Abstract
A novel vacuolar Na+/H+ exchanger, CgNHX1, was cloned from a halophytic species Chenopodium glaucum by using reverse transcriptase-polymerase chain reaction (RT-PCR) and rapid amplification of cDNA ends (RACE) technique. Sequence alignment and phylogenetic analysis of 22 NHX genes from GenBank as well as the new CgNHX1 gene indicate that NHX genes shared a great degree of similarity, regardless of their glycophytic or halophytic origin. Expression of the CgNHX1 gene was induced by NaCl and peaked at 400 mmol/L NaCl. Overexpression of NHX1 genes in rice enhanced their tolerance to salt stress. However, there is no significant difference in salt tolerance among the transgenic rice plants overexpressing the NHX1 genes from either glycophytic or halophytic species. The Na+ content of both the wild type (WT) and transgenic plants increased when exposed to 50 and 100 mmol/L NaCl, and the Na+ concentration in transgenic plants was marginally higher than that of WT. Our data demonstrate that the overexpression of the NHX1 gene from either glycophytic or halophytic species resulted in the enhanced tolerance to salt stress at a similar level, suggesting that NHX gene per se might not be the reason accounting for the difference in salt tolerance between glycophytes and halophytes.
Keywords: NHX gene, Rice transformation, Salt stress, Tolerance
INTRODUCTION
Accumulation of salts in irrigated soil is a primary factor depressing yield in crop production (Zhu, 2001). The detrimental effects of salt on plants are a consequence of osmotic stress and the toxicity of excess sodium ions to many critical biochemical processes (Jones, 1981). Based on the capacity to grow on a salinity environment, plants can be classified into glycophyte and halophyte. Most plant species are glycophytes, which are salt sensitive. In contrast, halophytes are able to grow in habitats excessively rich in salts, such as salt marshes, sea coasts, and saline or alkaline semideserts and steppes. The salt tolerance in halophytes may involve a range of adaptations, including ion compartmentalization, osmolyte production, germination responses, osmotic adjustment, succulence, selective transport and uptake of ions, enzyme responses, salt excretion, and genetic control (Flowers et al., 1977; 1986; Greenway and Munns, 1980). The molecular basis of the stress tolerance of halophyte, however, is still far from clear.
The Na+/H+ exchanger catalyzes the exchange of Na+ for H+ across membranes. It regulates internal pH, cell volume and sodium level in the cytoplasm, and maintains the intracellular ion homeostasis (Aronson, 1985; Orlowski and Grinstein, 1997). Reports showed that the exchanger plays an important role in plant salt tolerance (Qiu et al., 2002; Shi et al., 2000; 2002; Shi and Zhu, 2002). The Arabidopsis SOS1 gene, encoding a plasma membrane Na+/H+ antiporter, conferred salt tolerance in transgenic Arabidopsis (Qiu et al., 2002; Shi et al., 2000; 2002; 2003; Shi and Zhu, 2002). Transgenic Arabidopsis and tomato plants overexpressing AtNHX1 accumulated abundant quantities of the transporter in the tonoplast and exhibited substantially enhanced salt tolerance (Apse et al., 1999; Quintero et al., 2000; Zhang and Blumwald, 2001). Similar result was also achieved in transgenic rice expressing AgNHX1 gene (Ohta et al., 2002). These results implicate the pivotal function of the NHX family in vacuolar compartmentalization of Na+.
Genes encoding vacuole-type Na+/H+ antiporters have been isolated from numbers of plant species, including glycophytic species Arabidopsis thaliana (Apse et al., 1999; 2003; Gaxiola et al., 1999), Oryza sativa (Fukuda et al., 1999), Triticum aestivum (Brini et al., 2007) and Zea mays (Zorb et al., 2005), halophytic species Atriplex gmelini (Hamada et al., 2001), Mesembryanthemum crystallinum (Chauhan et al., 2000), Atriplex dimorphostegia (Li et al., 2003) and Suaeda salsa (Ma et al., 2004). Xiong and Zhu (2002) suggested that the difference in salt tolerance among plant species could result from differences in regulatory circuits or from gene alleles coding for key salt tolerance genes. We here reported that the molecular cloning and functional analysis of the newly isolated NHX1 genes from halophytes species Atriplex dimorphostegia and Chenopodium glaucum. Using the rice homology gene OsNHX1 as the representative of glycophytes, the study also assessed the difference in the regulation and function of NHX1 genes from halophytes and glycophytes. We observed that NHX1 genes from both types of plant species could enhance the salt tolerance in transgenic rice, suggesting that NHX1 gene per se might not be the reason accounting for the difference in salt tolerance between glycophytes and halophytes. The salt tolerance of halophytes might result from the different regulation systems of NHX1 gene expression or mechanisms other than vacuolar Na+ pump.
MATERIALS AND METHODS
Structure and phylogenetic analysis
The hydropathy plot analysis was carried out using the program of Protscale (http://us.expasy.org/tools/protscale.html). The membrane-spanning regions and their orientation were predicted by the method of Tmpred (http://www.ch.embnet.org/software/TMPRED_form.html). The phylogenetic tree was generated based on minimal evolution criterion using the neighbor-joining method with 1000 times of bootstrap. The following Na+/H+ antiporter genes were analyzed to assess their phylogenetic relationship: SsNHX1 (Suaeda salsa, AY261806), CgNHX1 (Chenopodium glaucum, AY371319), AdNHX1 (Atriplex dimorphostegia, AY211397), KfNHX1 (Kalidium foliatum, AY825250), KcNHX1 (Karelinia caspia, DQ304690), SeNHX1 (Salicornia europaea AY131235), AgNHX1 (Atriplex gmelini, AB038492), CrNHX1 (Citrus reticulate, AY607026), GhNHX1 (Gossypium hirsutu, AF515632), PtNHX1 (Populus tomentosa, AY660749), PhNHX1 (Petunia hybrida, AB051817), MsNHX1 (Medicago sativa, AY513732), BnNHX1 (Brassica napus, AA038856), OsNHX1 (Oryza sativa, AB021878), ZmNHX1 (Zea mays, AY270036), TaNHX1 (Triticum aestivum, AY040246), AtNHX1 (Arabidopsis thaliana, AF 510074), nhaA (Escherichia coli, AAL25795), nhaB (Escherichia coli, AAA24218), NHA1 (Saccharomyces cerevisiae, CAA97711), Sod2 (Schizosaccharomyces pombe, Z11736) and NHE8 (Homo sapiens, Q9Y2E8).
Plant materials
Seeds of Chenopodium glaucum were germinated at 37 °C for 3 d and grown at 26 °C with a photoperiod of 12 h. At the three-leaf stage, seedlings were treated with 400 mmol/L NaCl for 3 d before harvesting for RNA extraction.
Preparation of cDNA
Total RNA was extracted from either salt-treated or control seedlings using EZNA® Plant RNA Kit (Omega Bio-Tek, Doraville, GA, USA) according to the manufacturer’s instructions. The extracted total RNA was then treated with DNase to remove the possible DNA contamination. First strands of cDNAs were synthesized using TaKaRa One Step RNA PCR Kit (Takara, Shuzo, Tokyo, Japan) following the manufacturer’s protocol.
RT-PCR and 3′-RACE (rapid amplification of cDNA ends)
RT-PCR reactions were performed to amplify the most conservative regions of NHX1 gene from Chenopodium glaucum. Seven pairs of primers were designed based on the available sequences of NHX1 genes from GenBank. Among them, the primers, P1 (5′-ATG TGG TCA CAG TTA AGC TC-3′) and P2 (5′-CCA CGA CCT CCA AAG ACG GGT CGC AT-3′), were able to amplify a product. PCR reaction was performed as follows: initiated with denaturation for 2 min at 94 °C, followed by 35 cycles of denaturing at 94 °C for 30 s, annealing at 52.5 °C for 30 s, extension at 72 °C for 1.5 min, and a final extension step of 10 min at 72 °C was included. The amplified DNA fragment was purified and cloned into pMD18-T vector for sequencing.
The 3′-RACE was carried out to obtain the 3′-end of the CgNHX1 cDNA. The cDNA region corresponding to the 3′-end of CgNHX1 was amplified by two successive rounds of PCR using gene-specific 5′ primers P3 and P4 (5′-GAT CAC GAG CAC TAT AAC TAT TGT CC-3′ and 5′-GAG CTC CAT ATT GCT TGG TTT ACT CAT G-3′, respectively), together with the supplied 3′-Oligo dT-3 sites Adaptor Primer (Takara Shuzo, Tokyo, Japan). The second round of PCR yielded a product of approximate 850 bp, which was cloned into pMD18-T vector for sequencing.
Full length cDNA was amplified using primers corresponding to the sequence of the core and the 3′-ends (FP1: 5′-CTT GGA TCC ATG TGG TCA CAG TTA AGC TC-3′ and FP2: 5′-AGG ATG TCG ACG ATT TAC AAT TTT GCC ATT-3′, respectively). The PCR conditions were the same as the above.
The expression levels of different NHX1 genes (OsNHX1, AdNHX1 and CgNHX1) in transgenic rice plants were measured by RT-PCR using β-actin gene, OsACT, as internal control. PCR reaction of OsACT was performed as follows: initiated with denaturation for 4 min at 94 °C, followed by 26 cycles of denaturing at 94 °C for 30 s, annealing at 58 °C for 30 s, extension at 72 °C for 30 s, and a final extension step of 10 min at 72 °C was included. The RT-PCR reactions were performed to amplify the most conservative regions of NHX1 gene from 3 species. PCR reaction was performed as follows: initiated with denaturation for 4 min at 94 °C, followed by 30 cycles of denaturing at 94 °C for 30 s, annealing at 56 °C for 30 s, extension at 72 °C for 30 s, and a final extension step of 10 min at 72 °C was included.
DNA sequencing
DNA sequences of newly cloned NHX gene were determined using the DNA Sequencing Kit and ABI PRISM 377 DNA Sequencer (Perkin Elmer, Foster City, CA, USA). The nucleotide and amino acid sequences were then compared with those in GenBank database by using the GAPPED BLAST analysis program. The full-length cDNA sequence of CgNHX1 was deposited in the GenBank database (AY371319).
Real-time RT-PCR
Primers for the specific genes, CgNHX1 and OsACT, were designed and tested to ensure the specificity of the amplification. Primers were used for RT-PCR analysis of the CgNHX1 gene corresponding to a 460 bp cDNA fragment. The β-actin primers amplify a 150 bp cDNA fragment.
Two microlitres of each synthesized cDNA were used as templates for the real-time RT-PCR. The PCR reaction included 200 μmol/L of dNTPs, 0.8 μmol/L of forward and reverse primers, 2 units of Hot-Start Taq DNA polymerase, 0.4× SYBR Green I real-time PCR mix buffer (Biotium Inc., Hayward, CA, USA) and 1× reaction buffer. PCR reactions were performed using a Perkin Elmer PE 5700 Sequence Detector (Applied Biosystems, Foster City, CA, USA). The PCR reaction initiated by a Hot Start at 95 °C for 10 min, followed by 35 cycles of denaturing at 95 °C for 30 s, annealing at 55 °C for 30 s and extension at 72 °C for 40 s. The experiments were carried out in duplicate. A set of four standards containing purified target DNA (range from 700 copies to 7 000 000 copies, dilution factor of 10) were included for making the standard curve.
Rice transformation
The open reading frames (ORFs) of the OsNHX1, AdNHX1 and CgNHX1 were amplified using the corresponding cDNA as templates, and then ligated with a cauliflower mosaic virus 35S promoter and 3′-nopaline synthase (NOS) terminator. The constructed expressing cassettes were inserted into the binary plant vector pCAMBIA1301. The resulting plasmids 35S:OsNHX1 (35S-OS), 35S: AdNHX1 (35S-AD) and 35S:CgNHX1 (35S-CG) were then transformed into Agrobacterium tumefaciens strain EHA105 and used for plant transformation. Rice transformation was performed as previously described (Chen et al., 2003).
Plant growth and salt stress treatment
Both the wild type (WT) and transgenic rice plants were germinated and grown in nutrition solution (Xu et al., 2005) for three weeks. Rice plants with three or four true leaves were treated with the same nutrient solution supplemented with different concentrations of NaCl. Growth rates of the treated seedlings were calculated by measuring the seedling height. Dry weights of seedlings were determined by drying the seedlings in an oven at 60 °C for 72 h.
Measurement of Na+ content in leaves
After 6 d of salt treatment, leaves of the rice plants were harvested. The sodium contents of the leaves were determined using atomic absorption spectrophotometry (AAS) following the dry ash procedure. Briefly, leaf samples were dry-ashed for 16 h at 450 °C followed by solubilization in 1:1 HNO3. The sodium contents were determined by an atomic absorption spectrophotometer (AA-6650, Shimadzu Corporation, Kyoto, Japan).
RESULTS
Cloning and characterization of CgNHX1
Xinjiang Uygur Autonomous Region is one of the most arid areas in China. In the salt marsh area, only halophyte species can survive. Atriplex dimorphostegia and Chenopodium glaucum are two typical halophytes grown in the area. We have previously isolated the AdNHX1 gene from Atriplex dimorphostegia by RT-PCR method (Li et al., 2003). The AdNHX1 cDNA is 1700 bp long and shared 95% similarity to NHX1 gene from another Atriplex genus species, Atriplex gmelini.
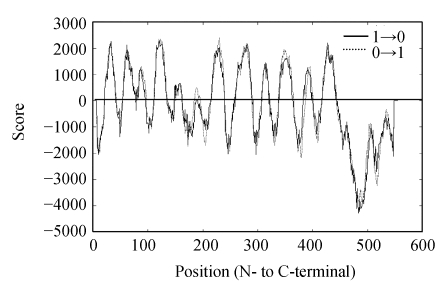
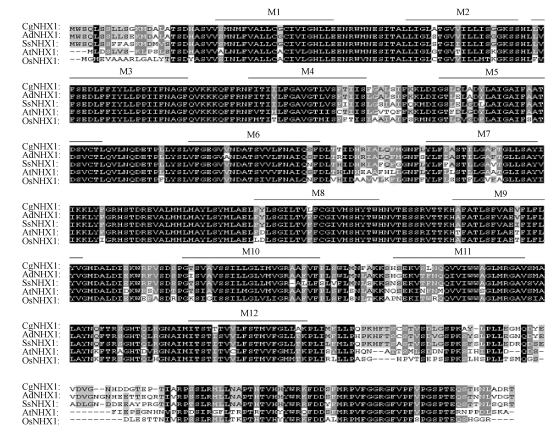
Based on the sequences of the NHX1 genes in GenBank, seven PCR primers were designed to amplify the conservative region of Chenopodium glaucum NHX1 genes. Among the PCR primers used, P1 and P2 primer pair (detailed in the section of Materials and Methods) was able to amplify a 1300 bp PCR product. Sequence analysis showed that the product contains the 5′-end of NHX1 genes, but not the 3′-end. Using 3′-RACE, we were able to amplify the 3′-end of the CgNHX1 gene. Combining the sequence information from 3′-RACE and 5′-end of CgNHX1 gene, we redesigned a pair of PCR primers and amplified the full length CgNHX1 gene. The resulted 2169 bp product was sequenced and deposited in GenBank (Y371319). The CgNHX1 cDNA contains a 1656 bp ORF and a 513 bp 3′-untranslated region. It encodes a protein of 551 amino acids with a calculated molecular mass of 60.8 kDa. Hydropathy analysis (Hofmann, 1993) showed that the N-terminal portion of CgNHX1 contains 12 hydrophobic transmembrane domains, while its C-terminal portion is a hydrophilic tail (Fig.1). Amino acid sequence of CgNHX1 shares 93%, 72% and 75% similarity to that of the AdNHX1, OsNHX1 and AtNHX1, respectively (Fig.2).
Fig. 1.
Hydrophobicity plot of CgNHX1 gene product
The hydrophobicity values were calculated by the program TMpred available at http://www.ch.embnet.org/software/TMPRED-form.html
Fig. 2.
Multiple sequence alignment of CgNHX1 with other putative Na+/H+ exchanger proteins
Putative membrane spanning domains of CgNHX1 (M1~M12) are indicated by overlines. Amino acid sequences were aligned using the Clustal X program. The accession numbers of the Na+/H+ antiporters are as follows: Chenopodium glaucum (CgNHX1, AY371319), Atriplex dimorphostegia (AdNHX1, AY211397), Suaeda salsa (SsNHX1, AY261806), Arabidopsis thaliana (AtNHX1, AF510074) and Oryza sativa (OsNHX1, AB021878). The black shading of the alignment indicates identical residues, dark gray and light gray shading indicates conserved residues and similar residues, respectively. The amiloride binding sites are enclosed with a box
According to the preliminary topological model, there are three potential N-glycosylation sites in the CgNHX1, located at Asn-36, -49 and -292. These sites correspond to the positions of the consensus N-glycosylation sites in human NHE1 (Counillon et al., 1994), suggesting that the CgNHX1 protein is glycosylated.
Phylogenetic analysis of NHXs from halophytes and glycophytes
The membrane-spanning regions are well conserved in eukaryotic Na+/H+ antiporters. Within these regions, CgNHX1 shares high degree of similarity with other vacuolar Na+/H+ antiporters such as AdNHX1, OsNHX1 and AtNHX1 (Fig.2). The protein sequence of 85-LFFIYLLPPI-94 in CgNHX1 is highly similar to those of AdNHX1, OsNHX1, AtNHX1 and mammalian NHEs. In mammals, this region was identified as the binding site of amiloride, which inhibits the eukaryotic Na+/H+ exchanger. These features confirm that the CgNHX1 is a vacuolar-type Na+/H+ antiporter.
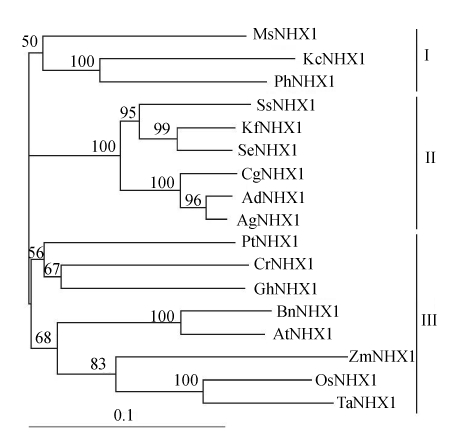
Sequences of 22 NHX1 genes from different species were obtained from the GenBank. Phylogenetic analysis showed that 17 plant-origin Na+/H+ antiporters fell into 3 distinct groups (Fig.3). Except for the gene KcNHX1 from Karelinia caspia, NHX1 genes from six halophytic species (Atriplex dimorphostegia, Atriplex gmelini, Chenopodium glaucum, Kalidium foliatum, Salicornia europaea and Suaeda salsa) all belong to Group II. And NHX1 genes from typical glycophytic species, just like Arabidopsis thaliana, Oryza sativa, Triticum aestivum, Zea mays, and so on, all belong to Group III. Phylogenetic analysis also showed that the NHX1 genes from yeast, E. coli, and human were distinct from those of plant species (data not shown), implying that plants may share a common ancestor.
Fig. 3.
Neighbor-joining phylogenetic tree of Na+/H+ antiporter protein sequences. Software packages, Clustalx and TreeView, were used to do multiple sequence alignment and generate phylogenetic trees. Bootstrap values (1000 replicates) for this tree are shown as the numbers. The accession numbers of Na+/H+ antiporters are: KfNHX1 (AY825250), KcNHX1 (DQ304690), SeNHX1 (AY131235), AgNHX1 (AB0384 92), CrNHX1 (AY607026), GhNHX1 (AF515632), PtNHX1 (AY660749), PhNHX1 (AB051817), MsNHX1 (AY513732), BnNHX1 (AA038856), ZmNHX1 (AY 270036) and TaNHX1 (AY040246). Bar=substitutions/site
Expression analysis of CgNHX1
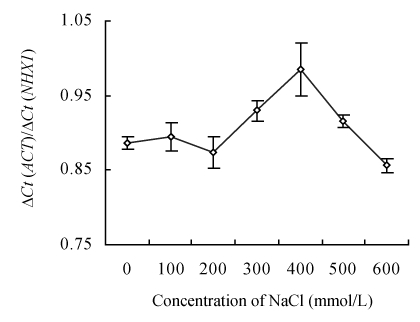
The expressions of CgNHX1 gene under different NaCl treatments were quantified with the purified CgNHX1 plasmid DNA as standards. RT-PCR conditions were optimized and the RT-PCR products were run at 1% agarose gel to ensure the elimination of primer-dimer. Expression levels of the CgNHX1 at different NaCl concentrations were calculated according to standard curves. The real-time RT-PCR results show that expression of the CgNHX1 reached at the maximum level when NaCl concentration was at 400 mmol/L (Fig.4).
Fig. 4.
Expression of CgNHX1 in Chenopodium glaucum under various concentrations of NaCl. The expression levels of CgNHX1 were quantified by real-time RT-PCR using actin gene as internal control to normalize the data. Ct: Threshold cycle
Overexpression of NHX1 genes from both halophytes and glycophytes enhanced the salt tolerance in transgenic rice
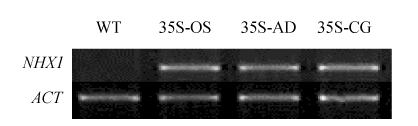
To study the effect of the NHX1 genes from different origins on the salt tolerance, three transgenic rice lines (35S-AD, 35S-CG and 35S-OS) were generated by Agrobacterium-mediated transformation method. All the three transgenes (AdNHX1, CgNHX1 and OsNHX1) were driven by Ca35S promoter, which is derived from the cauliflower mosaic virus. For each of the transgenic event, 20 transgenic plants were regenerated and propagated. PCR and RT-PCR analyses were performed on the T1 plants (the 1st generation of transgenic plants) to ensure the existence and expression of the transgenes (Fig.5).
Fig. 5.
The expression of different NHX1 genes in transgenic rice plants. The expression levels of NHX1 genes (OsNHX1, AdNHX1 and CgNHX1) were measured by RT-PCR using rice actin (OsACT) gene as internal control
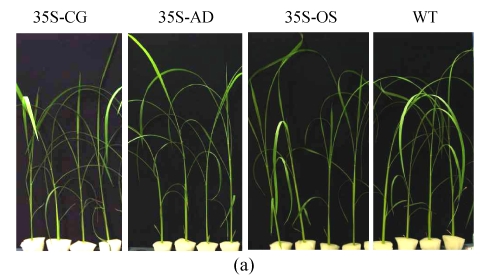
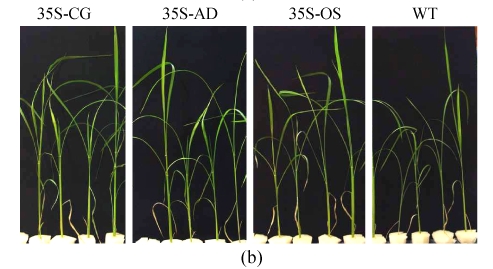
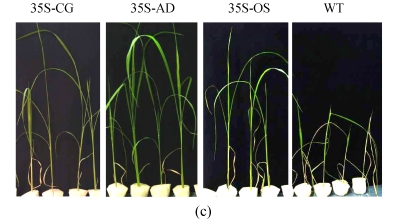
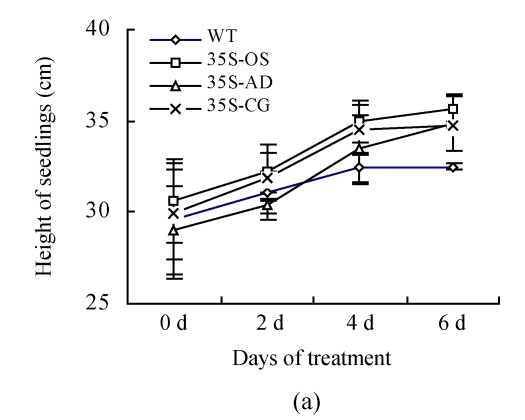
Two confirmed transgenic events (T2 homogenous seedlings) from constructs of 35S-AD, 35S-CG, 35S-OS and the WT (Nipponbare) were germinated and planted hydroponically in preparation for salt tolerance test. Three weeks after germination, the seedlings were transferred to the solution cultures containing 0 mmol/L, 50 mmol/L or 100 mmol/L NaCl. Ten seedlings were included in each event. The heights of seedlings, which were used as an indicator of salt tolerance, were measured every other day during the salt treatment. While the growth of both the transgenic and the WT rice plants was inhibited by NaCl treatment, the degree of inhibition was much lower in transgenic plants than that in the WT plants. Six days after salinity exposure at 50 mmol/L NaCl, the growth of the WT plants was severely inhibited, whereas that of the transgenic plants was not. The WT plants turned yellow and ceased growth in the 100 mmol/L NaCl treatment, whereas the transgenic plants were still green (Fig.6). The average relative growth rate of transgenic rice plants remained 3 cm/d, which was 3.1 times higher than that of the WT plants (Fig.7a).
Fig. 6.
The growth of WT (Nipponbare) and transgenic plants under various concentrations of NaCl. (a) Control, nutrient solution without NaCl; (b) 50 mmol/L NaCl; (c) 100 mmol/L NaCl
Fig. 7.
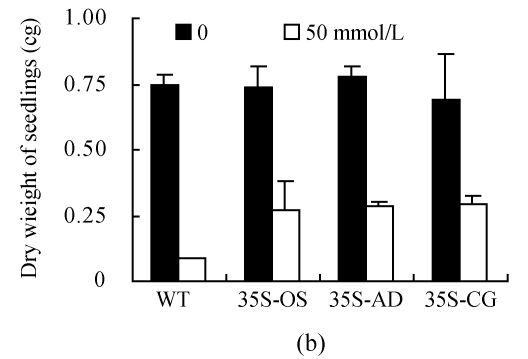
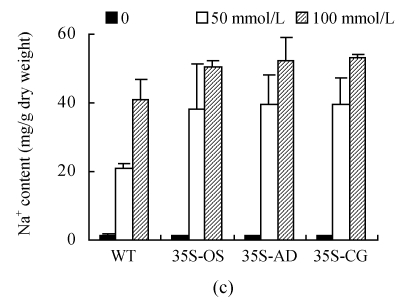
(a) Height of seedlings measured every 2 d during the treatment of 100 mmol/L NaCl; (b) Dry weight of seedlings measured after treatment with 50 mmol/L NaCl for 30 d; (c) Content of Na+ in leaves. All the rice plants were cultivated in hydroponic culture media for 3 weeks and then treated with various concentrations of NaCl. The WT and transgenic rice plants were planted with hydroponic culture media containing 50 or 100 mmol/L NaCl for 6 d. The Na+ contents were determined by atomic absorption spectrophotometer
After 30 d of 50 mmol/L NaCl treatment, the plants were harvested to measure their dry weights. As shown in Fig.7b, the dry weights of the WT plants were significantly decreased under salt stress compared to the transgenic plants; the dry weights of transgenic rice plants were 1.5~2.5 times greater than those of the control plants.
Transgenic plants accumulating more Na+ than WT plants
The Na+ contents of the transgenic and WT plants were analyzed 6 d after NaCl exposure. Without NaCl stress, the Na+ contents in the WT and transgenic plants were nearly the same. The Na+ contents increased after exposed to NaCl in both WT and transgenic rice plants (Fig.7c). The Na+ content in the transgenic plants was higher than that in the WT plants, but not statistically significant (P>0.05).
DISCUSSION
Using RACE method, we cloned a novel NHX1 gene from a halophytic species, Chenopodium glaucum. The newly cloned gene shared 92%, 84%, 69% and 67% sequence similarity to the NHX1 genes from Atriplex gmelini, Suaeda maritime, Arabidopsis thaliana and Oryza sativa, respectively. Phylogenetic analysis confirmed that the CgNHX1 belongs to the NHX1 gene family and is a close relative of the two other halophytic NHX1 genes, AgNHX1 and AdNHX1.
Salt-tolerant mechanism in plant involves Na+ extrusion and Na+ compartmentation. It has been shown that overexpression of AtNHX1, a vacuolar Na+/H+ transporter, conferred salt tolerance in A. thaliana and tomato (Apse et al., 1999; Zhang and Blumwald, 2001), suggesting the potential utility of this vacuolar compartmentation strategy on the improvement of plant salt tolerance. Recent studies demonstrated that transgenic rice plants containing 35S:AgNHX1 could survive after short period of high concentration salt exposure (Ohta et al., 2002). Halophyte could normally grow under the environment with up to 600 mmol/L of NaCl, while glycophyte species could not survive in the environment with Na+ concentration beyond 200 mmol/L. Although overexpression of both AtNHX1 and AgNHX1 enhanced salt tolerance in transgenic plants, it is not clear whether functions of NHX genes originated from glycophytes or halophytes for salt tolerance are different. In order to address this question, three constructs (35S:OsNHX1, 35S:AdNHX1 and 35S: CgNHX1) were introduced into rice plants and their functions were analyzed. The results demonstrate that overexpression of any of above transgenes enhanced salt tolerance in rice seedlings. The Na+ content, relative growth height and dry weight of the transgenic plants were higher than those of WT plants. Interestingly, overexpression of NHX1 genes from both glycophytic and halophytic species led to similar degree of salt tolerance in transgenic rice plants.
As mentioned earlier, NHX1 genes originated from either halophytes or glycophytes share highly similar DNA sequence and protein structure. We also knew that both types of NHX1 genes had similar function in enhancing salt tolerance. Taken together, we conclude that the NHX gene per se might not be the reason accounting for the difference in salt tolerance between glycophytes and halophytes. The better salt tolerance in halophytes might result from a different regulation system of NHX1 genes or mechanisms other than vacuolar Na+ pump. Further studies that elucidate the regulatory machinery of NHX genes might help us to understand the mechanism that confers better salt tolerance in halophytic species.
Acknowledgments
We thank Dr. Zhi-ning Wang of Walter Reed Army Institute of Research, USA and Dr. Xin Chen of Zhejiang University for critical reading of the manuscript.
Footnotes
Project supported by the National Natural Science Foundation of China (Nos. 30471118 and 30460015) and the Key Project of Education Ministry of China (No. 205178)
References
- 1.Apse MP, Aharon GS, Snedden WA, Blumwald E. Salt tolerance conferred by overexpression of a vacuolar Na+/H+ antiport in Arabidopsis . Science. 1999;285(5431):1256–1258. doi: 10.1126/science.285.5431.1256. [DOI] [PubMed] [Google Scholar]
- 2.Apse MP, Sottosanto JB, Blumwald E. Vacuolar cation/H+ exchange, ion homeostasis, and leaf development are altered in a T-DNA insertional mutant of AtNHX1, the Arabidopsis vacuolar Na+/H+ antiporter. Plant J. 2003;36(2):229–239. doi: 10.1046/j.1365-313X.2003.01871.x. [DOI] [PubMed] [Google Scholar]
- 3.Aronson PS. Kinetic properties of the plasma membrane Na+-H+ exchanger. Ann Rev Physiol. 1985;47(1):545–560. doi: 10.1146/annurev.ph.47.030185.002553. [DOI] [PubMed] [Google Scholar]
- 4.Brini F, Hanin M, Mezghani I, Berkowitz GA, Masmoudi K. Overexpression of wheat Na+/H+ antiporter TNHX1 and H+-pyrophosphatase TVP1 improve salt- and drought-stress tolerance in Arabidopsis thaliana plants. J Exp Bot. 2007;58(2):301–308. doi: 10.1093/jxb/erl251. [DOI] [PubMed] [Google Scholar]
- 5.Chauhan S, Forsthoefel N, Ran Y, Quigley F, Nelson DE, Bohnert HJ. Na+/myo-inositol symporters and Na+/H+-antiport in Mesembryanthemum crystallinum . Plant J. 2000;24(4):511–522. doi: 10.1046/j.1365-313x.2000.00903.x. [DOI] [PubMed] [Google Scholar]
- 6.Chen S, Jin W, Wang M, Zhang F, Zhou J, Jia Q, Wu Y, Liu F, Wu P. Distribution and characterization of over 1000 T-DNA tags in rice genome. Plant J. 2003;36(1):105–113. doi: 10.1046/j.1365-313X.2003.01860.x. [DOI] [PubMed] [Google Scholar]
- 7.Counillon L, Pouyssegur J, Reithmeier RA. The Na+/H+ exchanger NHE-1 possesses N- and O-linked glycosylation restricted to the first N-terminal extracellular domain. Biochemistry. 1994;33(34):10463–10469. doi: 10.1021/bi00200a030. [DOI] [PubMed] [Google Scholar]
- 8.Flowers TJ, Troke PF, Yeo AR. The mechanism of salt tolerance in halophytes. Annu Rev Plant Physiol. 1977;28(1):89–121. doi: 10.1146/annurev.pp.28.060177.000513. [DOI] [Google Scholar]
- 9.Flowers TJ, Haijibagheri MA, Clipson NJW. Halophytes. Quart Rev Biol. 1986;61(3):313–337. doi: 10.1086/415032. [DOI] [Google Scholar]
- 10.Fukuda A, Nakamura A, Tanaka Y. Molecular cloning and expression of the Na+/H+ exchanger gene in Oryza sativa . Biochim Biophys Acta. 1999;1446(1):149–155. doi: 10.1016/s0167-4781(99)00065-2. [DOI] [PubMed] [Google Scholar]
- 11.Gaxiola RA, Rao R, Sherman A, Grisafi P, Alper SL, Fink GR. The Arabidopsis thaliana proton transporters, AtNhx1 and Avp1, can function in cation detoxification in yeast. Proc Natl Acad Sci USA. 1999;96(4):1480–1485. doi: 10.1073/pnas.96.4.1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Greenway H, Munns R. Mechanisms of salt tolerance in non-halophytes. Ann Rev Plant Physiol. 1980;31(1):149–190. doi: 10.1146/annurev.pp.31.060180.001053. [DOI] [Google Scholar]
- 13.Hamada A, Shono M, Xia T, Ohta M, Hayashi Y, Tanaka A, Hayakawa T. Isolation and characterization of a Na+/H+ antiporter gene from the halophyte Atriplex gmelini . Plant Mol Biol. 2001;46(1):35–42. doi: 10.1023/A:1010603222673. [DOI] [PubMed] [Google Scholar]
- 14.Hofmann KSW. TM base—A database of membrane spanning proteins segments. Biol Chem Hoppe-Seyler. 1993;347:166–172. [Google Scholar]
- 15.Jones RGW. Salt Tolerance. In: Johnson CB, editor. Physiological Processes. Limiting Plant Productivity. Iondon: Butterworths; 1981. pp. 271–292. [Google Scholar]
- 16.Li JY, Zhang FC, Ma J, Cai L, Bao YG, Wang B. Using RT-PCR to amplify the NHX gene fragment in Atriplex dimorphostegia . Plant Physiol Commun. 2003;6(6):585–588. (in Chinese) [Google Scholar]
- 17.Ma XL, Zhang Q, Shi HZ, Zhu JK, Zhao YX, Ma CL, Zhang H. Molecular cloning and different expression of a vacuolar Na+/H+ antiporter gene in Suaeda salsa under salt stress. Biol Plantarum. 2004;48(2):219–225. doi: 10.1023/B:BIOP.0000033448.96998.44. [DOI] [Google Scholar]
- 18.Ohta M, Hayashi Y, Nakashima A, Hamada A, Tanaka A, Nakamura T, Hayakawa T. Introduction of a Na+/H+ antiporter gene from Atriplex gmelini confers salt tolerance to rice. FEBS Lett. 2002;532(3):279–282. doi: 10.1016/S0014-5793(02)03679-7. [DOI] [PubMed] [Google Scholar]
- 19.Orlowski J, Grinstein S. Na+/H+ exchangers of mammalian cells. J Biol Chem. 1997;272(36):22373–22376. doi: 10.1074/jbc.272.36.22373. [DOI] [PubMed] [Google Scholar]
- 20.Qiu QS, Guo Y, Dietrich MA, Schumaker KS, Zhu JK. Regulation of SOS1, a plasma membrane Na+/H+ exchanger in Arabidopsis thaliana, by SOS2 and SOS3. Proc Natl Acad Sci USA. 2002;99(12):8436–8441. doi: 10.1073/pnas.122224699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Quintero FJ, Blatt MR, Pardo JM. Functional conservation between yeast and plant endosomal Na+/H+ antiporters. FEBS Lett. 2000;471(2-3):224–228. doi: 10.1016/S0014-5793(00)01412-5. [DOI] [PubMed] [Google Scholar]
- 22.Shi H, Zhu JK. Regulation of expression of the vacuolar Na+/H+ antiporter gene AtNHX1 by salt stress and abscisic acid. Plant Mol Biol. 2002;50(3):543–550. doi: 10.1023/A:1019859319617. [DOI] [PubMed] [Google Scholar]
- 23.Shi H, Ishitani M, Kim C, Zhu JK. The Arabidopsis thaliana salt tolerance gene SOS1 encodes a putative Na+/H+ antiporter. Proc Natl Acad Sci USA. 2000;97(12):6896–6901. doi: 10.1073/pnas.120170197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shi H, Quintero FJ, Pardo JM, Zhu JK. The putative plasma membrane Na+/H+ antiporter SOS1 controls long-distance Na+ transport in plants. Plant Cell. 2002;14(2):465–477. doi: 10.1105/tpc.010371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shi H, Lee BH, Wu SJ, Zhu JK. Overexpression of a plasma membrane Na+/H+ antiporter gene improves salt tolerance in Arabidopsis thaliana . Nat Biotechnol. 2003;21(1):81–85. doi: 10.1038/nbt766. [DOI] [PubMed] [Google Scholar]
- 26.Xiong LM, Zhu JK. Salt Tolerance. In: Somerville CR, Meyerowitz EM, editors. The Arabidopsis Book. American Society of Plant Biologists. MD: Rockville; 2002. pp. 1–22. [DOI] [Google Scholar]
- 27.Xu M, Zhu L, Shou H, Wu P. A PIN1 family gene, OsPIN1, involved in auxin-dependent adventitious root emergence and tillering in rice. Plant Cell Physiol. 2005;46(10):1674–1681. doi: 10.1093/pcp/pci183. [DOI] [PubMed] [Google Scholar]
- 28.Zhang HX, Blumwald E. Transgenic salt-tolerant tomato plants accumulate salt in foliage but not in fruit. Nat Biotechnol. 2001;19(8):765–768. doi: 10.1038/90824. [DOI] [PubMed] [Google Scholar]
- 29.Zhu JK. Plant salt tolerance. Trends Plant Sci. 2001;6(2):66–71. doi: 10.1016/S1360-1385(00)01838-0. [DOI] [PubMed] [Google Scholar]
- 30.Zorb C, Noll A, Karl S, Leib K, Yan F, Schubert S. Molecular characterization of Na+/H+ antiporters (ZmNHX) of maize (Zea mays L.) and their expression under salt stress. J Plant Physiol. 2005;162(1):55–66. doi: 10.1016/j.jplph.2004.03.010. [DOI] [PubMed] [Google Scholar]