| Completeness†† | n.d. | n.d. | 100 (99.9) | n.d. | 98.2 (96.6) | n.d. |
| Rsym††‡‡ | n.d. | n.d. | 7.0 (43.6) | n.d. | 6.0 (48.2) | n.d. |
| Rp.i.m.††§§ | n.d. | n.d. | 2.2 (20.5) | n.d. | 2.9 (24.3) | n.d. |
x, possible screw axes could not be determined owing to insufficient data.
Unit-cell parameters, Matthews coefficient and related values and data-collection statistics correspond to the data sets at the two given temperatures.
Matthews coefficient (Matthews, 1968 ▶) and related values were calculated under the presumption that no DNA had bound to the protein.
Values derived from the presence of one DNA molecule per ASU in addition to protein.
Data-processing statistics are only given for synchrotron data collected at 100 K.
R
sym = 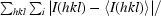
 .
.
R
p.i.m. = 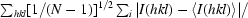
 . R
p.i.m. describes the precision of merged reflections by incorporation of the multiplicity N (Weiss, 2001 ▶).
. R
p.i.m. describes the precision of merged reflections by incorporation of the multiplicity N (Weiss, 2001 ▶).
Further screens at 292 K were set up for cocrystallization using a double-stranded blunt-ended DNA oligonucleotide (5′-CGGCTACCGC-3′ for a single strand) at a molar ratio of about 1:1. The DNA was designed to assure stable duplex formation owing to its GC-rich regions. It was purchased as single-strand dry solid from Thermo Electron GmbH (Ulm, Germany), dissolved in water and annealed to 1 mM concentration. Since cocrystallization was successful, no further DNA molecules were tried. At room temperature or in the absence of Mg2+ ions, DNA is not degraded by ExoAf (Miertzschke & Greiner-Stöffele, 2003 ▶) or ExoMt (Pfeifer & Greiner-Stöffele, 2005 ▶). These trials resulted in a couple of different crystallization hits for both nucleases. Improved crystallization conditions yielded rod-like or plate-shaped crystals of ExoAf (Figs. 1 ▶ b, 1 ▶ c and 1 ▶ d) using PEG 4000, PEG 4000 and Li2SO4, PEG 3350 or 2-propanol as precipitants (Table 1 ▶). Optimization of ExoMt hits in 24-well setups produced crystals exhibiting a compact olive-like or plate-like shape (Figs. 1 ▶ e, 1 ▶ f and 1 ▶ g) using various precipitants as listed in Table 1 ▶. Protein solutions were diluted with water to obtain the desired concentrations (Table 1 ▶). Crystals usually formed within a few days to one week.
2.3. Data collection and processing
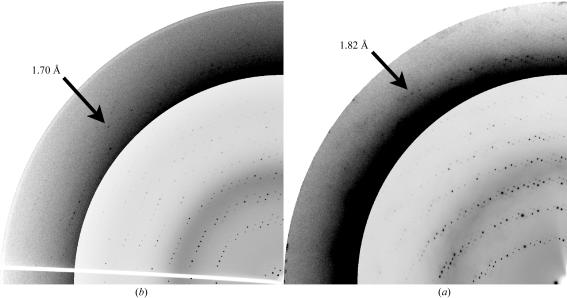
A specimen of each individual crystal form (Table 1 ▶) was first transferred to a quartz capillary (Müller, Schönwalde, Germany) in order to collect room-temperature data at in-house rotating-anode generators (MicroStar from Bruker-AXS, Karlsruhe, Germany or RU-H3R from Rigaku, Kent, England). For cryo data collection, crystals mounted in a rayon loop from Hampton Research (Aliso Viejo, CA, USA) were briefly incubated (1–2 s) in dry paraffin oil (Riboldi-Tunnicliffe & Hilgenfeld, 1999 ▶) or transferred to a buffer which had the same composition as the reservoir buffer used for crystallization plus an increasing concentration of ethylene glycol up to 25%(v/v). An appropriate fragment was manually dissected from intergrown crystals (Figs. 1 ▶ c and 1 ▶ e). Complete X-ray data sets were collected from crystal forms ExoAf-III and ExoMt-I (Figs. 1 ▶ c and 1 ▶ e, Table 1 ▶) at synchrotron beamlines BL14.1 or BL14.2 of BESSY and Free University Berlin at BESSY in Berlin (Germany) to maximum resolutions of 1.7 Å (ExoAf, Fig. 2 ▶ a) or 1.8 Å (ExoMt, Fig. 2 ▶ b). The resolution limit was chosen such that the signal-to-noise ratio was larger than 2 and R sym was lower than about 50% for the highest resolution shell. Processing and scaling (Table 1 ▶) of diffraction images was performed using programs from the HKL package (v.1.96.5 and v.1.97.2; Otwinowski & Minor, 1997 ▶) and the program RMERGE (Weiss, 2001 ▶). Further data reduction (Table 1 ▶) was conducted using the programs SCALEPACK2MTZ, CAD (Collaborative Computational Project, Number 4, 1994 ▶) and TRUNCATE (French & Wilson, 1978 ▶).
Figure 2.
X-ray diffraction patterns of crystals of (a) ExoAf and (b) ExoMt. Quarter wedges of diffraction images are shown with the high-resolution sections made darker so that reflections become clearly visible (as indicated by arrows with appropriate resolution).
3. Results and discussion
3.1. ExoAf
Purified ExoAf was first crystallized in a hexagonal space group at 292 K using PEG 8000 as precipitant in the presence of CaCl2 and at an acidic pH (Table 1 ▶). Large unit-cell parameters along two axes (a = b = 253 Å) caused overlapping reflections, thus preventing the collection of a sufficiently complete data set and determination of possible crystallographic screw axes. The addition of a double-stranded 10-mer DNA oligonucleotide at equimolar concentration yielded the new crystal forms II–IV, which could not be obtained in the absence of DNA (Table 1 ▶).
Crystal forms ExoAf-II and ExoAf-III may be related, since both have primitive monoclinic crystal lattices with one similar unit-cell parameters of about 78 Å. Data collection from an ExoAf-III crystal at 100 K was performed without additional cryoprotection by transferring the crystal directly from the crystallization drop to the cryo stream. The diffraction limit was 1.7 Å using synchrotron radiation (Fig. 2 ▶ a). However, the unit-cell c parameter and the β angle changed considerably upon crystal cooling (Table 1 ▶). Structure determination by molecular replacement showed that these crystals contain two protein molecules and one DNA molecule per asymmetric unit (data not shown). The 10-mer DNA is bound to both protein molecules. In solution, ExoAf is active as a monomer (unpublished results). For crystal form ExoAf-IV no suitable cryocondition has yet been determined.
3.2. ExoMt
ExoMt could only be crystallized in the presence of the DNA oligonucleotide. Crystal form ExoMt-I was grown from ethylene glycols of various molecular weights and also by using MPD as a precipitant (Table 1 ▶). In addition to the condition listed in Table 1 ▶, this crystal form was also obtained using 20% PEG 3350 and 0.2 M salt (NaF, NaCl, LiCl or KH2PO4) or 40–65% MPD and 0.1 M MES pH 6–7 as reservoir solution.
Successful cryoconditions were found for a crystal grown in the presence of PEG 8000 and ethylene glycol by increasing the ethylene glycol concentration to 25%. The diffraction limit extends to 1.8 Å resolution using synchrotron radiation (Fig. 2 ▶ b). The crystal unit cells did not show substantial contraction upon cryocooling. An almost complete data set was collected from an ExoMt-I crystal. Structure determination by molecular replacement shows that these crystals contain two protein molecules and one DNA duplex per asymmetric unit (data not shown). As in the case of ExoAf crystals, two ExoMt molecules bind to one 10-mer DNA. ExoMt is also active as a monomer (Miertzschke & Greiner-Stöffele, 2003 ▶). Tetragonal crystals (ExoMt-II) of various shapes were produced by using solvents that were more hydrophobic (Table 1 ▶). In addition to the condition listed in Table 1 ▶, these crystals were also obtained using 25–35% dioxane or 10–20% ethanol, 0.1 M MES or HEPES pH 5–6.5 as reservoir solution. ExoMt-II crystals diffract to a resolution of about 3 Å at the home source. Cryoconditions for ExoMt-II have yet to be established using common cryoprotectants and procedures.
4. Summary
The two thermophilic DNA-cleaving enzymes ExoAf and ExoMt were crystallized in various monoclinic or orthorhombic crystal forms. Addition of a 10-mer DNA oligonucleotide was essential to obtain crystals suitable for three-dimensional structure determination with resolution limits better than 2 Å. Based on the crystal structures determined using these crystals, it may be possible to understand the different catalytic properties of the thermophilic enzymes compared with the similar E. coli ExoIII and to obtain by rational design variants of either the thermophilic enzymes or E. coli ExoIII that possess the catalytic properties of the E. coli enzyme and the thermostability of ExoAf or ExoMt.
Acknowledgments
The authors would like to thank Uwe Müller and Martin Fieber-Erdmann from the Protein Structure Factory at BESSY (Berlin) for their help and assistance during synchrotron data collection.
References
- Bradford, M. M. (1976). Anal. Biochem.72, 248–254. [DOI] [PubMed] [Google Scholar]
- Brakmann, S. & Lobermann, S. (2002). Angew. Chem. Int. Ed. Engl.41, 3215–3217. [DOI] [PubMed] [Google Scholar]
- Collaborative Computational Project, Number 4 (1994). Acta Cryst. D50, 760–763. [Google Scholar]
- DiRita, V. J. & Gelvin, S. B. (1987). Mol. Gen. Genet.207, 233–241. [DOI] [PubMed] [Google Scholar]
- French, G. S. & Wilson, K. S. (1978). Acta Cryst. A34, 517–527. [Google Scholar]
- Henikoff, S. (1984). Gene, 28, 351–359. [DOI] [PubMed] [Google Scholar]
- Koltermann, A., Kettling, U. & Eigen, M. (2002). Patent No. DE19953854.
- Kow, Y. W. & Wallace, S. S. (1985). Proc. Natl Acad. Sci. USA, 82, 8354–8358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo, C. F., Mol, C. D., Thayer, M. M., Cunningham, R. P. & Tainer, J. A. (1994). Ann. NY Acad. Sci.726, 223–234. [DOI] [PubMed] [Google Scholar]
- Jancarik, J. & Kim, S.-H. (1991). J. Appl. Cryst.24, 409–411. [Google Scholar]
- Matthews, B. W. (1968). J. Mol. Biol.33, 491–497. [DOI] [PubMed] [Google Scholar]
- Miertzschke, M. & Greiner-Stöffele, T. (2003). Eur. J. Biochem.270, 1–12. [DOI] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol.276, 307–326. [DOI] [PubMed]
- Pfeifer, S. & Greiner-Stöffele, T. (2005). DNA Repair, 4, 433–444. [DOI] [PubMed] [Google Scholar]
- Puapaiboon, U., Jai-nhuknan, J. & Cowan, J. A. (2000). Anal Chem.72, 3338–3341. [DOI] [PubMed] [Google Scholar]
- Riboldi-Tunnicliffe, A. & Hilgenfeld, R. (1999). J. Appl. Cryst.32, 1003–1005. [Google Scholar]
- Trimble, W. S. & Hozumi, N. (1987). FEBS Lett.219, 70–74. [DOI] [PubMed] [Google Scholar]
- Weiss, M. S. (2001). J. Appl. Cryst.34, 130–135. [Google Scholar]