Abstract
In our 3D NOESY-[1H,15N,1H]-ZQ-TROSY experiment, the TROSY principle (transverse relaxation-optimized spectroscopy) is used in three-dimensional (3D) 15N-resolved nuclear Overhauser enhancement spectroscopy (NOESY), which enables resonance assignments by sequential nuclear Overhauser effects and the collection of structural constraints in large 15N- or 2H,15N-labeled proteins. Our experiment affords optimization of the transverse relaxation in all three frequency dimensions, provides suppression of the strong diagonal autorelaxation peaks, which otherwise tend to interfere with the analysis of nearby informative crosspeaks, and yields improved resolution for the entire spectrum when compared with conventional 3D 15N-resolved-[1H,1H]-NOESY, because of the narrower lineshapes along both proton dimensions. The key element of this experiment is an approach for correlating the 15N and 1H chemical shifts with two-dimensional ZQ-[15N,1H]-TROSY, where zero-quantum (ZQ) coherence is generated and the remote cross-correlation between the 1H and 15N chemical shift anisotropy interactions is used to reduce transverse relaxation during 15N evolution. Practical applications are illustrated with spectra of a protein with a molecular mass of 110,000 Da.
The introduction of transverse relaxation-optimized spectroscopy (TROSY) (1) has greatly expanded the molecular size range accessible for solution NMR experiments that correlate the chemical shifts of either 15N and 1H in N–H groups (2, 3) or 13C and 1H in aromatic C–H fragments (4) and for triple-resonance experiments used for sequential assignments of [2H, 13C, 15N]- or [13C,15N]-labeled proteins (5). With regard to extending NMR structure determination to larger molecular sizes, it is further of keen interest to improve sensitivity and resolution of nuclear Overhauser effect (NOE) experiments with the use of TROSY, which is the subject of this paper. NOE spectroscopy (NOESY) yields 1H–1H distance constraints that enable de novo determination of three-dimensional (3D) structures of biological macromolecules by NMR in solution (6).
In initial implementations of the TROSY principle in NOE experiments, [13C,1H]-TROSY and [15N,1H]-TROSY have been used in 3D 13C-resolved and 15N-resolved [1H,1H]-NOESY to resolve homonuclear 1H–1H NOEs along the heteronuclear dimension (7, 8). In these NOE experiments, the conventional [13C,1H]- or [15N,1H]-heteronuclear single-quantum coherence or heteronuclear multiple-quantum coherence schemes (9) were substituted by the corresponding single-quantum TROSY-type chemical shift correlation schemes (2, 4), whereby the desired transverse relaxation optimization was achieved along the 13C frequency axis (7) or in the 15N and 1HN dimensions (8), respectively. Because in 3D heteronuclear-resolved NOESY experiments the most interesting information resides in the [1H,1H]-spectral planes and high spectral resolution should therefore primarily be achieved along the two 1H dimensions, this approach is of only limited use. Here, we introduce a 3D 15N-resolved NOE experiment, 3D NOESY-[1H,15N,1H]-ZQ-TROSY, which affords optimization of the transverse relaxation in all three spectral dimensions. An additional important feature of this experiment is artifact-free suppression of the diagonal peaks, which are typically much stronger than the crosspeaks and tend to interfere with crosspeak identification and integration in NOESY spectra. A key element of 3D NOESY-[1H,15N,1H]-ZQ-TROSY is that the 15N and 1H chemical shifts are correlated by sensitivity-enhanced two-dimensional (2D) ZQ-[15N,1H]-TROSY steps, which use the remote cross-correlation between 1H chemical shift anisotropy (CSA) and 15N CSA interactions (10) to reduce the transverse relaxation rate of the [15N,1H]-ZQ coherence.
METHODS
The experimental schemes that are introduced in this paper (Fig. 1) are described by the product operator formalism (11) by using the single-transition basis operators Ii13 = 1/2 Ii + IiSi and Ii24 = 1/2 Ii − IiSi, where I and S represent the 1H and 15N spins, and i stands for “z,” “+,” or “−.” The abbreviations DQ± = I±S± and ZQ± = I∓S± are used for the double-quantum (DQ) and ZQ operators, respectively. For the 2D ZQ-[15N,1H]-TROSY element, two equivalent magnetization transfer pathways are given by Eqs. 1 and 2:
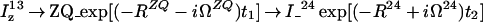 |
1 |
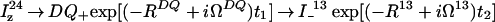 |
2 |
The initial steady-state 1H polarization at time point a (Fig. 1a), Iz is the sum of the single-transition polarization operators, Iz = Iz13 + Iz24, which can be considered separately. ΩI and ΩS are the chemical shifts relative to the carrier frequency of the spins I and S, J is the scalar coupling constant between I and S, ΩZQ = ΩI − ΩS, ΩDQ = ΩI + ΩS, Ω24 = ΩI − πJ, and Ω13 = ΩI + πJ. R24 and R13 are the previously described relaxation rates of the individual single-quantum transitions 2 → 4 and 1 → 3 (1, 4). RZQ and RDQ are the relaxation rates of the ZQ and DQ coherences given by Eqs. 3 and 4.
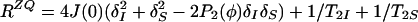 |
3 |
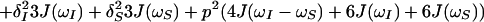 |
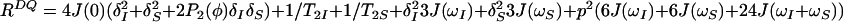 |
4 |
For rigid spheres, the spectral densities, J(ω), can be written as
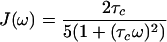 |
5 |
In Eqs. 3–5, ωS and ωI are the Larmor frequencies of the spins S and I, the dipole–dipole (DD) interaction is p2 = 1/8(γIγSћ/rIS3)2, the CSA interactions are δS2 = 1/18(ωSΔσS)2 and δI2 = 1/18(ωIΔσI)2, γI and γS are the gyromagnetic ratios of I and S, ћ is the Planck constant divided by 2π, rIS the distance between the nuclei S and I, Δσ = σzz − 0.5 (σxx + σyy), P2(φ) = (3 cos2 φ − 1)/2, where φ is the angle between the principal directions of the chemical shift tensors of the spins S and I, and 1/T2I and 1/T2S are the contributions to the transverse relaxation rates of the spins I and S because of DD coupling with remote protons (1).
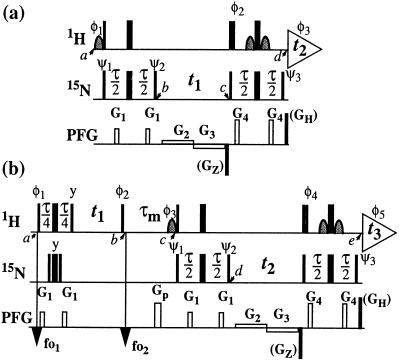
Figure 1.
Schemes for the experiments discussed in this paper. (a) 2D ZQ-[15N,1H]-TROSY. (b) 3D NOESY-ZQ-[1H,15N,1H]-TROSY with transverse relaxation optimization in all three dimensions and suppression of the diagonal peaks. On the lines marked 1H and 15N, narrow and wide bars stand for nonselective 90° and 180° rf pulses, respectively, and curved shapes represent water-selective 90° rf pulses. Water saturation is minimized by returning the water magnetization to the +z axis before data acquisition (15, 18). The time period τ is set to 5.4 ms. The line marked PFG indicates pulsed magnetic field gradients applied along the z axis. (a) The gradients are: G1 amplitude 30 G/cm, duration 1 ms; G2, 5 G/cm, 0.5⋅t1; G3, −5 G/cm, 0.5⋅t1; G4, 40 G/cm, 1 ms. The phases for the rf pulses are: φ1 = {−x}; φ2 = {x}; φ3 = {x, −x, −y, y}; ψ1 = {−x, x, −y, y}; ψ2 = {y, −y, x, −x}; ψ3 = {y}; x on all other pulses. To obtain a complex interferogram, a second free induction decay (FID) is recorded for each t1 delay, with the following different phases: φ1 = {x}, φ2 = {−x}, φ3 = {x, −x, y, −y}, ψ3 = {−y}. After Fourier transformation in the ω2 dimension, the complex interferogram is multiplied by exp[−iΩHt1], where ΩH is the offset in the ω2 dimension relative to the 1H carrier frequency in rad s−1. Further data processing following ref. 2. (b) The gradients are: G1, 30 G/cm, 1 ms; Gp, 40 G/cm, 2 ms; G2, 5 G/cm, 0.5⋅t2; G3, −5 G/cm, 0.5⋅t2; G4, 40 G/cm, 1 ms. The phases of the rf pulses are: φ1 = {4(45°), 4(225°)}; φ2 = {x}; φ3 = {−x}; φ4 = {x}; φ5 = {x, −x, −y, y, −x, x, y, −y}; ψ1 = {−x, x, −y, y}; ψ2 = {y, −y, x, −x}; ψ3 = {y}; x on all other pulses. The 1H carrier frequency offsets are set to 8.7 ppm at time point fo1 and to 4.8 ppm at fo2. Quadrature detection in ω1 is achieved by the States time-proportional phase incrementation method (9) applied with the phase φ2. For each t2 increment, a second FID is recorded with the following different phases: φ3 = {x}, φ4 = {−x}, φ5 = {x, −x, y, −y, −x, x, −y, y}, ψ3 = {−y}. Further data processing as described for a. As an alternative, in both schemes the coherence selection can be supported by addition of the PFGs GZ (−50 G/cm, 1.59 ms) and GH (50 G/cm, 0.177 ms). GZ is then inverted in concert with the phase shifts used to obtain a complex interferogram, and the nonzero initial values for the evolution times need to be taken into account.
In the slow tumbling limit, only terms in J(0), 1/T2I and 1/T2S need to be retained in Eqs. 3 and 4. For backbone 15N–1H amide moieties in proteins, where δS ≈ δI and the unique axes of the 1H and 15N CSA tensors are nearly parallel (12, 13), P2(φ) ≈ 1 and the transverse relaxation caused by 1H CSA and 15N CSA interactions is largely suppressed for ZQ coherence (Eq. 3). Because 15N–1H DD relaxation is negligible for ZQ coherence (11) and the acquired signal comes from the relaxation-optimized single-transition 2 → 4 (2), Eq. 1 represents a way for obtaining TROSY-type 2D [15N,1H]-correlation spectra with the transverse relaxation optimized in both dimensions. The unfavorable pathway of Eq. 2 is suppressed by phase cycling, pulsed field gradient (PFGs), or a combination of both (Fig. 1a).
The ZQ-[15N,1H]-TROSY experiment contains a sensitivity-enhanced quadrature detection scheme that requires only a single insensitive nuclei enhanced by polarization transfer (INEPT) element for the transfer of both precessing orthogonal components of the ZQ coherence to the observable signal. When compared with the serial arrangement of two INEPT elements in the standard single transition-to-single transition polarization transfer enhancement scheme (2), a single INEPT element results in a smaller number of 1H and 15N pulses and an overall shorter pulse sequence, with concomitant improvement in sensitivity. With the use of the PFGs for coherence pathway selection according to Eq. 1, only a one-step phase cycle is required for the collection of a phase-sensitive sensitivity-enhanced 2D spectrum. These features make 2D ZQ-[15N,1H]-TROSY an attractive building block in more complex NMR experiments (see below).
Relaxation caused by remote DD coupling is not affected by TROSY, either in the ZQ variant or in the standard version with single-quantum coherence optimized by cross-correlation of DD coupling and CSA interactions (1, 2). The best results with the experiments of Fig. 1 will therefore be obtained in uniformly 2H,15N-labeled proteins, where 1/T2I and 1/T2S are affected only by interactions between closely spaced 15N–1H groups.
In the two independent polarization transfer pathways of the experiment of Fig. 1a (Eqs. 1 and 2), the initial spin-polarizations associated with the individual transitions 1 → 3 and 2 → 4 are not mixed during the experiment, so that they can be separately detected as single transitions. Because the signal detected through the 2 → 4 transition (Eq. 1) is also independent of the initial spin-polarization of the 2 → 4 transition, the scheme of Fig. 1a can be used as a building block in the 3D NOESY-ZQ-[1H,15N,1H]-TROSY experiment (Fig. 1b), where the diagonal autorelaxation peaks are suppressed, and the transverse relaxation is optimized in all three spectral dimensions. In the following, the experiment of Fig. 1b is analyzed for a combination of two 2-spin systems representing two 15N–1H groups interacting by 1H–1H DD coupling. The two spin systems are distinguished by additional superscripts, (1) and (2). Only the relevant magnetization transfer pathways are considered, and relaxation during the polarization transfer steps is neglected. Between the timepoints a and b (Fig. 1b), the equilibrium density matrix, δa = Iz(1) + Iz(2), is transformed into a density matrix that contains the relevant terms given by Eq. 6.
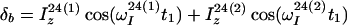 |
6 |
Because of the S3E element (14) at the beginning of the pulse sequence (Fig. 1b), the steady-state magnetizations associated with the 1 → 3 proton transitions of both amide moieties are not 1H frequency labeled during t1; they are kept transverse during the NOE mixing time τm and eventually dephased by the gradient Gp (Fig. 1b). During τm, the 1 → 3 transitions can be repolarized by DD cross relaxation, with rates σ, and by longitudinal autorelaxation, with rates ρijkl, resulting in exchange of magnetization between the i → j and k → l transitions as given by Eq. 7.
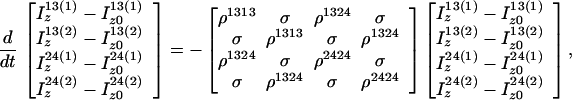 |
7 |
with
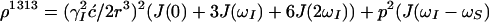 |
8 |
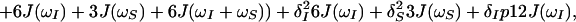 |
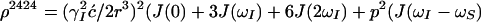 |
9 |
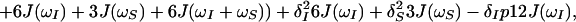 |
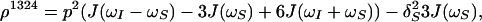 |
10 |
and
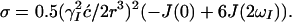 |
11 |
The time evolution of I13(1) is obtained by integration of Eq. 7 with the initial conditions Iz13(1)(0) = 0, Iz13(2)(0) = 0, Iz24(1)(0) = I0/2 cos(ωI24(1)tI) and Iz24(2)(0) = I0/2 cos(ωI24(2)t1), where the steady-state proton magnetization I0 was arbitrarily set to 2 when the following equations were written. The resulting expression for Iz13(1)(τm) can be simplified, for the limit of slow tumbling (ωIτc ≫ 1), with the initial buildup rate approximation (στm ≪ 1) (11), which results in a density submatrix describing the magnetization of the spin I(1) at time c:
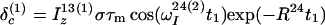 |
12 |
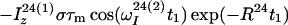 |
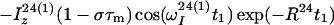 |
δc(2) can be obtained by permuting the labels (1) and (2) in Eq. 12. The condition στm ≪ 1 can readily be fulfilled. For example, with τc = 70 ns, r = 0.29 nm and τm = 50 ms, στm has the value 0.13.
After the time point c in Fig. 1b, the 1H longitudinal polarizations, Iz13 and Iz24, are converted to heteronuclear ZQ and DQ coherences, respectively, of which only the ZQ component is retained:
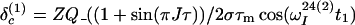 |
13 |
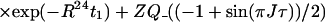 |
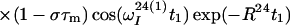 |
The first and second terms in Eq. 13 represent the crosspeaks and the diagonal peaks, respectively, as detected through the spin I(1). From the second term, it is seen that for the INEPT condition of τ = 1/(2J), the diagonal peak vanishes if J ≡ 1J(15N,1H).
Finally, the transfer function for the experimental scheme of Fig. 1b within the chosen approximations is
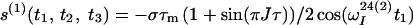 |
14 |
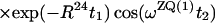 |
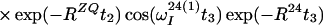 |
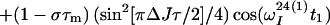 |
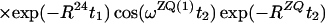 |
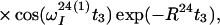 |
where ΔJ is the difference between the actual coupling constant, 1J(15N, 1H), and J. The first term in Eq. 14 represents the NOESY crosspeak connecting the chemical shifts of the 2 → 4 transitions of 1H(1) and 1H(2). Considering the small variation of 1J(15N,1H) observed along polypeptide chains in proteins, the residual intensity of the diagonal peaks should, on average for all residues, be less than 3% of the full intensity for any reasonably chosen set of experimental conditions. To facilitate practical applications, the experiment was designed so that residual diagonal peaks will always have opposite sign to the crosspeaks (Eq. 14). Loss in signal amplitude for kinetically labile 15N–1H groups by transfer of saturation from the solvent is minimized by aligning a large part of the water magnetization along +z before data acquisition (15). During τm radiation damping is used to accelerate the return of the phase-coherent water magnetization to thermal equilibrium (16, 17). For the remainder of the pulse sequence, the water magnetization is aligned along +z by using standard procedures (18, 19).
RESULTS
The experiments introduced in this paper are of interest primarily for studies of large proteins. All the illustrations shown in this section have therefore been recorded with a protein of molecular mass 110,000, the 7,8-dihydroneopterin aldolase (DHNA) from Staphylococcus aureus (20). For the present NMR experiments, this protein was labeled in the extent of 70% with 2H and uniformly with 15N, and it was dissolved in a mixed solvent of 95% 1H2O/5% 2H2O at 25°C and pH 6.0. DHNA is a homooctamer protein containing 8 subunits of 121 aa residues each. Because of the high symmetry, the complexity of the NMR spectra is manageable in spite of the large size.
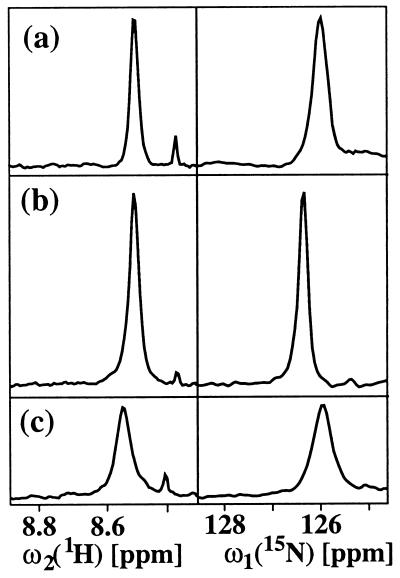
The ZQ coherence-based 2D ZQ-[15N,1H]-TROSY experiment of Fig. 1a is evaluated in Fig. 2 by comparison with the single-quantum coherence-based [15N,1H]-TROSY experiment of ref. 2 and a water-flip-back version of the conventional single-quantum coherence-based [15N,1H]-COSY experiment (9), which both also correlate the 1H and 15N resonances of 15N–1H moieties. The figure shows cross-sections along both the 15N and 1H chemical shift axes of a well-separated cross peak. In a and b, only the upfield component of the 1H doublet is observed, so that the peak positions along ω2 (1H) differ from that observed for the same 15N–1H fragment in the conventional [15N,1H]-COSY experiment by about 45 Hz (1). A similar shift is seen for the peak position of b along ω1 (15N) when compared with c. The ω1 (15N) positions in a and c are identical, because in a the 1H frequency was subtracted from the ZQ precession frequency (see Fig. 1 legend). As expected from the experimental procedures used (Fig. 1a and ref. 2; the spectra of Fig. 2a and 2b both show transverse relaxation-optimized narrow line shapes along ω2 (1H). Only the single-quantum coherence-based [15N,1H]-TROSY experiment (Fig. 2b) provides an appreciably narrower line width along the 15N dimension, so that this scheme also yields the highest signal amplitude. Overall, the presently introduced 2D ZQ-[15N,1H]-TROSY scheme (Fig. 1a) provides better sensitivity and resolution than the conventional [15N,1H]-COSY experiment (Fig. 2c), but the [15N,1H]-TROSY experiment of ref. 2 clearly gives the best result (Fig. 2b). As we will see in the following, the ZQ-based scheme is nonetheless a preferred element in 3D heteronuclear-resolved NOESY experiments.
Figure 2.
Comparison of three [15N,1H]-correlation experiments. (a) 2D ZQ-[15N,1H]-TROSY recorded with the scheme of Fig. 1a. (b) 2D single-quantum-[15N,1H]-TROSY recorded according to ref. 2. (c) Water-flip-back version of the conventional [15N,1H]-HSQC experiment (9). The experiments were performed at a 1H frequency of 750 MHz with a solution of [70% 2H,u15N] DHNA from S. aureus (20) in 95% 1H2O/5% 2H2O at 25°C and pH = 6.0. DHNA is a homooctamer protein of molecular mass 110,000; the concentration in the NMR sample was 0.4 mM as calculated per subunit. The measuring time and the experimental setup were identical for all three spectra. For a well-separated cross peak, cross sections along the 1H and 15N dimensions are shown.
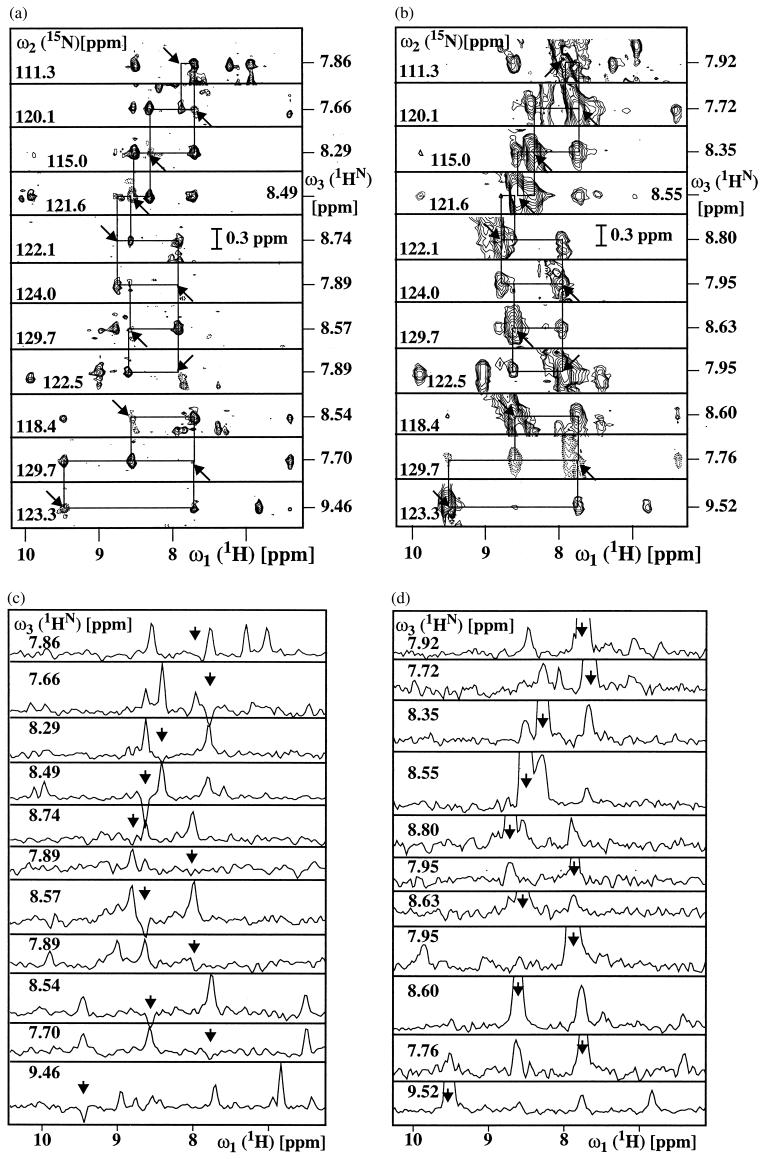
In 3D 15N-resolved experiments, the line width in the 15N dimension is usually, for practical reasons, determined by the relatively short maximal 15N chemical shift evolution time rather than by the effective transverse relaxation time. The narrow 15N line width obtained with single-quantum coherence-based [15N,1H]-TROSY (Fig. 2b) would therefore usually be masked by the experimental conditions used. As a consequence, the comparatively small number of polarization transfer delays, the absence of loss of magnetization during the 15N quadrature detection, and the individual single-transition-to-individual multiple-quantum coherence transfer scheme used (see Methods) make 2D ZQ-[15N,1H]-TROSY attractive as a building block in 3D 15N-resolved experiments such as 3D NOESY-[1H,15N,1H]-ZQ-TROSY (Fig. 1b). Fig. 3 a and c present a spectrum of the protein DHNA recorded with the experiment of Fig. 1b by a set of strips, i.e., 2D ω1 (1H)/ω3(1HN) contour plots approximately centered about the ω3(1HN) shift corresponding to the diagonal peak position of interest and taken at the chemical shift of the directly bound 15N nucleus in the ω2-dimension, and the corresponding one-dimensional cross sections taken along the ω1 dimension at the ω3(1HN) positions of the diagonal peaks identified by the arrows. As a reference for evaluation of the new experiment, a similar presentation of the same regions of a conventional 3D 15N-resolved [1H,1H]-NOESY spectrum (16) of DHNA is displayed in Fig. 3b and d. Along both 1H frequency axes, corresponding peak positions in the two spectra differ by approximately 45 Hz (1). Comparison of the two spectra shows that an important advantage of the 3D NOESY-[1H,15N,1H]-ZQ-TROSY experiment results from the fact that the diagonal peaks are either completely suppressed or have very small residual negative intensity (arrows in Fig. 3). The traces in Fig. 3c show that the scheme of Fig. 1b yields artifact-free suppression of the diagonal peaks, so that the crosspeaks near the diagonal become amenable for detailed analysis. The two spectra further demonstrate that significantly narrower crosspeak line widths were achieved when using TROSY, which in turn also yielded higher signal amplitudes.
Figure 3.
Comparison of corresponding spectral regions in a 3D NOESY-ZQ-[1H,15N,1H]-TROSY spectrum recorded with the scheme of Fig. 1b (a, c) and a conventional 3D 15N-resolved [1H,1H]-NOESY spectrum recorded according to ref. 16 (b, d). Both experiments were recorded with the sample of DHNA described in Fig. 2, using a Bruker DRX-750 spectrometer. Data size 128(t1)*30(t2)*1024(t3) complex points, t1max = 24 ms, t2max = 15 ms and t3max = 105 ms. Eight scans per increment were acquired, resulting in a measuring time of 34 h per spectrum. The spectra were processed with the program prosa (22). (a and b): Contour plots of [ω1(1H),ω3(1H)] strips. NOE connectivities are shown with thin horizontal and vertical lines. (c and d): Cross-sections taken along ω1(1H) at the ω3(1H) positions of the diagonal peaks in the strips a and b, respectively. In a–d, the positions of the diagonal peaks are indicated by arrows.
DISCUSSION
This paper describes an extension of the principle of transverse relaxation-optimization (1) in three ways. Firstly, crosscorrelation between 15N CSA and 1HN CSA is used to reduce transverse relaxation during the 15N evolution time. So far, cross-correlated relaxation was used in TROSY schemes only between 15N–1HN DD coupling and 15N CSA for optimization during the 15N evolution, between aromatic 13C–1H DD coupling and 13C CSA for optimization during the 13C evolution (4, 7) and between 15N–1HN DD coupling and 1HN CSA for optimization during 1HN acquisition (1), and in cross-correlated relaxation-enhanced polarization transfer (CRINEPT) between 15N–1H DD coupling and 15N CSA for optimization during magnetization transfers (21). Secondly, compared to previous TROSY implementations in 3D heteronuclear-resolved NOE experiments (7, 8), where transverse relaxation was optimized during the evolution of the heteronuclear spin and the directly observed 1H dimension, the experiment of Fig. 1b affords optimization in all three dimensions and thus yields narrower lines and concomitantly increased peak heights in both dimensions of the all-important [1H,1H]-NOESY planes. Thirdly, the scheme of Fig. 1b results in high-quality suppression of the diagonal peaks, which enables a quantitative analysis of NOESY crosspeaks in spectral regions that would otherwise be only hardly accessible.
The newly introduced 3D NOESY-[1H,15N,1H]-ZQ-TROSY experiment (Fig. 1b) uses two different types of the aforementioned cross-correlation effects. The cross-correlation between 15N–1HN DD coupling and 1HN CSA interactions is used to compensate transverse relaxation during the 1H chemical shift evolution period t1 and the 1HN signal acquisition period, t3 (1, 2). Thereby the magnitude of the DD/CSA cross-correlation effect depends strongly on the strength of the polarizing magnetic field, B0 (1, 3). Although very good results were obtained at the presently used field strength corresponding to a 1H resonance frequency of 750 MHz (Fig. 3), further improvement is anticipated at higher fields, with an optimum between 1.0 and 1.1 GHz (1, 3). The remote cross-correlation between 15N CSA and 1HN CSA within the 15N–1HN moieties is used to compensate transverse relaxation during the 15N chemical shift evolution period t2. The CSA/CSA cross-correlation effect is determined exclusively by the magnitudes and mutual orientations of the 1H and 15N CSA tensors and is therefore independent of the field strength B0, which is in contrast with DD/CSA cross-correlation. Because in proteins the principal axes of the axially symmetric CSA tensors for 15N and 1H are both nearly parallel to the 15N–1HN bond (12, 13), and the contributions of 1H CSA and 15N CSA to transverse relaxation are comparable in magnitude, the presently described use of CSA/CSA cross-correlation for TROSY-type relaxation compensation becomes possible. The use of CSA/CSA cross-correlation requires the generation of 15N–1HN ZQ coherence. Although the original single-quantum coherence-based scheme using DD/CSA cross-correlation (2) provides intrinsically better compensation than the ZQ-coherence-based scheme (Fig. 2), this potential advantage can hardly be exploited in 3D heteronuclear-resolved experiments (see Results), and we obtained good quality artifact-free data only with the use of the ZQ-coherence-based scheme. Further, the desired transfer function (Eq. 14) can be obtained with a lesser number of INEPT-type transfer steps than would be required with alternative single-quantum coherence-based schemes, and in this way signal loss during the magnetization transfer periods can be minimized. In addition, the resonances of the 15N–1H2 groups of Gln and Asn residues are not suppressed by the ZQ-TROSY correlation scheme, so that NOE crosspeaks with these groups are also observed in 3D NOESY-[1H,15N,1H]-ZQ-TROSY spectra.
In the experiment of Fig. 1b, the suppression of the strong diagonal peaks that are typically obtained with conventional NOESY schemes (6, 11, 16) may turn out to be of considerable interest in practice. A comparison of the two top slices in Fig. 3c with the corresponding slices in Fig. 3d shows that some 1HN–1HN cross-peaks can be resolved in the 3D NOESY-[1H,15N,1H]-ZQ-TROSY spectrum that are totally obscured by strong diagonal peaks in the corresponding conventional NOESY experiment. Diagonal peak suppression is achieved through selective saturation of the 1 → 3 transitions in all amide groups at the beginning of the experimental scheme of Fig. 1b and repolarization via NOE cross-relaxation with the magnetization of the 2 → 4 transitions. If both polarization transfer pathways of Eqs. 1 and 2 were recovered, two 3D 15N-resolved NOESY spectra could be obtained, with and without diagonal peaks, respectively.
Acknowledgments
Financial support was obtained from the Schweizerischer Nationalfonds (project 31.49047.96). We thank Dr. Hans Senn for a gift of 2H,15N-labeled DHNA and Mrs. M. Geier and Mrs. E. Ulrich for secretarial assistance.
ABBREVIATIONS
- 2D
two-dimensional
- 3D
three-dimensional
- DQ
double-quantum
- ZQ
zero-quantum
- DD
dipole–dipole
- CSA
chemical shift anisotropy
- TROSY
transverse relaxation-optimized spectroscopy
- NOE
nuclear Overhauser effect
- NOESY
NOE spectroscopy
- INEPT
insensitive nuclei enhanced by polarization transfer
- DHNA
7,8-dihydroneopterin aldolase
References
- 1.Pervushin K, Riek R, Wider G, Wüthrich K. Proc Natl Acad Sci USA. 1997;94:12366–12371. doi: 10.1073/pnas.94.23.12366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pervushin K V, Wider G, Wüthrich K. J Biomol NMR. 1998;12:345–348. doi: 10.1023/A:1008268930690. [DOI] [PubMed] [Google Scholar]
- 3.Wüthrich K. Nat Struct Biol. 1998;5:492–495. doi: 10.1038/728. [DOI] [PubMed] [Google Scholar]
- 4.Pervushin K, Riek R, Wider G, Wüthrich K. J Am Chem Soc. 1998;120:6394–6400. [Google Scholar]
- 5.Salzmann M, Pervushin K, Wider G, Senn H, Wüthrich K. Proc Natl Acad Sci USA. 1998;95:13585–13590. doi: 10.1073/pnas.95.23.13585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wüthrich K. NMR of Proteins and Nucleic Acids. New York: Wiley; 1986. [Google Scholar]
- 7.Brutscher B, Boisbouvier J, Pardi A, Marion D, Simorre J P. J Am Chem Soc. 1998;120:11845–11851. [Google Scholar]
- 8.Zhu G, Kong X, Sze K. J Biomol NMR. 1999;13:77–81. doi: 10.1023/A:1008398227519. [DOI] [PubMed] [Google Scholar]
- 9.Cavanagh J, Fairbrother W J, Palmer III A G, Skelton N J. Protein NMR Spectroscopy. Principles and Practice. New York: Academic; 1996. [Google Scholar]
- 10.Kumar P, Kumar A. J Magn Reson. 1996;A119:29–37. [Google Scholar]
- 11.Ernst R R, Bodenhausen G, Wokaun A. The Principles of Nuclear Magnetic Resonance in One and Two Dimensions. Oxford: Clarendon; 1987. [Google Scholar]
- 12.Hiyama Y, Niu C-H, Silverton J V, Bavoso A, Torchia D A. J Am Chem Soc. 1988;110:2378–2383. [Google Scholar]
- 13.Gerald R, Bernhard T, Haeberlen U, Rendell J, Opella S J. J Am Chem Soc. 1993;115:772–782. [Google Scholar]
- 14.Sørensen M D, Meissner A, Sørensen O W. J Biomol NMR. 1997;10:181–186. [Google Scholar]
- 15.Grzesiek S, Bax A. J Biomol NMR. 1993;3:627–631. doi: 10.1007/BF00198368. [DOI] [PubMed] [Google Scholar]
- 16.Talluri S, Wagner G. J Magn Reson. 1996;A112:200–205. doi: 10.1006/jmrb.1996.0132. [DOI] [PubMed] [Google Scholar]
- 17.Sobol A, Wider G, Iwai H, Wüthrich K. J Magn Reson. 1998;150:262–271. doi: 10.1006/jmre.1997.1287. [DOI] [PubMed] [Google Scholar]
- 18.Piotto M, Saudek V, Sklenar V J. J Biomol NMR. 1992;2:661–665. doi: 10.1007/BF02192855. [DOI] [PubMed] [Google Scholar]
- 19.Sklenar V. J Magn Reson. 1995;A114:132–134. [Google Scholar]
- 20.Hennig M, D’Arcy A, Hampele I C, Page M G P, Oefner C, Dale G. Nat Struct Biol. 1998;5:357–362. doi: 10.1038/nsb0598-357. [DOI] [PubMed] [Google Scholar]
- 21.Riek R, Wider G, Pervushin K, Wüthrich K. Proc Natl Acad Sci USA. 1999;96:4918–4923. doi: 10.1073/pnas.96.9.4918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Güntert P, Dötsch V, Wider G, Wüthrich K. J Biomol NMR. 1992;2:619–629. [Google Scholar]