Abstract
Study Objectives:
The dentate gyrus (DG) of the adult hippocampus contains progenitor cells, which have potential to differentiate into neurons. Previously we reported that 96 hours of total sleep deprivation reduces neurogenesis in the DG of adult rats. Loss of either non-rapid eye movement (NREM) or rapid eye movement (REM) sleep could have contributed to the effect of total sleep deprivation. The present study assessed the effect of 4 days of REM sleep deprivation (REMD) on neurogenesis.
Design:
REMD was achieved by brief treadmill movement initiated by automatic online detection of REM sleep. A yoked-control (YC) rat was placed in the same treadmill and experienced the identical movement regardless the stage of the sleep-wake cycle. The thymidine analog 5- bromo- 2′- deoxy-uridine and the intrinsic proliferation marker, Ki-67, were both used to label proliferating cells.
Setting:
Basic neurophysiology laboratory.
Participants:
Male Sprague-Dawley male rats (300 – 320 g)
Results:
REM sleep was reduced by 85% in REMD rats and by 43% in YC, compared with cage control animals and by 79% in REMD rats compared with YC. NREM sleep and slow wave activity within NREM did not differ in REMD and YC groups. Cell proliferation was reduced by 63 % in REMD compared with YC rats, and by 82% and 51%, respectively, in REMD and YC rats compared with cage controls. Across all animals, cell proliferation exhibited a positive correlation with the percentage of REM sleep (r = 0.84, P < 0.001). Reduced cell proliferation in REMD rats was confirmed with the intrinsic proliferation marker, Ki-67. REMD also reduced the percentage of proliferating cells that later expressed a mature neuronal marker.
Conclusions:
The present findings support a hypothesis that REM sleep-associated processes facilitate proliferation of granule cells in the adult hippocampal DG.
Citation:
Guzman-Marin R; Suntsova N; Bashir T; Nienhuis R; Szymusiak R; McGinty D. Rapid eye movement sleep deprivation contributes to reduction of neurogenesis in the hippocampal dentate gyrus of the adult rat. SLEEP 2008;31(2):167–175.
Keywords: adult neurogenesis, hippocampus, dentate gyrus
THE SUBGRANULAR CELL LAYER IN THE DENTATE GYRUS (DG) OF THE ADULT HIPPOCAMPUS CONTAINS PROGENITOR CELLS, WHICH HAVE THE POTENTIAL TO proliferate and differentiate into neurons. These progenitors mature locally into granule cells of the DG, sending axonal projections to area CA3 and dendrites into the molecular layer.1.2 Adult neurogenesis has been demonstrated in birds and several mammals, including humans. The processes of cell proliferation, migration, maturation, and survival are all subject to modulation by experiential events.3 Stress is an important negative regulator of cell proliferation.4,5
Previously we reported that 96 hours of total sleep deprivation (TSD) affects neurogenesis in the rat DG by reducing cell proliferation in the subgranular cell layer, and the percentage of new cells later expressing a neuronal marker.6,7 The inhibitory effects of extended sleep deprivation have been confirmed.8 However, mammalian sleep is physiologically heterogeneous. The two primary stages of mammalian sleep, non-rapid eye movement (NREM) and rapid eye movement (REM) sleep, have very different, even opposite, electrophysiologic and metabolic properties, compared with waking, and could have different effects on neurogenesis. Revealing the relative impact of REM and NREM sleep on neurogenesis is a necessary step in understanding the mechanisms underlying suppression of neurogenesis in response to sleep loss. The aim of the present study was to assess the effect of REM sleep deprivation (REMD) on neurogenesis in the DG of the adult rat. To achieve REM deprivation, rats lived on a treadmill that was briefly activated when REM was detected by fast Fourier transform analysis of the electroencephalogram (EEG) and electromyogram (EMG) activity. Yoked control (YC) animals lived on the same treadmill and were subjected to the same treadmill movements.
MATERIALS AND METHODS
All experiments were conducted in accordance with the National Research Council Guide for the Care and Use of Laboratory Animals. All protocols were reviewed and approved by the institutional Animal Care and Use Committee. Sprague-Dawley male rats (300 – 320 g) were used for this experiment. Animals were housed individually in Plexiglas cages (27×29×30 cm) in a 12:12 light:dark cycle with access to water and food ad libitum. Under deep anesthesia (ketamine 80 mg/kg, intraperitoneal + xylazine 10 mg/kg, intraperitoneal) and aseptic conditions, rats were surgically prepared for chronic EEG and EMG recording as described previously.7 Rats were allowed a 7-day recovery period following surgery.
Sleep Recordings
The recording chamber consisted of a Plexiglas enclosure (28 cm×28 cm×40 cm), which is fixed and suspended 0.6 cm above the vinyl belt of a treadmill, the belt forming the floor of the cage. For several days, rats were acclimated to the enclosure and recording cable. Food and water were available at all times while the animals were on the treadmill. Between each use, the treadmill belt and cage were scrubbed with detergent, wiped with a bleach solution, and rinsed with water.
Two channels of EEG and 1 channel of EMG activity were digitized at 128 Hz each on Cambridge Electronic Design hardware (Cambridge, UK). Sleep-state analysis and REM detection were based on a Power Pass++ program developed by R. Nienhuis derived from Bergmann et al.9 REM detection was based on fast Fourier transform EEG analysis from 1 to 20 Hz, yielding power in delta (1–4 Hz), sigma (10–16 Hz), and theta (6–8 Hz) bands plus integrated EMG activity in each 2-second epoch (cosine-tapered and 5% overlap). Criteria for state were individualized for each animal by setting thresholds using frequency histograms of each EEG band and EMG parameters derived from 24-hour baseline recordings. State was identified for each 10 seconds when 3 out of 5 two-second epochs within the 10-second period were in agreement, simulating the usual manual scoring procedure. REM sleep was identified when theta power was high and delta and sigma power and EMG amplitude were low. Delta power (1–4 Hz) derived from the fast Fourier transform was determined for all NREM sleep and quiet waking (QW) epochs and averaged per 24 hours for each of these states. The percentage change in delta power, herein called slow-wave activity (SWA), from baseline levels and the ratio of NREM sleep to QW delta power (normalized measure of SWA) were determined for each experimental day.
REM Sleep Deprivation
When REM sleep was detected automatically, the treadmill was activated for 5 seconds, causing the animal to step to avoid being carried into the wall of the chamber. The treadmill speed was set at 10 cm per second. A yoked control (YC) rat was placed in the same treadmill and experienced the identical movement regardless of stage of the sleep-wake cycle. Continuous recordings of EEG and EMG activity were used to verify the effectiveness of REMD procedure. Off-line visual scoring of sleep-wake cycle states was based on the predominant state within each 10-second epochs. Agreement of manual and automated REM detection was greater than 95%. The false positive rate was less than 1 epoch per hour.
Experimental Procedures
Experiment 1
To evaluate the effect of REMD on cell proliferation, 24 male rats were divided into 3 groups (n = 8): REMD, YC, and cage control (CC). All were surgically implanted for polysomnographic recordings, and REMD and YC treatments were applied, as described above. At the end of the fourth day of experimental manipulations, at Zeitgeber time 0, all animals were injected with the thymidine analog, 5- bromo- 2′- deoxy-uridine (BrdU) (intraperitoneal 300 mg/kg dissolved in 0.9% NaCl) and perfused 2 hours later.
Experiment 2
To examine the phenotype of the newly generated cells, 18 male rats were prepared for REM sleep deprivation, as in Experiment 1. After four days of experimental manipulations, a single dose of BrdU (300 mg/kg, i.p.) was given, and rats were returned to their home cages. After 3 weeks animals were perfused.
Experiment 3
We examined the stress response induced by the treadmill REM sleep deprivation method. In separate groups of REMD and YC rats (n = 4 per group), blood samples were collected at the end of the fourth day of deprivation. Animals were rapidly killed by decapitation. Blood was collected from the trunk into cooled polyethylene tubes with heparin as the anticoagulant and centrifuged immediately for 10 minutes and stored at −20°C. The elapsed time from removal from the treadmill until blood collection was less than 1 minute. Serum corticosterone was measured by radioimmunoassay (Esoterix, Agoura Hills CA).
Perfusion and immunohistochemistry
Subjects from all groups in experiments 1 and 2 were deeply anesthetized (nembutal 100 mg/kg), perfused transcardially with 0.1 mol phosphate buffer followed by ice cold paraformaldehyde (4%) and 10% and 30% sucrose solution; brains were removed and stored in 30% sucrose at 4°C until they sank. Brains were cut in 40-μm coronal sections. Sections were preserved in a cryoprotectant solution.
Immunohistochemistry was performed simultaneously on sections from YC, REMD, and CC rats to maximize the reliability of comparisons across groups. Tissue from all groups was treated with aliquots from the same batch of antibodies.
BrdU immunostaining was performed on a 1-in-6 series of free-floating sections as described previously.6,7 For DNA denaturation, sections were pretreated with 2 mol HCl at 37°C for 30 minutes followed by 10 minutes in borate buffer (pH 8.5). Tissue was rinsed in 0.1 mol Tris-buffered saline (TBS). Sections were incubated with a mouse anti-BrdU primary antibody (1:400, Novocastra, Norwell, MA) for 48 hours. Sections were subsequently incubated with a biotinylated horse anti-mouse IgG (1:200, Vector Laboratories), then reacted with avidin-biotin complex (1:100, Vector Elite) and developed with diaminobenzidine tetrahydrochloride (DAB, Sigma, San Louis, MO). Omission of the primary antibody resulted in an absence of specific nuclear staining.
A separate set of sections from most (n = 6 in each group) of the same brains used for BrdU immunostaining were processed for Ki-67 immunostaining in slide-mounted sections using rabbit anti-Ki-67 (1:1000, Novocastra) primary antibody and the peroxidase method to visualize the Ki-67 expression.5
BrdU- and Ki-67-positive cells were counted using a 40× objective (Nikon E800, Melville, NY) throughout the rostrocaudal extent of the granule cell layer. The optical fractionator method was used for counting, as has been previously described.6 The Stereo Investigator program calculated the total number of BrdU- or Ki-67-positive cells per DG.
To assess the phenotype of surviving BrdU-positive cells, a 1-in-12 series of sections were processed for immunofluorescent triple labeling for BrdU, NeuN, and S100β, as has been described previously.7 After pretreatment and blocking with goat serum and triton-X 10% in TBS, sections were incubated in a mixture of rat anti-BrdU (1:100; Accurate, Westbury, NY), mouse anti-NeuN (1:300, Chemicon, Temecula, CA) and rabbit anti-S100β (1:2500, Swant, Bellinoza, Switzerland) primary antibodies in TBS for 3 days at 4°C. After rinsing with TBS and blocking for 1 hour, sections were incubated in a secondary antibody mixture (all at 1:300) of Alexa 488 Goat anti-rat, Alexa 567 Goat anti-mouse, and Alexa 633 Goat anti-rabbit (Molecular Probes, Carlsbad, CA) in TBS for 2 hours. Sections were mounted and cover slipped with Vectastain (Vector, Burlingame, CA) mounting medium.
BrdU-positive cells within the granule cell layer were analyzed using a Leica TCS SP MP (Leica, Bannockburn, IL)fixed-stage upright confocal microscope for co-expression with NeuN and S100β using argon, krypton, and helium-neon lasers. Analysis was performed in sequential scanning mode, in which only 1 laser and its respective detection line (488, 567 or 633 nm) are active at a time to exclude cross bleeding between the 2 or 3 different channels. Colocalization was confirmed in 50 cells per animal by z-series through the cell nucleus and 3-dimensional reconstruction (z-step, 1 μm). An individual blinded to experimental conditions did all counting.
Statistical Analysis
Differences between groups in sleep parameters, SWA, and cell counts were assessed with analysis of variance (ANOVA) followed by Bonferroni or Fisher LSD posthoc tests. Correlations between the percentages of NREM and REM sleep and BrdU- and Ki-67-positive cells were calculated using the Pearson product-moment correlation. A P value of < 0.05 was adopted for significance
RESULTS
To achieve REM sleep deprivation with minimal disturbance of NREM sleep, we employed on-line automated detection of REM, based on EEG and EMG parameters, to initiate brief movement of a treadmill at a walking rate. We compared proliferation and neurogenesis in CC, REMD, and YC animals. Pairs of YC and REMD animals lived on the same treadmill and therefore experienced the identical physical stimulation (Figure 1). The treadmill was inactive during periods of spontaneous waking and NREM sleep in the REMD animals.
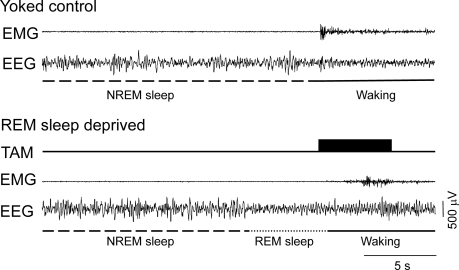
Figure 1.
Simultaneous polysomnographic recording from a rapid eye movement (REM) sleep-deprived (REMD) rat and its yoked control (YC). Note that the treadmill was automatically activated for 5 seconds (dark rectangle) after the online scoring system detected REM sleep in the REMD rat. EEG refers to electroencephalogram; EMG, electromyogram; TAM, treadmill activation mark; NREM, non-rapid eye movement sleep.
Experiment 1
Sleep-wake parameters and SWA
The mean sleep-wake parameters in REMD, YC, and CC groups before initiation of treatments and during experimental manipulations are summarized in Table1. During the baseline, there were no significant differences in the mean percentages of waking, total sleep, NREM sleep, and REM sleep among the groups (P > 0.05, 1-way ANOVA). During experimental manipulations, the mean percentages of each sleep-wake state differed significantly among the groups (Table 1). The mean percentage of REM sleep was reduced by 85% and 33%, respectively, in the REMD and the YC group compared with the CC and by 79% in the REMD group compared with the YC (for all comparisons P < 0.001, Bonferroni t-test).
Table 1.
Sleep-Wake Cycle Parameters During 24-Hour Baseline and 96-Hour Experimental Recording Sessions
| Wake | NREM sleep | REM sleep | Total sleep | |
|---|---|---|---|---|
| 24-h baseline | ||||
| CC | 49.7 ± 1.6 | 40.8 ± 1.7 | 9.5 ± 0.9 | 50.3 ± 1.6 |
| YC | 50.9 ± 2.2 | 38.7 ± 1.5 | 10.4 ± 1.2 | 49.1 ± 2.2 |
| REMD | 49.3 ± 1.8 | 41.6 ± 1.3 | 9.1 ± 0.8 | 50.7 ± 1.7 |
| 96-h experiment | ||||
| CC | 48.1 ± 1.2 | 42.8 ± 1.3 | 9.1 ± 0.3 | 51.9 ± 1.2 |
| YC | 56.0 ± 2.2a | 37.9 ± 2.1 | 6.1 ± 0.4a | 44.0 ± 2.2a |
| REMD | 63.3 ± 1.7a,b | 35.4 ± 1.6a | 1.3 ± 0.2a,b | 36.7 ± 1.7a, b |
| F2,21 = 19.6*** | F2,21 = 5.1* | F2,21 = 185.3*** | F2,21 = 19.6*** |
The table shows the mean percentages of time spent in different sleep–wake states. CC refers to the cage control group; YC, yoked control group; REMD, rapid eye movement sleep deprived group. Data are means ± SEM for groups of 8 rats each.
***P < 0.001 and *P < 0.05, 1-way analysis of variance.
P < 0.05 vs CC and bP < 0.05 vs YC, Bonferroni t-test.
Differences in the percentages of NREM sleep between REMD versus YC group and in the YC versus CC group were not significant. NREM sleep was reduced by 17% in REMD rats compared to CC (P < 0.05, Bonferroni t-test). Therefore, REMD rats exhibited a dramatic reduction of REM sleep compared with both CC and YC animals and a small reduction of NREM sleep compared to CC rats.
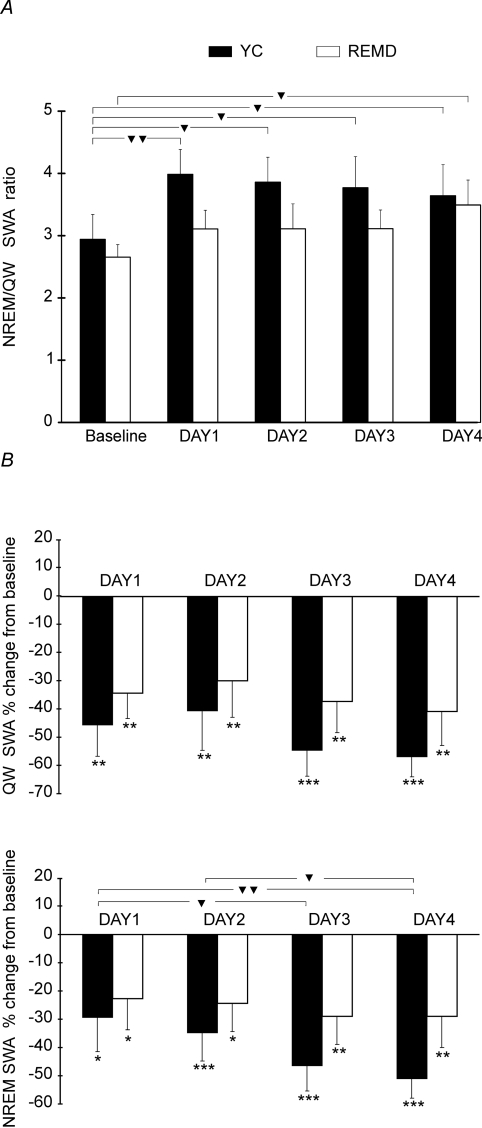
Additionally, we quantified EEG SWA in REMD and YC groups during NREM sleep and QW for the 24-hour baseline period and each day of treatment. A 2-way ANOVA analysis performed for the normalized measure of SWA (NREM sleep/QW delta power ratio) with “condition” as between-subjects factor with 2 levels (YC and REMD) and repeated measures on second factor (time) revealed that YC and REMD rats showed the absence of a significant main effect of the factor “condition” (F1,14 = 1.2, P = 0.3). The NREM sleep-QW delta power ratios (Figure 2, A) did not differ significantly between YC and REMD groups during the baseline or treatment days. The analysis revealed the presence of significant main effect of the factor “time” (F4,56 = 3.8, P = 0.009). Pairwise posthoc comparisons showed that YC animals had significantly higher NREM sleep-QW delta power ratios during each treatment day compared to the baseline. In REMD animals, the increase in the ratio compared with the baseline was significant only on the day 4. The differences in ratios between the days of treatment were insignificant within both REMD and YC groups. Two-way ANOVA showed the absence of significant interaction between the factors “condition” and “time” (F4,56= 0.8, P = 0.5). Both QW and NREM sleep SWA were significantly reduced during each day of treatment compared to the baseline in YC and REMD animals (1-way ANOVA followed by Fisher LSD posthoc test) (Figure 2,B). A 2-way ANOVA, structured as described above, revealed that YC and REMD rats did not differ significantly in percentage decrease in either QW or NREM SWA from baseline levels (F1,14 = 1.0, P = 0.3 and F1,14 = 1.2, P = 0.3, respectively). The percentage decrease in NREM sleep SWA from baseline levels significantly increased across treatment days (F3,42 = 3.2, P = 0.03). The same tendency was found for the percentage change in QW SWA (F3,42 = 2.3, P = 0.09). However, posthoc pairwise comparisons revealed significant differences between treatment days only for NREM sleep SWA in YC rats (Figure 2, B). No significant interactions between the factors were found (F3,42 = 0.2, P = 0.9 and F3,42 = 0.9, P = 0.5 for percentage change in QW and NREM delta power, respectively). Therefore, REMD and YC rats exhibited similar reductions in both NREM and QW delta power during treatment days, compared with the baseline, and did not differ in NREM sleep-QW SWA ratios during the baseline and treatment days.
Figure 2.
Non-rapid eye movement (NREM) sleep/quiet waking (QW) delta power ratios (A) and percentage change from the baseline in QW and NREM sleep electroencephalogram (EEG) slow-wave activity (SWA) (B) in yoke-control (YC) and rapid eye movement-deprived REMD rats for each 24-h period. A: For the baseline period and each treatment day the NREM/QW SWA ratios were calculated. The triangles indicate the significant differences between the ratios: ▼ P < 0.05; ▼▼ P < 0.01 (2-way analysis of variance [ANOVA] with a “condition” as a between-subject factor and repeated measures on factor “time” followed by Fisher least square difference [LSD] posthoc test). Note that REMD and YC rats did not differ significantly in NREM sleep/QW SWA ratios. B: For each treatment day, the SWA was calculated as a percentage change from corresponding baseline levels for YC and REMD groups (mean ± SEM). The stars indicate significant changes in absolute values of EEG SWA during treatment days compared to the baseline:*P < 0.05, **P < 0.01, ***P < 0.001 (1-way repeated measures ANOVA followed by Fisher LSD posthoc test). The triangles indicate significant differences in percentage change in SWA: ▼ P < 0.05; ▼▼ P < 0.01 (2-way ANOVA with a “condition” as a between-subject factor and repeated measures on factor “time” followed by Fisher LSD posthoc test). Note that REMD and YC rats did not differ significantly in the percentage change of both QW and NREM sleep SWA from baseline levels.
Cell Proliferation
The mean number of BrdU-positive cells in the subgranular cell layer of the DG differed significantly among the groups (Table 2). The REMD group exhibited 63% reduction in the number of BrdU-positive cells when compared with the counterpart YC group (P < 0.05, Bonferroni t-test) (Table 2, Figure 3, A-B). REMD and YC animals had 82% and 51% fewer BrdU-labeled cells, respectively, when compared to the CC (P < 0.001, Bonferroni t-test).
Table 2.
Cell Proliferation and Phenotypes of BrdU-Positive Cells
| BrdU + cells | Ki-67 + cells | % BrdU/NeuN | % BrdU/S100β | |
|---|---|---|---|---|
| CC | 3437 ± 387 | 5094 ± 281 | 76.4 ± 2.5 | 7.3 ± 2.1 |
| YC | 1702 ± 220a | 2557 ± 297a | 59.4 ± 3.7a | 5.5 ± 2.0 |
| REMD | 636 ± 131a,b | 1104 ± 203a,b | 53.3 ± 4.1a | 6.8 ± 2.5 |
| F2,21 = 27.9*** | F2,21 = 58.5*** | F2,15 = 11.6*** | F2,15 = 0.2 |
Cell proliferation was assessed by counting the total number of BrdU(5- bromo- 2′- deoxy-uridine)- or Ki-67-positive cells per dentate gyrus. Phenotypes of the surviving cells were determined by immunofluorescent triple labeling for BrdU, NeuN (neurons) and S100β (glia) 3 weeks after the administration of BrdU. The percentages of BrdU-positive cells colabeled for either NeuN or S100β are presented. Data are means ± SEM.
***P < 0.001, 1-way analysis of variance.
P < 0.01 vs cage controls, bP < 0.01 vs yoked controls, Bonferroni t-test.
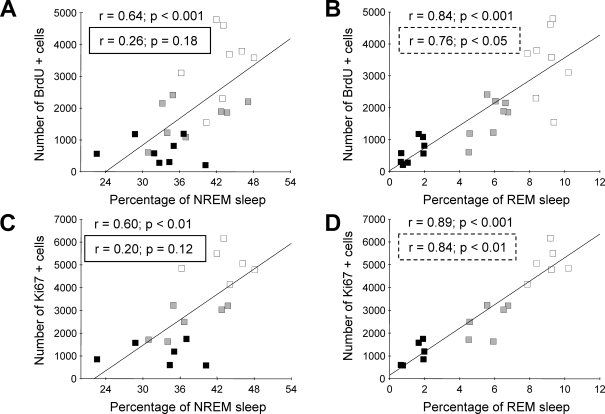
Figure 3.
Scatter plots and regression lines for the number of BrdU (5- bromo- 2′- deoxy-uridine) and Ki-67 positive cells vs percentages of non-rapid eye movement (NREM) (A, C) and rapid eye movement (REM) sleep (B, D) for the entire recording session. Note that the percentages of both NREM and REM sleep were significantly correlated with the number of BrdU and Ki-67 positive cells. However, as described in the text, the correlation between the number of proliferating cells and the percentage of NREM sleep becomes nonsignificant when controlling for the percentage of REM sleep (see r values inside rectangles A,C). The correlation between the numbers of proliferating cells and the percentage of REM sleep remained significant when controlling for the percentage of NREM sleep (see r values in dashed rectangles B,D). The dark, gray and white squares represent individual REM-deprived (REMD), yoke-control (YC), and cage-control animals.
A different set of sections from most brains used for BrdU immunohistochemistry was processed for Ki-67 immunohistochemistry; Ki-67 is an endogenous marker of cell proliferation. The differences in the mean number of Ki-67-positive cells among the groups were statistically significant (Table 2). REMD animals had 57% fewer Ki-67-positive cells compared to YC (P < 0.01, Bonferroni t-test). The REMD group and the YC groups exhibited approximately 80% and 50% reductions in the numbers of Ki-67-positive cells, respectively, compared with the CC group (P < 0.001, Bonferroni t-test).
We conducted correlation analyses between the percentages of NREM and REM sleep and the number of BrdU-positive cells, combining REMD, YC, and CC groups (Figure 3). Across all animals, the number of BrdU-positive cells was significantly (P < 0.001) correlated with the percentages of both NREM (r = 0.84) and REM sleep (r = 0.64) (Figure 3, A, B). A partial correlation analysis showed that the correlation between the percentage of NREM sleep and BrdU-positive cell counts was secondary to the contribution of REM sleep. When percentage of REM sleep was used as the controlling variable, the correlation between the number of BrdU-positive cells and NREM sleep was no longer significant (r = 0.26, P = 0.18). However, when the analysis controlled for the percentage of NREM sleep, the correlation between BrdU-positive cell counts and the percentage of REM sleep remained high and significant (r = 0.76, P < 0.05).
The number of Ki-67-positive cells also significantly correlated with the percentages of both NREM (r = 0.60, P < 0.01) and REM sleep (r = 0.89; P < 0.001) (Figure 3C and D). The partial correlation analysis revealed that controlling for the percentage of NREM sleep had little effect on the correlation between the number of Ki-67-positive cells and the percentage of REM sleep (r = 0.84; P < 0.01). However, when the contribution of REM sleep was removed, the correlation between the number of Ki-67-positive cells and the percentage of NREM sleep was no longer significant (r = 0.20, P = 0.12).
Experiment 2
Cell Maturation
The phenotype of the surviving BrdU-positive cells was evaluated 3 weeks after the administration of BrdU by immunofluorescent triple labeling for BrdU, the neuronal marker NeuN, and the glial marker S100β (Figure 4, C). The mean percentages of the BrdU/NeuN-positive cells differed significantly among the groups (Table 2). The percentages of BrdU-positive cells colabeled with NeuN in REMD and YC animals were 30% and 22% lower, respectively, than in CC animals (P < 0.001 and P < 0.05, respectively, Bonferroni t-test), but the REMD and YC groups did not differ significantly. The localization of BrdU/NeuN-positive cells was similar in all groups. There were no differences between the groups with respect to the percentage of BrdU-positive cells that expressed the glial phenotype (Table 2). The percentages of cells that did not co-localize BrdU with either NeuN or S100β did not differ significantly between REMD and YC animals and but were higher in these groups compared to CC (P < 0.001, Bonferroni t-test).
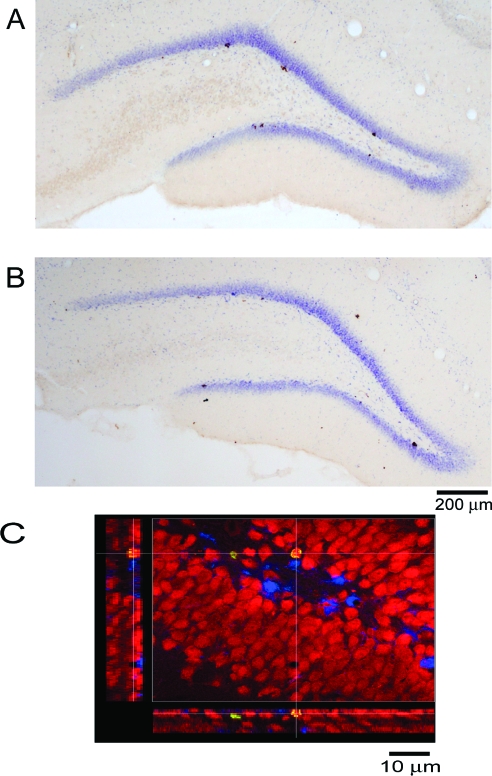
Figure 4.
BrdU (5-bromo- 2′- deoxy-uridine) labeling in the dentate gyrus (DG) of the hippocampus. A and B: representative photomicrographs of sections from yoke-control (YC) and REM-deprived (REMD) animals, respectively. C: A 3-week-old BrdU-positive cell in the DG colocalizing with NeuN shown in an orthogonal view.
Experiment 3
Corticosterone Levels
To investigate whether corticosterone may play a role in the effect of the REMD procedure, we compared plasma corticosterone in REMD and YC groups. We found no significant differences in the corticosterone levels between groups (15.43 + 4.02 μg/dL REMD vs 20.97 + 10.7 μg/dL YC; P = 0.64).
DISCUSSION
In the present report, we showed that 4 days' exposure to a REMD procedure reduced cell proliferation in the DG of the hippocampus of the adult rat. REMD animals exhibited 81% and 63% reductions in the number of BrdU-positive cells, compared with the CC and YC rats, respectively. We also used an endogenous proliferation marker, Ki-67, to rule out the possibility that changes in the number of BrdU-positive cells could be related to differences in distribution or availability of BrdU during the REMD procedure. The changes observed with Ki-67 staining closely paralleled those obtained with BrdU staining.
Pairs of REMD and YC animals were exposed to the same experimental environment and experienced the identical physical stimulation. The amount of NREM sleep did not differ significantly between these groups. Since brief 2-hour REM sleep deprivation using brain stimulation reduced concomitant SWA,10 we considered the possibility that SWA within NREM could be differentially affected in REMD and YC groups. REMD and YC rats exhibited similar reductions in both NREM and QW SWA during treatment days, compared with the baseline, and did not differ in NREM sleep-QW SWA ratios during the baseline and treatment days. Therefore, suppression in cell proliferation in REMD animals compared with YC was most likely to be attributable to REM sleep loss (79% in REMD vs YC group). However, although the experiment was well controlled, we cannot exclude the possibility that the REMD procedure could cause changes in other NREM sleep parameters or in waking behaviors that may be essential for proliferative process.
A decrease in proliferation in the YC, compared with CC, animals could have resulted from effects of either the procedure itself or partial REMD or both. Differences in experimental milieu and mild reduction of NREM sleep could contribute to reduction in cell proliferation in REMD compared with CC animals.
We found that the numbers of both BrdU-positive and Ki-67-positive cells were highly positively correlated with the percentages of both REM sleep and NREM sleep across individual animals. The results of partial correlation analyses showed that correlation between the percentage of REM sleep and rate of proliferation was not secondary to correlation with NREM sleep because it remained significant while controlling for the percentage of NREM sleep. These findings strengthen the argument that REM sleep-associated processes are critical for cell proliferation. Pollack et al (personal communication) also concluded that REM sleep contributes to the stimulation of proliferation in DG, using a different REMD method. However, we do not exclude the possibility that NREM sleep is also essential to the proliferative process. By simple analogy, the output of an electronic device can be prevented both by reduction of a power source and by disturbance of complex frequency-dependent operations. Similarly, NREM and REM sleep may both have critical roles in cell proliferation and maturation.
Given that cell proliferation can be suppressed by either acute11 or chronic elevation of glucocorticoids,4 and a recent study using a different method of sleep deprivation, concluded that the effects of sleep deprivation on proliferation could be attributed to elevated corticosterone,12 we measured corticosterone immediately following the interval of BrdU exposure. In the present study, corticosterone levels in the REMD group did not differ from the YC group subjected to the same experimental conditions. This indicates that the reduction in proliferation in REMD animals compared with YC was not due to elevated corticosterone at the time of BrdU administration. This study does not exclude the possibility that stress-related processes occurring earlier in the procedure may have influenced the proliferative process. However, we emphasize that we adapt animals to our deprivation procedures, to minimize stress responses. Animals were not restrained within typical cage-sized environments and experienced no treadmill movement during all periods of spontaneous waking and NREM sleep in REMD animals. Thus, our procedure, like some TSD methods,6,13 does not differentially elevate corticosterone in experimental animals compared with YC controls. Corticosterone levels in both REMD and YC groups were slightly higher than those found in basal conditions14 and in our previous TSD study.6 We cannot exclude the possibility that mild elevations in corticosterone levels could contribute to a decrease in cell proliferation in REMD and YC compared with CC. However, mildly elevated corticosterone levels are not always associated with suppression of proliferation. Voluntary wheel running, which is accompanied by increased corticosterone,15 also increases proliferation.16
Mirescu et al12 have reported that the antiproliferative effects of 3 days of sleep deprivation using the single small-pedestal-over-water method could be prevented by adrenalectomy with basal corticosterone replacement in drinking water. This method suppresses REM sleep but is not selective for REM.17 There are several differences between Mirescu et al's and our study. The duration of deprivation, 3 days versus 4 days in our study, is one of the differences between studies. The effects of sleep deprivation/REM deprivation may be cumulative (see below), and stress responses may diminish with time. In addition, Mirescu et al compared proliferation in adrenalectomized sleep-deprived/REMD animals and adrenalectomized CC, rather than pedestal controls. Some uncontrolled effects of the procedure, possibly including changed corticosterone levels associated with circadian patterns of water consumption, altered circadian rhythms, water bath exposure, or associated body temperature changes, could have affected proliferation in REMD animals, masking the effects of REMD. Another difference between studies was the timing of BrdU administration, zeitgeber time 0 in our study, zeitgeber time 7 in Mirescu et al. Additional work is needed to determine what procedural details account for the different findings. We have recently found that a 4-day sleep-fragmentation procedure that markedly suppressed REM, but not NREM sleep, also suppresses proliferation in adrenalectomized rats,18 supporting the view that elevated corticosterone does not account for effects of sleep loss on proliferation.
Both REMD and YC rats showed a lower percentage of BrdU-positive cells that later exhibited a neuronal phenotype. In the CC group, the majority (76%) of the newly formed cells in the DG expressed the mature neuronal marker NeuN, a finding that is consistent with other studies.19 However, in the YC and REMD groups, only 59% and 53% of BrdU-positive cells, respectively, colocalized NeuN. The localization of BrdU/NeuN-positive cells within the DG was similar in all groups. These findings suggest that REMD does not interfere with the subsequent migration of new granule cells. The percentages of BrdU/S100β-positive cells did not differ significantly between the groups, whereas the percentages of cells that were not stained for either glial or neuronal markers were more than 2-fold higher in REMD and YC animals, compared with CC. These unidentified cells may have a delayed maturation course or may have a permanently impaired capacity for maturation. Effects of the procedure, including mild stress, could have contributed to this deficit.
Our original descriptions of effects of TSD have been replicated. Tung et al8 reported a 36% reduction in proliferation after 54 hours of TSD using the disc-over-water method. This group also showed that the suppression of proliferation persisted after 8 hours of recovery sleep. Eight days of sleep restriction using a rotating drum also reduced proliferation in mice.20 Moreover, repeated 6-hour periods of sleep restriction have been shown to abolish the neurogenic effects of hippocampal-dependent learning, as well as the survival of proliferating cells.21 In all of these studies, both NREM and REM sleep were suppressed. However, Pollack et al (personal communication) showed that 4 days of REMD using the multiple-pedestal method, suppressed proliferation. It should be noted that these antiproliferative effects depend on sustained sleep or REM deprivation. It has been reported that short-term TSD (12 hours) by gentle handling does not change22 or increases proliferation,23 and 24 hours of deprivation does not change proliferation12 or reduces proliferation only in the hilus.20
The present findings suggest that REM sleep is critical for the proliferative process. A REM-like state is elevated in older infants,24 and it has been proposed that REM sleep plays a role in the development of the nervous system. Studies at electrophysiologic and molecular levels support a hypothesis that REM sleep plays a role in brain plasticity during development.25 The impact of REM sleep on adult neurogenesis may be an extension of these developmental processes.
REM sleep either may directly enable concurrent proliferation or may facilitate molecular processes that enable proliferation in subsequent states. REM sleep exhibits circadian rhythmicity, and there is some evidence that proliferation does not.20, 26–28 However, we have recently found that proliferation is strongly increased in the light phase of the circadian cycle, when total sleep and REM sleep amounts are highest.29 However, direct evidence that proliferation is enhanced during REM sleep is not yet available. The fact that short-term sleep deprivation does not diminish proliferation,22, 23 and that proliferation does not recover after 6 to 8 hours of recovery sleep,8,12 support a hypothesis that REM sleep enables cumulative molecular processes that support subsequent proliferation. Several mechanisms could underlie cumulative effects of sleep or REM sleep loss. It has been shown that brain protein synthesis is increased in sleep.30 Protein synthesis is required for cellular proliferation and growth. Moreover, sleep deprivation increases markers of oxidative stress in the hippocampus, as well as message for proinflammatory cytokines.31 Inflammatory processes suppress adult neurogenesis.32 Sleep deprivation also dramatically decreases levels of serum insulin-like growth factor-1,13 which facilitates proliferation,33 but may have a delayed action. Direct effects of sleep on hippocampal plasticity are well documented. Following 48 hours of TSD, hippocampal expression of plasticity-related genes, including brain-derived neurotrophic factor, is reduced, and the decrement is correlated with REM sleep loss.34 Brain-derived neurotrophic factor also facilitates DG cell proliferation.35 The relative contribution of REM and NREM sleep to these processes has not been evaluated.
In summary, the present study compared REMD and YC animals to assess a hypothesis that REM sleep-associated processes facilitate proliferation and maturation of granule cells in the adult hippocampal DG. Our findings strongly support the hypothesis that REM sleep is essential to the proliferative process. Although NREM sleep amounts and SWA were not differentially changed in REMD and YC animals, we cannot exclude the possibility that subtle changes in NREM sleep induced by the REMD procedures may have affected proliferation. The role of NREM in the proliferative process was not assessed in this study. In addition, REMD and YC procedures may have differentially affected waking behaviors that account for differences between groups. We also showed that corticosterone levels were not different in REMD and YC animals at the time of proliferative cell labeling, but this study does not rule out the possibility that stress-related processes occurring earlier in the procedure may have influenced the proliferative process. Further work is needed to show conclusively that REM directly facilitates proliferation and to determine the role of NREM sleep in hippocampal neurogenesis.
ACKNOWLEDGMENTS
We gratefully acknowledge Feng Xu and Keng-Tee Chew for their excellent assistance. This research was made possible by a grant from the American Sleep Medicine Foundation, a foundation of the American Academy of Sleep Medicine to RGM. This research was also supported by the US Department of Veterans Affairs Medical Research service and US National Institutes of Health grants MH 075076 and HL 60296.
Footnotes
Disclosure Statement
This was not an industry supported study. Dr. Szymusiak has received research support from Sepracor. Dr. McGinty has received research support from Sepracor. The other authors have indicated no financial conflicts of interest.
REFERENCES
- 1.Kuhn GH, Dickinson-Anson H, Gage FH. Neurogenesis in the dentate gyrus of the adult rat: age-related decrease of neuronal progenitor proliferation. J Neurosci. 1996;16:2027–33. doi: 10.1523/JNEUROSCI.16-06-02027.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Markakis EA, Gage FH. Adult-generated neurons in the dentate gyrus send axonal projections to field CA3 and are surrounded by synaptic vesicles. J Comp Neurol. 1999;406:449–60. [PubMed] [Google Scholar]
- 3.Lledo PM, Alonso M, Grubb M. Adult neurogenesis and functional plasticity in neuronal circuits. Nat Rev Neurosci. 2006;7:179–93. doi: 10.1038/nrn1867. [DOI] [PubMed] [Google Scholar]
- 4.Cameron HA, Gould E. Adult neurogenesis is regulated by adrenal steroids in the dentate gyrus. Neuroscience. 1994;61:203–9. doi: 10.1016/0306-4522(94)90224-0. [DOI] [PubMed] [Google Scholar]
- 5.Heine V, Maslam S, Joels M, Lucassen P. Suppressed proliferation and apoptotic changes in the rat dentate gyrus after acute and chronic stress are reversible. Eur J Neurosci. 2004;19:131–44. doi: 10.1046/j.1460-9568.2003.03100.x. [DOI] [PubMed] [Google Scholar]
- 6.Guzman-Marin R, Suntsova N, Stewart DR, Szymusiak R, McGinty D. Sleep deprivation reduces proliferation of cells in the dentate gyrus of the hippocampus in rats. J Physiol. 2003;549:563–71. doi: 10.1113/jphysiol.2003.041665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guzman-Marin R, Suntsova N, Methippara M, Szymusiak R, McGinty D. Sleep deprivation suppresses neurogenesis in the adult hippocampus of rats. Eur J Neurosci. 2005;22:2111–6. doi: 10.1111/j.1460-9568.2005.04376.x. [DOI] [PubMed] [Google Scholar]
- 8.Tung A, Takase L, Fornal C, Jacobs B. Effects of sleep deprivation and recovery sleep upon cell proliferation in adult rat dentate gyrus. Neuroscience. 2005;134:721–3. doi: 10.1016/j.neuroscience.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 9.Bergman BM, Mistlberger RE, Rechtschaffen A. Period-amplitude analysis of rat electroencephalogram: stage and diurnal variations and effects of suprachiasmatic nuclei lesions. Sleep. 1987;10:523–36. [PubMed] [Google Scholar]
- 10.Benington JH, Woundenberg MC, Heller HC. REM-sleep propensity accumulates during 2-h REM-sleep deprivation in the rest period in rats. Neurosci Lett. 1994;180:76–80. doi: 10.1016/0304-3940(94)90917-2. [DOI] [PubMed] [Google Scholar]
- 11.Gould E, McEwen BS, Tanapat P, Galea L, Fuchs E. Neurogenesis in the dentate gyrus of the adult tree shrew is regulated by psychosocial stress and NMDA receptor activation. J Neurosci. 1997;17:2492–8. doi: 10.1523/JNEUROSCI.17-07-02492.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mirescu C, Peters JD, Noiman L, Gould E. Sleep deprivation inhibits adult neurogenesis in the hippocampus by elevating glucocorticoids. Proc Natl Acad Sci USA. 2006;103:19170–5. doi: 10.1073/pnas.0608644103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Everson CA, Crowley WR. Reductions in circulating anabolic hormones induced by sustained sleep deprivation in rats. Am J Physiol Endocrinol Metab. 2004;286:E1060–70. doi: 10.1152/ajpendo.00553.2003. [DOI] [PubMed] [Google Scholar]
- 14.Sapolsky RM, Krey LC, McEwen BS. Prolonged glucocorticoid exposure reduces hippocampal neuron number: implications for aging. J Neurosci. 1985;5:1222–7. doi: 10.1523/JNEUROSCI.05-05-01222.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Droste SK, Gesing A, Ulbricht S, Muller MB, Linthorst AC, Reul JM. Effects of long-term voluntary exercise on the mouse hypothalamic-pituitary-adrenocortical axis. Endocrinology. 2003;144:3012–23. doi: 10.1210/en.2003-0097. [DOI] [PubMed] [Google Scholar]
- 16.van Pragg H, Christie BR, Sejnowski TJ, Gage FH. Running enhances neurogenesis, learning, and long-term potentiation in mice. Proc Natl Acad Sci USA. 1999;96:13427–31. doi: 10.1073/pnas.96.23.13427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Machado RB, Hipolide DC, Benedito-Silva AA, Tufik S. Sleep deprivation induced by the modified multiple platform technique: quantification of sleep loss and recovery. Brain Res. 2004;1004:45–51. doi: 10.1016/j.brainres.2004.01.019. [DOI] [PubMed] [Google Scholar]
- 18.Guzman-Marin R, Bashir T, Suntsova N, Szymusiak R, McGinty D. Adult hippocampal neurogenesis is reduced by sleep fragmentation in the adult rat. Neuroscience. 2007;148:325–333. doi: 10.1016/j.neuroscience.2007.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Malberg JE, Eisch AJ, Nestler EJ, Duman RS. Chronic antidepressant treatment increases neurogenesis in adult rat hippocampus. J Neurosci. 2000;20:9104–10. doi: 10.1523/JNEUROSCI.20-24-09104.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roman V, Van der Borght K, Leemburg SA, Van der Zee EA, Meerlo P. Sleep restriction by forced activity reduces hippocampal cell proliferation. Brain Res. 2005;1065:53–9. doi: 10.1016/j.brainres.2005.10.020. [DOI] [PubMed] [Google Scholar]
- 21.Hairston IS, Little MT, Scanlon MD, Palmer TD, Sapolsky RM, Heller HC. Sleep restriction suppresses neurogenesis induced by hippocampus-dependent learning. J Neurophysiol. 2005;91:1586–95. doi: 10.1152/jn.00218.2005. [DOI] [PubMed] [Google Scholar]
- 22.Van der Borght K, Ferrari F, Klauke K, Roman V, Havekes R, Sgoifo A, van der Zee EA, Meerlo P. Hippocampal cell proliferation across the day: increase by running wheel activity, but no effect of sleep and wakefulness. Behav Brain Res. 2006;167:36–41. doi: 10.1016/j.bbr.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 23.Grassi Zucconi GG, Cipriani S, Balgkoiranidou I, Scattoni R. ‘One night’ sleep deprivation stimulates hippocampal neurogenesis. Brain Res Bull. 2006;69:375–81. doi: 10.1016/j.brainresbull.2006.01.009. [DOI] [PubMed] [Google Scholar]
- 24.Roffwarg H, Muzio J, Dement W. Ontogenetic development of the human sleep cycle. Science. 1996;152:604–19. doi: 10.1126/science.152.3722.604. [DOI] [PubMed] [Google Scholar]
- 25.Shaffery JP, Sinton CM, Bissette G, Roffwarg H, Marks GA. Rapid eye movement sleep deprivation modifies expression of long-term potentiation in visual cortex of immature rats. Neuroscience. 2002;110:431–43. doi: 10.1016/s0306-4522(01)00589-9. [DOI] [PubMed] [Google Scholar]
- 26.Ambrogini P, Orsini L, Mancini C, Ferri P, Barbanti I, Cuppini R. Persistently high corticosterone levels but not normal circadian fluctuations of the hormone affect cell proliferation in the adult rat dentate gyrus. Neuroendocrinology. 2002;76:366–72. doi: 10.1159/000067581. [DOI] [PubMed] [Google Scholar]
- 27.Holmes MM, Galea LA, Mistlberger RE, Kempermann G. Adult hippocampal neurogenesis and voluntary running activity: circadian and dose-dependent effects. J Neurosci Res. 2004;76:216–22. doi: 10.1002/jnr.20039. [DOI] [PubMed] [Google Scholar]
- 28.Kochman LJ, Weber ET, Fornal CA, Jacobs BL. Circadian variation in mouse hippocampal cell proliferation. Neurosci Lett. 2006;406:256–9. doi: 10.1016/j.neulet.2006.07.058. [DOI] [PubMed] [Google Scholar]
- 29.Guzman-Marin R, Suntsova N, Bashir T, Szymusiak R, McGinty D. Cell proliferation in the dentate gyrus of the adult rat fluctuates with the light-dark cycle. Neurosci Lett. 2007;422:198–201. doi: 10.1016/j.neulet.2007.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ramm P, Smith CT. Rates of cerebral protein synthesis are linked to slow wave sleep in the rat. Physiol Behav. 1990;48:749–753. doi: 10.1016/0031-9384(90)90220-x. [DOI] [PubMed] [Google Scholar]
- 31.McEwen BS. Sleep deprivation as a neurobiologic and physiologic stressor: Allostasis and allostatic load. Metabolism. 2006;55:S20–S23. doi: 10.1016/j.metabol.2006.07.008. [DOI] [PubMed] [Google Scholar]
- 32.Ekdahl CT, Claasen JH, Bonde S, Kokaia Z, Lindvall O. Inflammation is detrimental for neurogenesis in adult brain. Proc Natl Acad Sci USA. 2003;100:13632–7. doi: 10.1073/pnas.2234031100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Carro E, Nunez A, Busiguina S, Torres-Aleman I. Circulating insulin-like growth factor I mediates effects of exercise on the brain. J Neurosci. 2000;20:2926–33. doi: 10.1523/JNEUROSCI.20-08-02926.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guzman-Marin R, Ying Z, Suntsova N, Szymusiak R, Gomez-Pinilla F, McGinty D. Suppression of hippocampal plasticity-related gene expression by sleep deprivation. J Physiol. 2006;575:807–19. doi: 10.1113/jphysiol.2006.115287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Scharfman H, Goodman J, Macleod A, Phani S, Antonelli C, Croll S. Increased neurogenesis and the ectopic granule cells after intrahippocampal BDNF infusion in adult rats. Exp Neurol. 2005;192:348–56. doi: 10.1016/j.expneurol.2004.11.016. [DOI] [PubMed] [Google Scholar]