Summary
In this manuscript we demonstrate that the combination of LIF removal and retinol or 4-oxoretinol treatment results in dose dependent, very large increases in CRABPI and CRABPII mRNA levels in embryonic stem cells. Transient transfection experiments demonstrate that the 7.8 kb CRABPI promoter does not contain the transcriptional regulatory elements responsible for this increase, though the CRABPI mRNA increase occurs at the transcriptional level. CRABPI and CRABPII do not bind 4-oxoretinol, however. This research is significant because it shows for the first time that LIF inhibits CRABPI and CRABPII gene expression in murine embryonic stem cells, and demonstrates a novel role for retinol in ES cells.
Murine embryonic stem (ES) cells cultured without leukemia inhibitory factor (LIF) or with retinoids differentiate and concomitantly metabolize retinol (vitamin A) to 4-oxoretinol. Our objective was to examine the effects of retinol or 4-oxoretinol on cellular retinoic acid binding protein (CRABP) I and II mRNA levels and retinol metabolism. ES cells were cultured with or without LIF, and with various doses of all-trans retinol, all-trans 4-oxoretinol, or all-trans retinoic acid (RA). In ES cells treated with retinol or 4-oxoretinol in the absence of LIF the CRABP-I (Crabp1, NM_013496; GI:7304974) and CRABP-II (Crabp2, NM_007759; GI:33469074) mRNA levels at 72 h were 66 ± 4 and 413 ± 6 fold higher, respectively, than the levels in control ES cells cultured without retinoids and in the presence of LIF. The increase in CRABPI mRNA occurred through an increase in CRABPI gene transcription. CRABPI protein was also increased by >50-fold in cells treated with retinol in the absence of LIF. However, [3H]4-oxoretinol does not bind to murine CRABPI or CRABPII. CYP26A1 mRNA levels and [3H]4-oxoretinol production from [3H]retinol increased in cells cultured without LIF and with exogenous retinoids. The enormous increases in CRABPI and II transcripts (~60 and 400 fold, respectively) in the absence of LIF may regulate aspects of the ES cell differentiation program in response to retinol.
Keywords: embryonic stem cells, retinoic acid binding proteins, LIF, retinol metabolism, cell differentiation, cytochrome P450
INTRODUCTION
The retinoids are a group of compounds with structures similar to that of vitamin A (retinol) which have diverse effects on several biological phenomena, including cell differentiation, embryonic development, vision, and reproduction. 4-Oxoretinol is a retinol metabolite produced by F9 teratocarcimona cells, MCF-7 and T47D human breast carcinoma cell lines, NB-4 human promyelocytic leukemia cells, and embryonic stem (ES) cells (Achkar et al., 1996; Chen et al., 1997; Faria et al., 1998; Lane et al., 1999). 4-Oxoretinol is capable of activating gene transcription via the retinoic acid receptors (RARs), but not the retinoid X receptors (RXRs) (Achkar et al., 1996). Exogenous 4-oxoretinol causes axial truncations in Xenopus embryos similar to those induced by RA (Achkar et al., 1996; Blumberg et al., 1996), and 4-oxoretinol is present at high levels in Xenopus oocytes (Blumberg et al., 1996). We have also shown that 4-oxoretinol is produced in concert with ES cell differentiation (Lane et al., 1999). 4-Oxoretinol is found in the serum after a dose of vitamin A (Penniston and Tanumihardjo, 2005).
Murine embryonic stem cell differentiation is regulated by the cytokine leukemia inhibitory factor (LIF). Physiologically, LIF is required for blastocyst implantation in mouse embryos (Bhatt et al., 1991; Nichols et al., 1996; Stewart et al., 1992). Leukemia inhibitory factor also maintains the pluripotency of cultured murine ES cells (Rathjen et al., 1990). Retinoids can override the LIF signal to cause differentiation of murine ES cells (Chen and Gudas, 1996; Tighe and Gudas, 2004). When LIF is removed from the culture medium, ES cells differentiate in the absence of pharmacological doses of exogenous retinoids, although there is retinol present in the serum (Lane et al., 1999). We have previously shown that the production of 4-oxoretinol from retinol occurs in ES cells during this differentiation process. Removal of LIF, in the presence of serum containing retinol, results in the induction of the cytochrome P540 enzyme, CYP26A1, which can catalyze the conversion of retinol to 4-oxoretinol (Lane et al., 1999). CYP26A1 also catalyzes the conversion of all trans-retinoic acid (RA) to 4-oxoRA (White et al., 1997). However, in ES cells retinol is not metabolized to RA during the differentiation process associated with LIF removal (Lane et al., 1999). Rather, 4-oxoretinol, generated from retinol, is the only RAR ligand detected in these differentiating ES cells (Lane et al., 1999). Therefore, we hypothesized that after LIF removal 4-oxoretinol is the retinoid which induces ES cell differentiation (Lane et al., 1999).
Proteins with a high binding affinity for retinoids exist in the cytosol, namely cellular retinol binding proteins (CRBPs) -I and II and cellular retinoic acid binding proteins (CRABPs) I and II. These proteins bind retinol and RA, respectively. Cellular retinol binding proteins function in part to direct the metabolism of retinol into retinyl esters (Ong et al., 1994). The functions of the CRABPs are not resolved, and CRABPI may function differently from CRABPII. Cellular retinoic acid binding proteins I and II share approximately 75% amino acid homology and are the same molecular weight (Giguere, 1994). There is evidence that CRABPs can perform each of the following three functions. First, because CRABPs bind to RA they may act to lower active intracellular RA concentrations, protecting cells from the differentiation-inducing effects of RA. Second, CRABPs may lower active, intracellular RA concentrations by modulating metabolism of RA to 4-oxoRA via CYP26A1. The third function of CRABPs may be as shuttles to move RA through the aqueous cytosol to the nucleus, where RA can interact with RARs and initiate gene transcription.
In support of the first two functions, overexpression of CRABPI in F9 teratocarcinoma cells increased the rate of 4-oxoRA formation and decreased the sensitivity of these cells to RA-induced differentiation (Boylan and Gudas, 1991; Boylan and Gudas, 1992). Also, holo-CRABPI has been shown to lower active intracellular RA concentrations by enhancing RA metabolism to 3, 4-didehydro, 4-hydroxyl, 4-oxo-, 16-hydroxy-4-oxo and 18-hydroxy-RA via members of the CYP26 family of cytochrome P450 enzymes (Fiorella et al., 1993; Napoli et al., 1991). Thus, CRABPI appears to function to decrease cellular responses to RA by catalyzing its metabolism. Cellular retinoic acid binding protein I is expressed in a variety of tissues. CRABPI is excluded from nuclei in spermatogonia (Zheng et al., 1996). CRABPI is associated with mitochondria in some cell types (Ruff and Ong, 2000).
The third potential function of CRABP is indicated by the presence of CRABP II in cell nuclei, where the presumed function is to deliver RA to RARs (Cornic et al.; Dong et al., 1999; Noy, 2000; Sessler and Noy, 2005). Kinetic studies of the movement of RA to RARs showed that CRABPI is a passive vehicle, binding and releasing its ligand depending on concentration gradients. In contrast, CRABPII was shown to deliver RA to RARs in a direct collisional process (Dong et al., 1999). In support of this model, overexpression of CRABPII, but not CRABPI, stimulated the transcription of a reporter gene controlled by an upstream RARE (Delva et al., 1999; Dong et al., 1999). Also, CRABPII is often expressed in cells that synthesize high levels of RA, where CRABPII was shown to shuttle RA to RARs and affect gene transcription (Delva et al., 1999; Dong et al., 1999). Therefore, in contrast to CRABPI, CRABPII increases RA-mediated gene transcription, sensitizing at least some cell types to the effects of RA. However, CRABPII is excluded from the nuclei of cells in the ovary (Bucco et al., 1995) and uterus (Bucco et al., 1996). The CRABPII gene is differentially expressed during development due to the presence of an RARE in its 5′ regulatory region (Astrom et al., 1994; Durand et al., 1992). The CRABPII gene is also regulated post-transcriptionally in F9 cells (MacGregor et al., 1992).
In the present study we show that CRABPI and II transcripts are greatly increased by either treatment with exogenous retinol or 4-oxoretinol in ES cells induced to differentiate by the removal of LIF. We also demonstrate that although 4-oxoretinol greatly increases CRABPI and II mRNA levels in ES cells, 4-oxoretinol does not bind to either CRABPI or CRABPII.
MATERIALS AND METHODS
Cell Culture
Murine CCE-WT, AB1, or J1 embryonic stem cell lines were grown in monolayer culture and maintained as described (Lane et al., 1999). At the start of each experiment cells were trypsinized and seeded with or without LIF at the appropriate density to provide 70–80 % confluence at the time of harvest (approximately 5 × 104 cells/60 mm dish for the 96 h time point). New LIF was added to the “+LIF” cultures every 24 h. The retinoids all trans-RA, retinol, or 4-oxoretinol were added 24 h after plating. This is referred to as “time zero”. All retinoids were dissolved in 100% ethanol. The 4-oxoretinol was synthesized as described (Achkar et al., 1996) and stored under nitrogen at −70°C prior to use. All retinoid manipulations were performed under subdued light.
Transient Transfections
Transient transfections of murine ES cells were performed by the calcium phosphate method according to (Means et al., 2000). To each plate of 5 × 105 cells, 10μg of the CRABP 7.8 exon3 βgal plasmid (Means and Gudas, 1997), and 2μg of the Renilla luciferase plasmid were added. At the appropriate times cells were harvested and assayed for βgal activity. The βgal activity was normalized to the luciferase activity in each sample to control for variations in transfection efficiency. The transfection efficiency was ~10–15% in these experiments.
RNA Isolation and Northern Blot Analysis
Total RNA was isolated from cells cultured with or without LIF and with or without 1 μM all trans-RA, retinol, and 4-oxoretinol at 24, 72 and 96 h after retinoid addition. (For the 72 and 96 h samples, retinoids and LIF were re-added at 48 h). To mimic the treatment received by the cells used in the [3H]-retinol metabolism assays, 16 h before harvest the culture medium was removed from all plates and replaced with medium containing 5% FCS and 50 nM unlabeled all trans-retinol. Total RNA was isolated using RNA Stat-60 (Tel-Test, Friendswood, TX), as described previously (Li et al., 2004). Northern blot analysis was performed and quantitated as described previously (Lane et al., 1999).
Western Analysis
For Western analyses cells were plated at 1 × 105 cells per plate and cultured for various times under the conditions indicated in the figure legend. Cells were harvested directly into final sample buffer, after rinsing the plates twice with ice-cold phosphate buffered saline. Protein assays were performed, and 40 μg protein/lane was loaded on a 10% acrylamide gel. After electrophoresis, the proteins were transferred to Bio-Rad Trans-Blot® Pure Nitrocellulose Membrane (Bio-Rad Laboratories, Hercules, CA) using 100V for 1 h at 4°C. The membrane was washed with phosphate buffered saline with 0.1% Tween-20 (cat#P9416, Sigma-Aldrich, St. Louis, MO), and Ponceau S staining was performed (10 ml used, cat#P7170, Sigma-Aldrich, St. Louis, MO). Membranes were blocked for 1 h using PBS + Tween + 5% Blotto (cat#SC-2325, Santa Cruz Biotechnology, Santa Cruz, CA), and incubated for 24 h with shaking at 4°C with the primary antibody to CRABPI at a 1:500 dilution (cat#MA3-813, Affinity BioReagents, Golden, CO). The secondary antibody was added at a 1:5000 dilution (donkey anti-mouse IgG-HRP, cat#SC-2318, Santa Cruz Biotechnology, Santa Cruz, CA) for 1 h at room temperature. After washing in phosphate buffered saline plus Tween, an ECL kit was used to develop the signal (Pierce SuperSignal® West Pico Chemiluminescent Substrate, cat#34080, Pierce Biotechnology, Rockford, IL). The blots were quantitated using a PhosphorImager and ImageQuant software.
Retinoid Metabolism, Extraction, and HPLC Analysis
After 24, 48, 72, or 96 h, the culture medium was removed from the ES cells and radiolabeling medium, consisting of DME plus 5% FCS and 50 nM [3H]retinol (specific activity, 35.2 μCi/nmol; 1 Ci = 37 GBq) was added. A control of radiolabeling medium without cells was also included. Cell and media samples (0.5 ml) were harvested after 16 h as described previously (Guo and Gudas, 1998). All samples were frozen at −70°C until extraction. Cell number for each treatment and time point was determined from duplicate dishes counted during the incubation period.
Retinoids were extracted as described previously (McClean et al., 1982) and separated using a Waters Millennium System as described (Achkar et al., 1996; Lane et al., 1999). Recovery of retinoids from the samples was assessed by the addition of nonradiolabeled retinoid standards at the beginning of the extraction process; recovery was very similar in all samples. The identification of the retinoids was determined by coelution of the radiolabeled samples with the known retinoid standards included in the samples and by UV spectra. To assess retinol metabolism quantitatively, each peak (retinol) or group of peaks (polar metabolites) was integrated and the total retinoid concentration was calculated as described (Guo and Gudas, 1998). Significant differences in [3H]retinol metabolism were determined by student’s t-test comparing control cells cultured in the presence of LIF will each other treatment.
[3H] 4-Oxoretinol synthesis and CRABP binding assays
[15-3H]-4-oxoretinol was prepared by reduction of 4-oxoretinaldehyde with sodium borotride ([3H]NaBH4 dissolved in 0.01 N NaOH) following the literature procedure (Boehm et al., 1990). [15-3H]-4-oxoretinol was analyzed for purity and specific activity by HPLC. The peak corresponding to [3H] 4-oxoretinol, determined using a Packard FloOne scintillation counter in line with a PDA detector in the HPLC system, was purified by HPLC and used to determine the ability of [3H] 4-oxoretinol to bind CRABPI or CRABPII.
The ability of [3H] 4-oxoretinol to bind to CRABPI and II was measured as described previously (Grippo and Gudas, 1987). COS-7 cells were plated at a density of 2 × 106 cells per 100 mm dish. Twenty-four hours later the cells were transiently transfected by the calcium phosphate method with 20 μg empty pSG5 vector, pSG5 containing cDNA encoding full-length murine CRABPI, or pSG5 containing cDNA encoding full-length murine CRABPII. Cells were harvested 48 h after transfection and the cytosolic fraction isolated by differential centrifugation as described previously (Grippo and Gudas, 1987). Cells were scraped into ice cold PBS and centrifuged at 600 × g for 5 min at 4°C. Following centrifugation, the cells were disrupted by Dounce homogenation (40 strokes) in one packed cell volume of 10 mM Tris buffer, pH 7.4, containing 7 nM 2-mercaptoethanol. Following homogenization, the cell extracts were centrifuged at 100,000 × g for 60 min at 4°C. A clear supernatant, the cytosol, was removed from beneath the fatty layer. This served as the source of CRABPI or II for the binding assays. The COS-7 cytosols prepared using this method contained between 3 and 9 mg/ml of protein, as determined using the BioRad protein assay (Hercules, CA). Cytosolic extracts were stored at −70°C until use. Duplicate plates were harvested for confirmation of CRABPI and II mRNA content to confirm successful transfection.
To determine the amount of [3H] 4-oxoretinol bound to CRABPI or CRABPII, 150 μg of cytosolic protein from each transfection was incubated with 2 mM dithiothreitol, 10 mM Tris buffer, pH 7.4, and 10–100 nM [3H] 4-oxoretinol or [3H] retinoic acid (positive control) in a total volume of 50 μl for at least 5 h at 4°C. Excess [3H] 4-oxoretinol or [3H] retinoic acid was removed by extraction with 100 μl of a suspension of 2.5% activated charcoal and 0.2% dextran sulfate. Non-specific [3H] 4-oxoretinol and [3H] retinoic acid binding was determined by incubation in the presence of a 100-fold molar excess of unlabeled 4-oxoretinol or retinoic acid, respectively. Following removal of the charcoal by centrifugation, 40 μl of clear supernatant was counted by liquid scintillation. Duplicate samples were assayed for both total binding (binding detected in the absence of excess unlabeled 4-oxoretinol or retinoic acid) and non-specific binding (binding detected in the presence of 100-fold or 1,000-fold molar excess unlabeled 4-oxoretinol or retinoic acid). Specific [3H] 4-oxoretinol or [3H] retinoic acid binding was determined by subtracting nonspecific from total binding.
RESULTS
Time Course of Increases in CRABPI and II mRNA Levels in ES Cells Cultured in the Presence of Exogenous Retinol or 4-Oxoretinol
To determine if exogenous retinol or 4-oxoretinol can affect CRABPI and II mRNA levels, Northern blot analysis was performed on ES cells treated with or without LIF and with or without various concentrations of retinol, 4-oxoretinol, or RA for various times (24, 72 or 96 h). CRABPI and II transcripts were absent in vehicle, control cells regardless of the presence or absence of LIF (Figure 1A–C). In the presence of LIF, CRABPI and II mRNAs were induced only slightly by treatment with 1 M RA for 72 and 96 h (Fig. 1A–C). In contrast, in the absence of LIF CRABPI and II mRNAs were greatly increased in response to all retinoid treatments (Fig. 1A–C).
Figure 1. Time course and dose-response analyses of ES cells cultured ± LIF and ± RA, retinol or 4-oxoretinol.
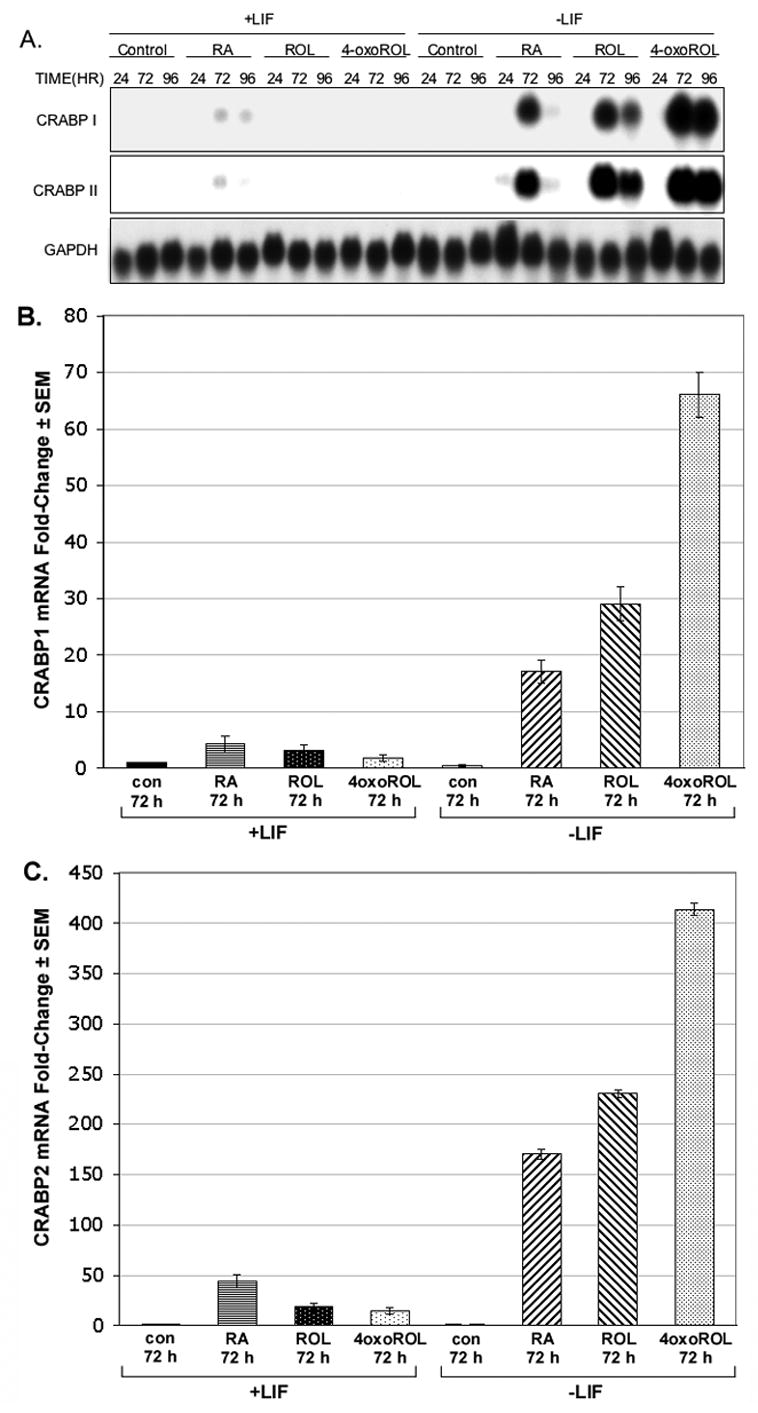
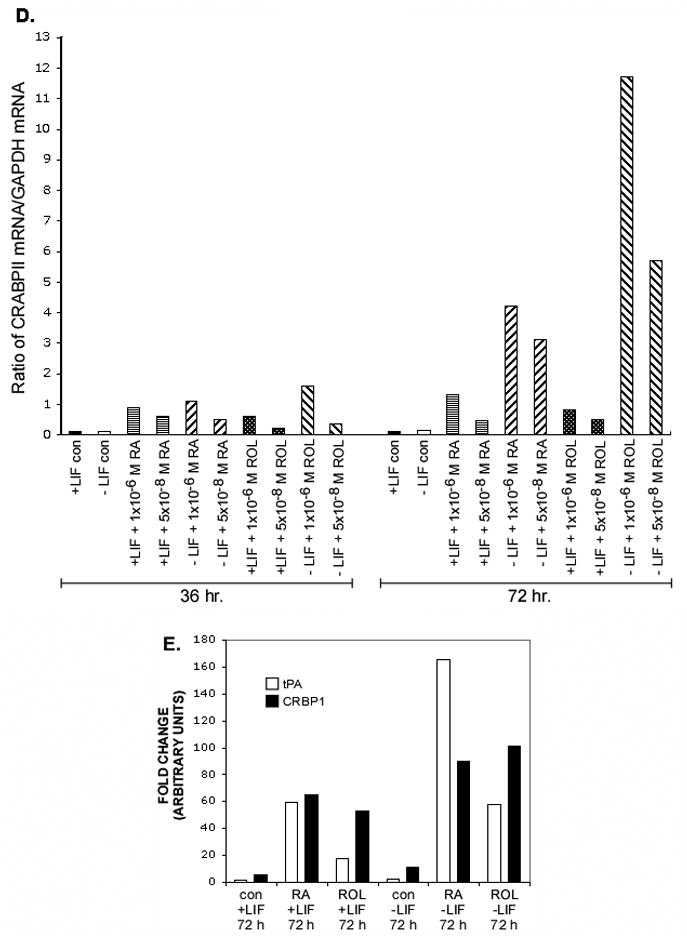
Northern blot analysis of CRABPI, CRABPII, and GAPDH mRNA levels in CCE-WT cells (A). CCE-WT cells were cultured for 24, 72, and 96 h plus or minus 1000 U/ml of LIF in the presence of 1 μM retinoic acid (RA), 1 μM retinol (ROL), or 1 μM 4-oxoretinol (4-oxoROL). Fresh LIF was added to the “+LIF” cultures daily. Northern blots were quantitated using a PhosophorImager. CRABPI or CRABPII mRNA levels are expressed in arbitrary units, normalized to the GAPDH mRNA level. Northern analyses were performed seven times with seven different RNA preparations from ES cells with very similar results. The quantitation of several blots like the one shown in (A) for CRABPI (B), and CRABPII (C) mRNA is shown. The CRABPI or II transcript levels were normalized to GAPDH mRNA and then fold change was plotted using the control, +LIF 24 hr. sample set as 1 (means ± SEM). A dose-response experiment showing the effect of both 1 × 10−6 M and 5 × 10−8 M retinoids on CRABPII mRNA levels is shown (D). This dose-response experiment was repeated. The (D) y-axis, arbitrary units. Quantitative data for tissue plasminogen activator (tPA) and cellular retinol binding protein 1 (CRBP1) mRNA for comparison (E).
The patterns of expression for both CRABPI and II were similar. The highest levels of CRABPI and CRABPII mRNA were seen after 72 h of treatment with 4-oxoretinol in the absence of LIF; CRABPI mRNA levels were increased by 17 ± 2 (RA), 29 ± 3 (retinol), and 66 ± 4 (4-oxoretinol) fold over control, +LIF treated ES cells, while the levels of CRABPII mRNA were 170 ± 5 (RA), 230 ± 4 (retinol), and 413 ± 6 (4-oxoretinol) fold higher than the control, +LIF levels (Fig. 1A–C). The CRABPII transcripts increased in response to increasing doses of the various retinoids (Northern blot, quantitation only, Fig. 1D).
Other RA regulated genes, such as CRBP1, tissue plasminogen activator, and laminin B1 ((Vasios et al., 1989) not shown) did not show such dramatic increases in transcript levels in response to retinol and LIF removal (Fig. 1E). The levels of RARγ transcripts did not change by more than two-fold under the same treatment conditions. In the absence of LIF, RARγ transcript levels were 3-fold lower than in the presence of LIF (data not shown). While only the data from the CCE ES line are shown in Figure 1, the ES lines AB1 and J1 were also tested and exhibited very similar, large increases in CRABPI and CRABPII mRNA levels after 4-oxoretinol addition (not shown), indicating that this is a general characteristic of murine ES cells.
The Increase in CRABPI mRNA Levels Occurs Through Increased Transcription of the CRABPI Gene
CRABPII gene transcription is regulated by an RARE in the promoter of the CRABPII gene (Durand et al., 1992). As we have shown previously, 4-oxoretinol can induce gene transcription via an RARE (Achkar et al., 1996). However, the CRABPI gene has no known RARE (Means and Gudas, 1997). Therefore, to determine whether the large increases in CRABPI mRNA levels following the removal of LIF and the addition of retinol or 4-oxoretinol to the ES cells occurred via an increase in the rate of transcription of the CRABPI gene, we performed an experiment in which cells were cultured with or without retinol and with or without actinomycin D, an inhibitor of RNA synthesis. Actinomycin D blocked the large increase in CRABPI mRNA that occurs between 48 and 80 h after retinol addition, indicating that this increase in CRABPI mRNA is mediated at least in part by an increase in transcriptional activation (Fig. 2A). The late time course (between 48–72 h) of CRABPI and CRABPII mRNA increases after retinol treatment in the absence of LIF suggests that this is not a primary response, but rather a secondary transcriptional response dependent on the earlier activation or repression of genes in response to LIF removal and retinol treatment.
Figure 2. A. Northern blot analysis of CRABPI and CRABPII mRNA in the presence of actinomycin D.
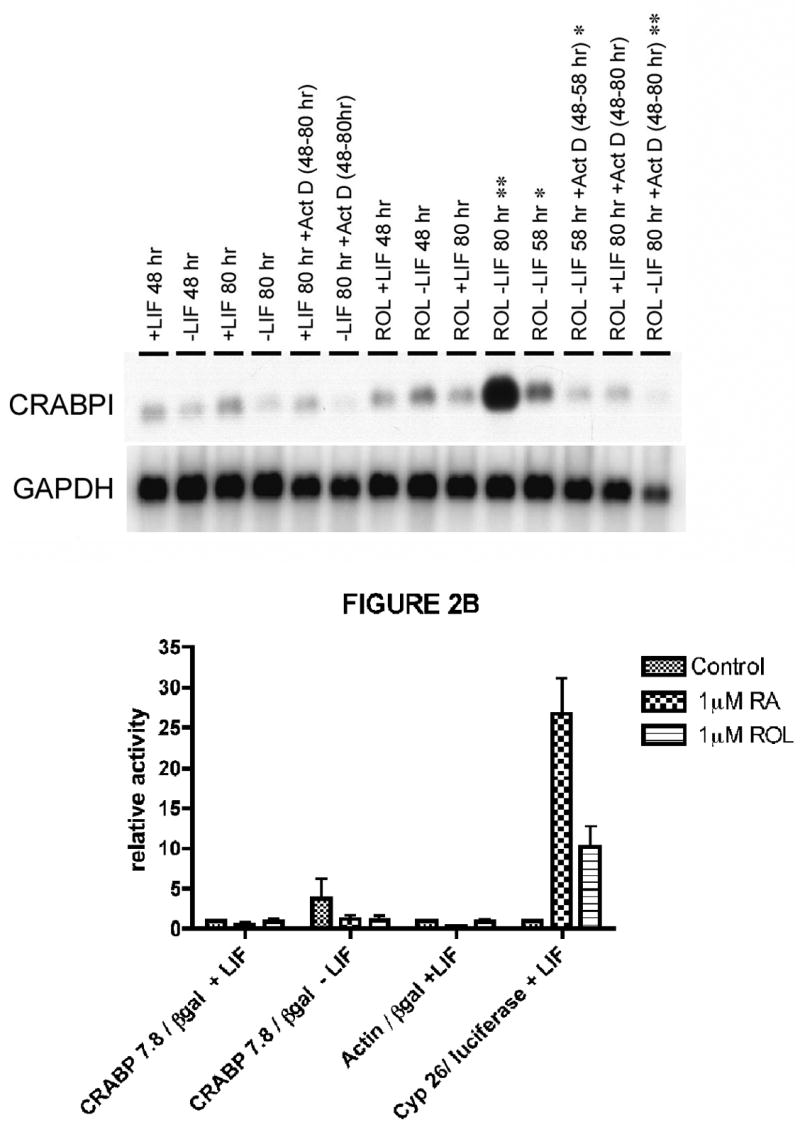
ES cells were cultured as in Fig. 1, except that actinomycin D (2 μg/ml) was added to some dishes at 48 h. Some cell samples were harvested at 48 hr., some at 58 h, and remainder at 80 h. The Northern blot was hybridized to a CRABPI cDNA probe, and then to a GAPDH cDNA. The hybridization data were quantitated as in Fig. 1. This experiment was performed three times, starting with fresh cells, with very similar results. One experiment is shown. The lanes to be compared are + and − actinomycin D at 58 h (Θ), and at 80 h (ΘΘ). (p value < 0.05). B. Transient transfection of J1 murine ES cells with the murine CRABPI promoter driving expression of the β-galactosidase reporter gene. On day one, cells were plated at 5 × 105 cells per dish in ES medium with or without LIF, as appropriate. Thirty-six hours later, the cells were transiently transfected using the calcium phosphate method. Sixteen hours later, the dishes were washed and medium was added, again with or without LIF as appropriate. Four hours later, 1 μM retinol or 1 μM retinoic acid was added to the appropriate plates. Forty-eight hours later, the cells were harvested, and the protein extracts were assayed for β-galactosidase activity and luciferase activity. The β-galactosidase activity, and the firefly luciferase activity driven by the CYP26 promoter were assayed, and the activities were normalized for transfection efficiency to the Renilla luciferase plasmid which was also transiently transfected in each dish. The CRABP 7.8 exon3/β-galactosidase plasmid was employed at 10 μg DNA per dish, and the Renilla luciferase plasmid was added to each dish at 2 μg of DNA per dish. The actin/β-galactosidase plasmid (Vasios et al., 1989) was added at 10 μg DNA per dish. The murine CYP26 promoter/firefly luciferase reporter vector was added at 10 μg DNA per dish. The β-galactosidase was measured according to (Vasios et al., 1989). The firefly luciferase and the Renilla luciferase activities were assayed according to the manufacturer’s instructions using a luminometer (Turner Designs, Sunnyvale, CA). The experiments were performed three independent times. The mean values and SEM error bars as indicated were calculated using the GraphPad Prism software program.
To delineate the promoter region involved we then performed transient transfection experiments using a construct which contained 7.8 kb of the murine CRABP1 promoter driving the expression of the β-galactosidase (lacZ) reporter gene. This is the same plasmid we used previously to generate transgenic mice which expressed lacZ in the same regions of the developing embryo in which the endogenous CRABPI gene is expressed (Means and Gudas, 1997). We did not observe an increase in the β-galactosidase activity driven by the CRABPI promoter in embryonic stem cells [AB1, CCE, or J1 (shown)] cultured either plus or minus LIF in the presence of 1 μM RA or 1 μM retinol versus those cultured in the presence or absence of LIF without exogenous retinoids (Fig. 2B).
As a positive control for β-galactosidase expression, we also used a plasmid in which a truncated β-actin promoter drives expression of the β-galactosidase reporter gene. As expected, no change in the activity of β-galactosidase driven by the actin promoter was observed in cells cultured +LIF minus retinol or RA versus those cultured +LIF plus retinol or RA (Fig. 2B). As a positive control for the effects of retinoids on transcription of retinoid responsive genes, we employed the murine CYP2A1 promoter driving the expression of the luciferase reporter gene; 9-fold and 26-fold increases in luciferase activity were observed in the retinol and RA treated cells, respectively, versus the control ES cells.
Thus, we conclude that the increased levels of CRABPI transcripts observed following culture of ES cells in retinol in the absence of LIF result from increased transcriptional activation of the CRABPI gene, and that the regulatory DNA elements involved in this activation are not present in the 7.8 kb promoter we tested in our transient transfection assays.
CRABPI Protein Levels are Elevated in ES Cells Cultured Without LIF in the Presence of Retinol
We performed Western blot analyses to determine whether the large increases in CRABPI transcripts in cells cultured in the presence of retinol in the absence of LIF corresponded to increases in CRABPI protein. There was over a fifty-fold increase in CRABPI protein level in ES cells cultured in the presence of retinol minus LIF over the level in the presence of LIF only (control) (Fig. 3A), and this large increase was not seen in cells cultured in the presence of all-trans RA minus LIF (Fig. 3A). Ponceau S staining and actin (Fig. 3B) showed that the protein loaded per lane was similar. Thus, the increase in CRABPI transcripts was associated with a corresponding increase in CRABPI protein, although the time course kinetics of the increase in the CRABPI protein were delayed relative to the CRABPI transcript increases.
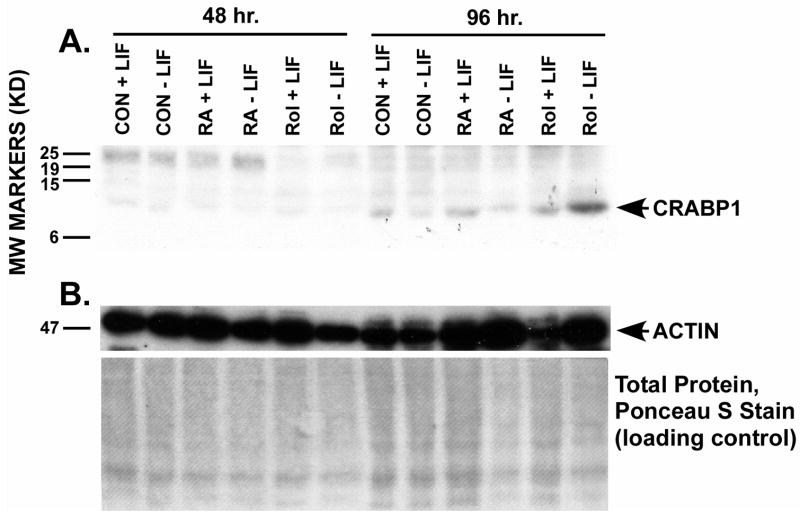
Figure 3. Western analysis of CRABPI protein levels.
ES cells were cultured with or without 1 μM all-trans RA, with or without 1 μM all-trans retinol, and in the absence or presence of LIF for 48 or 96 h (We did not test 4-oxoretinol in this experiment because of the limited supply of this drug.) Cells were harvested and Western analyses were performed as described in the Materials and Methods section. The CRABPI specific signal is indicated (panel A). The signal was greatest (quantitated by PhosphorImager, arbitrary units) when ES cells were cultured for 96 h in retinol minus LIF. Control, +LIF 96 hr, 3 arbitrary units; −LIF, 96 hr, 1; +RA, +LIF, 96 hr, 3; +RA, −LIF, 96 hr, 1; +Rol, +LIF, 96 hr, 4: +Rol, −LIF, 96 hr, 53. The loading of protein per lane was assessed by Ponceau S staining of total proteins on the blot and by actin (panel B) after electrophoretic protein transfer from the gel to the membrane. This experiment was repeated, starting with fresh ES cell cultures, with identical results. One experiment is shown here.
Retinol Metabolism to 4-Oxoretinol is Correlated with the Increases in CRABPI and CRABPII mRNA Levels
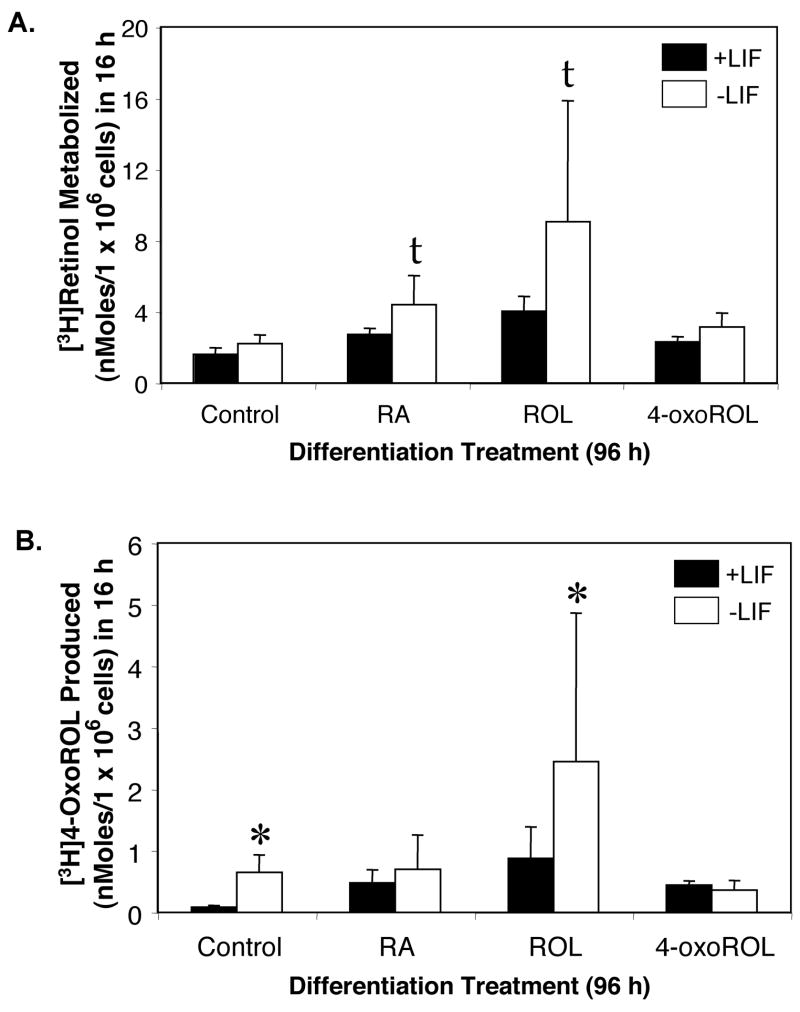
To examine the relationships among retinol metabolism, endogenous 4-oxoretinol production from retinol, and CRABPI and CRABPII mRNA levels, ES cells cultured with LIF were harvested 16 h after the addition of 50 nM [3H]retinol. This time point was chosen to ensure the maximum production of 4-oxoretinol (Lane et al., 1999). CCE-WT ES (Fig. 4) and AB1 ES cells (not shown) metabolized [3H]retinol to [3H]4-oxoretinol. Culture with RA (p = 0.09) or retinol (p = 0.06) in the presence of LIF also tended to increase [3H]retinol metabolism when compared to control +LIF cells. Culture in the absence of LIF tended to increase the amount of [3H]retinol metabolized, when compared to control +LIF cells, for cells treated with RA (p = 0.18) and 4-oxoretinol (p = 0.15) (Figure 4A). The absence of LIF tended to increase polar metabolite production in vehicle control cells when compared to vehicle control cells cultured with LIF (p = 0.11) (Figure 4B). Culture with RA (p = 0.13) and LIF also tended to increase polar metabolite production when compared to control +LIF cells. Culture in the absence of LIF and the presence of 4-oxoretinol also tended (p = 0.16) to increased the production of [3H]4-oxoretinol from [3H]retinol, when compared to control +LIF cells. Culture in the presence of LIF and 4-oxo-retinol significantly increased polar metabolite production but due to the small amount of increase, this may not be biologically significant. In general, these data indicate that ES cells cultured in the absence of LIF and the presence of exogenous retinoids (retinol or RA) metabolize more [3H]retinol to [3H]4-oxoretinol than ES cells cultured in the presence of LIF. Similarly, higher CRABPI and CRABPII transcript levels are observed in cells cultured in the presence of these retinoids (RA, retinol or 4-oxoretinol) minus LIF (Fig. 1).
Figure 4. A,B. [3H]Retinol metabolism by CCE-WT ES cells.
CCE-WT ES cells were cultured in DMEM supplemented with 10% FCS (Lane et al., 1999). Cells were cultured for 96 h with or without 1,000 units/ml of LIF (Life Technologies) and with no or 1 μM of non-radiolabeled RA, retinol or 4-oxoretinol. LIF was added every 24 h to the +LIF samples. After 96 h, the medium was removed and new medium, containing 5% FCS and 50 nM [3H]retinol, was added for 16 h [The concentration of nonradioactive retinol in this 5% FCS medium was ~ 50 nM (X. Guo and L. Gudas, unpublished data)]. The retinoids were extracted and quantitated by HPLC as decribed in the Methods section. Total amounts of [3H]retinol metabolized (A) or [3H]4-oxoretinol produced (B) by CCE-WT cells first cultured for 96 h with (■) or without (□) LIF and with or without 1 μM RA, retinol, or 4-oxoretinol to induce differentiation. Fifty nM [3H]retinol was then added for 16 h. Data were normalized both for recovery of [3H] retinoids and for cell number in each sample. Data shown are the means ± SEM of three experiments. Note the y-axis scale differences between (A) and (B). (Θ, p value < 0.05; t, p value between 0.06 and 0.10, versus +LIF vehicle control).
Exogenous Retinol Induces Retinoid-Responsive Gene Expression, Including CYP26A1
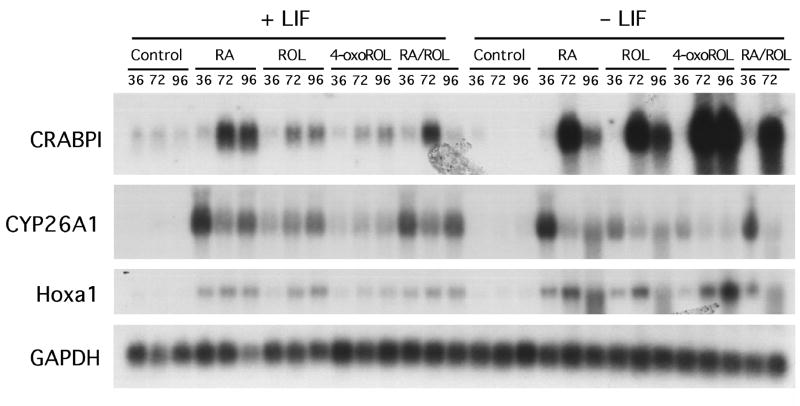
We hypothesize that endogenous 4-oxoretinol, derived from the metabolism of retinol by the cells, induces the differentiation of ES cells cultured in the absence of LIF. Therefore, we performed Northern analysis to determine the effect of exogenous retinol on the expression of additional retinoid-responsive genes associated with retinoid signaling and cellular differentiation. After 96 h, both retinol and 4-oxoretinol increased Hoxa1 transcript levels (Langston and Gudas, 1992) in CCE-WT cells when compared to control cells (Fig. 5). Additionally, the levels of Hoxa1 transcripts were greater in retinol and 4-oxoretinol treated cells cultured in the absence of LIF than in retinol and 4-oxoretinol treated cells cultured in the presence of LIF (Fig. 5). Treatment with 4-oxoretinol for 96 h resulted in the greatest increases in HoxA1 mRNA levels relative to control cells (cultured in the presence of LIF) (Fig. 5). These data indicate that both retinol and 4-oxoretinol treatment of ES cells can increase the transcript levels of Hoxa1, a key “early response” gene which mediates aspects of the differentiation response in ES cells (Martinez-Ceballos et al., 2005). CYP26A1 mRNA was induced by all three exogenously added retinoids, retinoic acid, retinol, and 4-oxoretinol (Fig. 5). CYP26A1 mRNA levels were higher at 36 h than at later time points and did not vary with LIF levels (Fig. 5).
Figure 5. Northern blot analysis of CRABPI, CYP26A1, Hoxa1, and GAPDH mRNA levels in CCE-WT cells.
CCE-WT cells were cultured for 36, 72, and 96 h plus or minus 1000 U/ml of LIF in the presence of exogenously added 1 μM RA, 1 μM retinol (ROL), or 1 μM 4-oxoretinol (4-oxoROL). RA/ROL indicates treatment with both 0.5 μM RA and 0.5 μM ROL. Fresh LIF was added to the “+LIF” cultures daily. Northern blots were quantitated using a PhosophorImager. Northern analyses were performed seven times with seven different RNA preparations from ES cells with very similar results (within 10%). One representative blot of the CRABPI, CYP26A1, Hoxa1, and GAPDH mRNA levels is shown.
4-Oxoretinol Does Not Bind to CRABPs
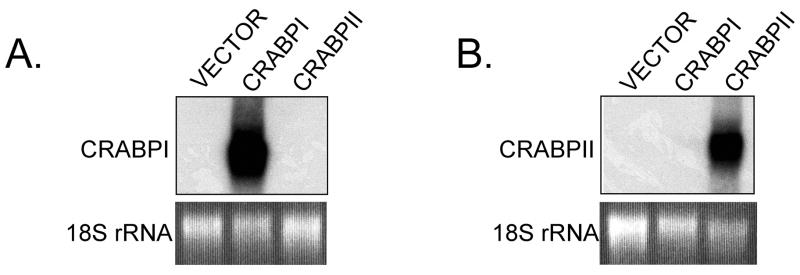
Because 4-oxoretinol increased CRABPI and CRABPII mRNA levels, we wanted to determine if 4-oxoretinol could bind to either of these proteins. To examine 4-oxoretinol binding, COS7 cells were transiently transfected with plasmids containing either full-length murine CRABPI or CRABPII cDNA. COS7 cells were chosen because they do not express detectable endogenous CRABPI or CRABPII. Additionally, COS7 cells are mammalian and therefore, the posttranslational modifications of the CRABPI or CRABPII proteins should be identical to those in ES cells. Transfection of COS7 cells with these constructs resulted in very high levels of CRABPI and CRABPII mRNA, respectively (Fig. 6). Extracts from cells transfected with CRABPI did not express CRABPII mRNA, and vise versa. The short, 4 h, exposure time of the Northern blot to film reflects the high levels of expression of the CRABP transcripts in the COS cells (Fig. 6).
Figure 6. Cellular retinoic acid binding protein I and II mRNAs in transiently transfected COS 7 cells.
COS 7 cells were transiently transfected with pSG5 vector with either the CRABPI or CRABPII full-length cDNA, or pSG5 vector alone. Forty-eight hours later duplicate samples were harvested for protein isolation and Northern blot analysis. Duplicate RNA samples were hybridized separately with full-length CRABPI (A) or CRABPII (B) cDNA probes. 18S rRNA was included as a loading control. Northern blots were exposed to film for 4 h.
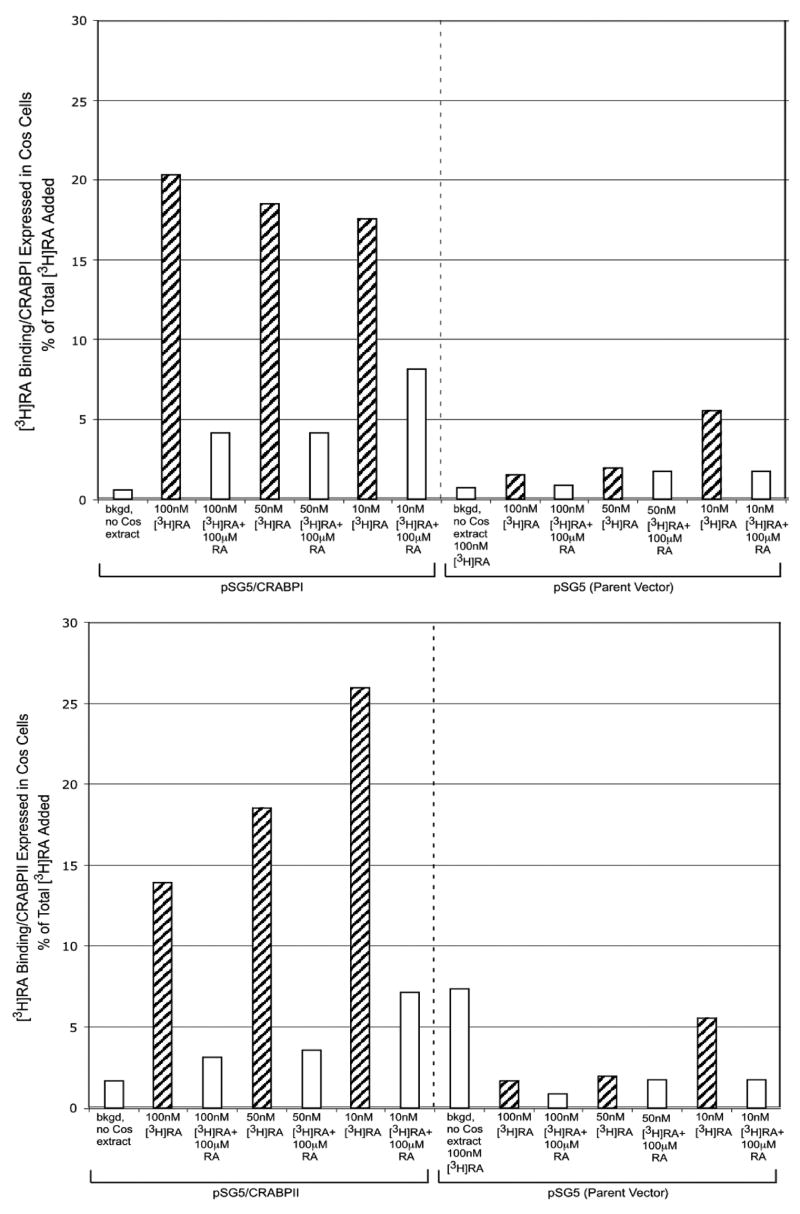
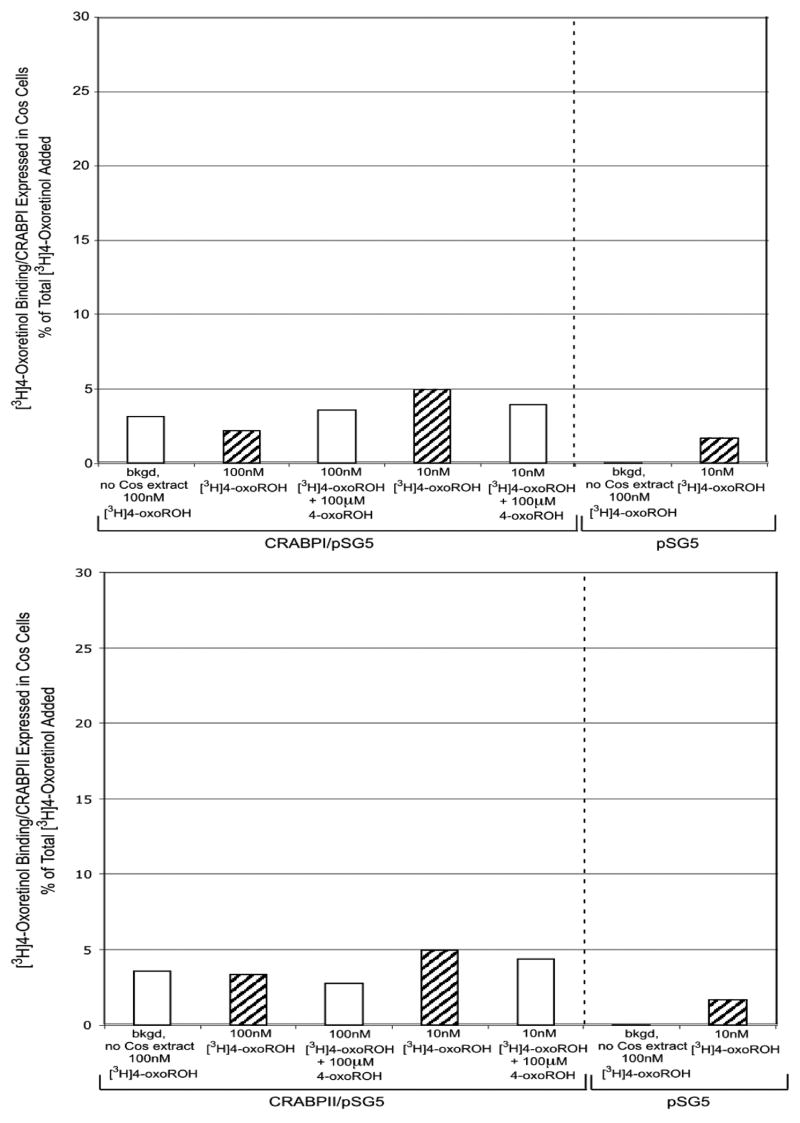
The [3H]4-oxoretinol synthesized from 4-oxoretinaldehyde exactly matched the absorption spectrum of the unlabeled 4-oxoretinol standard (Fig. 7). The specific activity of the [3H]4-oxoretinol was 0.35 mCi/mmole. The ability of CRABPI and II to bind [3H]RA was used as a positive control for the integrity of the binding proteins (Fig. 8A). The protein extract containing the overexpressed CRABPI bound to approximately 17% to 20% of the [3H]RA added, and this binding was successfully competed with excess, nonradioactive RA, indicating that [3H]RA bound specifically to CRABPI (Fig. 8A, upper panel). The protein extract from cells which overexpress CRABPII bound approximately 14 to 26% of the [3H]RA added, and this binding was decreased with the addition of excess, nonradiolabeled RA, indicating ligand specificity (Fig. 8A, lower panel). In contrast, very little [3H]RA binding was exhibited by lysates from COS7 cells transfected with the empty parent vector, pSG5 (Fig. 8A, upper and lower panels). [3H]4-Oxoretinol did not bind to CRABPI or CRABPII (Fig. 8B, upper and lower panels). These data indicate that although 4-oxoretinol treatment of ES cells increases the levels of CRABPI and CRABPII transcripts, it does not bind to these proteins under the assay conditions used.
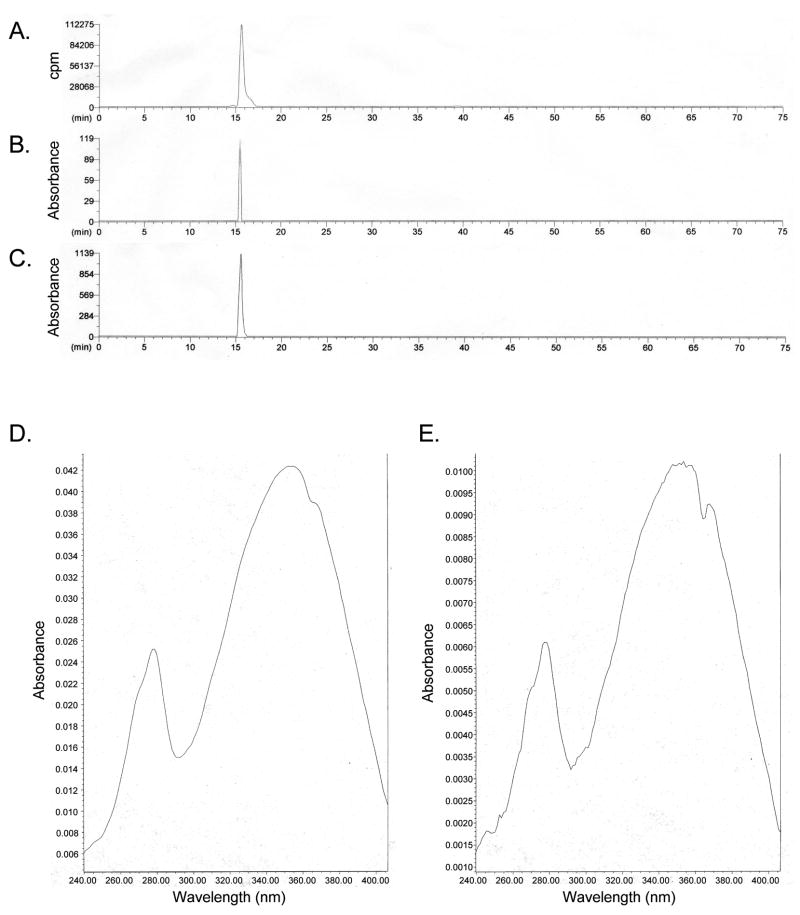
Figure 7. [3H]4-Oxoretinol purification and quantitation.
[3H]4-Oxoretinol, with the [3H] on C15, was synthesized from 4-oxoretinaldehyde as described in the Materials and Methods. Two μl of synthesized [3H]4-oxoretinol was assayed by reverse-phase HPLC. (A) Sample labeling (cpm) as detected by a liquid scintillation counter in-line with the HPLC. Elution of the [3H]4-oxoretinol was detected via absorption at 340 nm. The elution time of the synthesized [3H]4-oxoretinol (B) matched that of the 4-oxoretinol standard (C) (2 μl of a 1 mM 4-oxoretinol standard was loaded on the column). The absorption spectrum of the 4-oxoretinol standard (D) versus synthesized [3H]4-oxoretinol (E).
Figure 8. A. [3H]RA binding assays.
Protein (150 μg) extracts from COS cells transiently transfected with pSG5 containing CRABPI (upper panel), pSG5 possessing CRAPBII (lower panel) or pSG5 (both panels) were incubated for 6 h at 4°C in a total volume of 50 μl with increasing amounts of [3H]RA in the presence or absence of 100 μM of unlabeled RA, and then the extracts were charcoal treated as described under “Materials and Methods”. [3H]RA binding is expressed as % of total [3H]RA added, normalized to the amount of protein extract assayed. B. [3H]4-Oxoretinol binding assays. Protein (150 μg) extracts from COS cells transiently transfected with pSG5 containing CRABPI (upper panel), pSG5 possessing CRAPBII (lower panel) or pSG5 (both panels) were incubated for 6 h at 4°C in a total volume of 50 μl with increasing amounts of [3H]4-oxoretinol in the presence or absence of 100 μM of unlabeled 4-oxoretinol, and then the extracts were charcoal treated as described under “Materials and Methods”. [3H]4-oxoretinol binding is expressed as % of total [3H]4-oxoretinol added, normalized to the amount of protein extract assayed. This experiment was repeated twice with similar results. One representative experiment is shown.
DISCUSSION
Retinol and 4-Oxoretinol Increase CRABPI and CRABPII mRNA Levels
4-Oxoretinol is produced from retinol when ES cells are induced to differentiate by the removal of the cytokine LIF (Lane et al., 1999) (Fig. 4B). Previous studies have shown that 4-oxoretinol mediates gene transcription via RARs (Achkar et al., 1996). In this study we show that retinol and 4-oxoretinol are more effective than all-trans-RA at increasing the levels of both CRABPI and CRABPII transcripts in the absence of LIF, whereas RA is more effective in the presence of LIF (Fig. 1). Our data indicate that this occurs via a transcriptional mechanism, at least for the increase in CRABPI mRNA (Fig. 2). Of the three retinoids examined, 4-oxoretinol caused the greatest increase in CRABPI and II mRNA levels when compared to control, followed by retinol and RA (Fig. 1). 4-Oxoretinol treatment also resulted in a sustained increase in CRABPI and CRABPII transcripts, as compared to the more transient increases caused by RA and retinol (Fig.1). The reason for this most likely is that RA and retinol are both metabolized by CYP26A1, whereas 4-oxoretinol is more stable in cells (Achkar et al., 1996; Lane et al., 1999). We also observed similar fold increases in CRABPI protein levels by Western analysis (Fig. 3). It is well established that RA, a high affinity ligand for the CRABPs, can induce CRABPI and II gene transcription in some cell types (for reviews see: (Means and Gudas, 1995; Ong et al., 1994)). However, as shown previously (Lane et al., 1999), CCE-WT ES cells do not metabolize retinol to RA.
Our data indicate that retinol regulates CRABPI mRNA levels primarily at the transcriptional level (Fig. 2). Interestingly, the promoter region of the CRABPI gene has not been reported to contain an RARE. Previous studies have shown that P19 embryonal carcinoma and J1 ES cells exhibit increases in CRABPI mRNA levels in response to RA treatment (Chen and Gudas, 1996; Means et al., 2000). RA did not induce the transcription of a reporter construct downstream of a 7.8 kb regulatory region of the CRABPI gene in P19 cells, while this same 7.8 kb regulatory region was active during development (Means and Gudas, 1997; Means et al., 2000). In this study we show that treatment with exogenous retinol or RA, even in ES cells cultured without LIF, was unable to induce transcription of a reporter construct driven by 7.8 kb of the CRABPI promoter (Fig. 2B). Thus, the DNA elements required for the transcriptional activation of the CRABPI gene under these culture conditions may reside either in a more 5′ region or 3′ of the gene.
In contrast to CRABPI, the CRABPII gene promoter region contains an RARE, indicating that CRABPII gene expression is regulated by RA in some cells (Astrom et al., 1994; Astrom et al., 1992; Durand et al., 1992). Previous research in our laboratory has shown that 4-oxoretinol can induce gene transcription via a RARE (Achkar et al., 1996). Thus, although we did not determine if such a transcriptional mechanism is responsible for the increase in CRABPII mRNA levels displayed by ES cells treated with exogenous 4-oxoretinol in the absence of LIF, we hypothesize that the endogenous 4-oxoretinol made in response to the removal of LIF and culture with retinol induces the transcription of CRABPII (Fig. 1).
Retinol Metabolism to 4-Oxoretinol by CYP26A1
In a previous study we showed that retinol is converted to 4-oxoretinol by the enzyme CYP26A1 (Lane et al., 1999). [3H]Retinol metabolism and [3H]4-oxoretinol production were increased in the absence of LIF and the presence of retinoids (Fig. 4). It is important to note that in this study the ES cells were cultured for the retinoid metabolism assay with only 50 nM [3H]retinol, and therefore, the levels of [3H]4-oxoretinol produced were in the nanomolar range. 50 nM of [3H]retinol was used to avoid the addition of high concentrations of [3H]retinol, which can cause differentiation, in control, +LIF cells. Previously, we cultured CCE-WT ES cells in the presence of 1 μM retinol (100 nM [3H]retinol and 900 nM unlabeled retinol) (Lane et al., 1999). This earlier experimental protocol resulted in much larger increases in the amounts of both retinol used and 4-oxoretinol produced, as well as an increase in CYP26A1 mRNA.
4-Oxoretinol Increases Retinoid-Responsive Gene mRNA Levels and May Interact with the LIF Signaling Pathway to Regulate Cell Differentiation
In addition to the CRABPs, other RA-responsive genes, such as RARβ and Hoxa1, were induced by exogenous 4-oxoretinol treatment (Fig. 5 and data not shown). Exposure of ES cells to RA also induces RARβ2 (Chen and Gudas, 1996). The ability of 4-oxoretinol to induce RARβ2 and Hoxa1 gene expression in ES cells in the absence of RA (Fig. 5) indicates a potential mechanism by which 4-oxoretinol can influence early embryonic development. This study is the first to show the induction of CRABPI and CRABPII gene expression by 4-oxoretinol (Figs. 1, 5). Other non-retinoid signaling molecules have also been shown to increase CRABP expression. For example, Li and Ong (Li and Ong, 2003) administered estrogen to ovariectomized rats and found that estrogen, but not RA, stimulated CRABPII expression in this tissue. Thyroid hormone can also induce CRABPI expression (Park et al., 2005).
When embryonic stem cells are cultured as aggregates in the absence of LIF, embryoid bodies form which contain various early embryonic cell lineages. Murray and Edgar (Murray and Edgar, 2001) have shown that LIF inhibits the development of visceral and parietal endoderm in these embryoid bodies, but not the development of primitive endodermal cell precursors. Primitive ectodermal cell differentiation was also inhibited by LIF (Murray and Edgar, 2001). Moreover, important differences in gene expression are observed in ES cells cultured with vs. without LIF (Martinez-Ceballos et al., 2005; Sauter et al., 2005). Recently, Martin-Ibanez et al., 2007 (ref = J. Neurosci Res, in press) showed that in the absence of LIF, murine ES cells, cultured as embryoid bodies and treated with RA, differentiate to neuronal cells with primarily a GABAergic phenotype. They also showed that the addition of LIF blocks the ability of RA to induce the neuronal differentiation of ES cells. In our experiments, LIF blocked the large increases in CRABPI and CRABPII transcripts in response to retinol and 4-oxoretinol (Fig. 1). Thus, we speculate that these large increases in CRABPI and II transcripts may be important for neuronal differentiation to take place. LIF is a major factor influencing both the identities and levels of genes expressed in response to retinoids. We also hypothesize that the differentiation states of ES cells are determined by their responses to both LIF and 4-oxoretinol, as reflected by the vastly different CRABPI and CRABPII transcript levels exhibited by cells cultured with versus without LIF (Figs. 1, 5).
4-Oxoretinol Does Not Bind to CRABPI or CRABPII
Both CRABPI and CRABPII bind RA, 4-oxo-RA, 4-hydoxy-RA, 9-cis-RA and 13-cis-RA, but not retinol (Fiorella et al., 1993; Fogh et al., 1993). In this study we produced high levels of CRABPI or CRABPII protein in COS 7 cells by transiently transfecting COS 7 cells with full-length cDNAs encoding either of these binding proteins. Although 4-oxoretinol increases the levels of both CRABPI and II transcripts (Fig. 1), it does not bind to either protein under the assay conditions employed (Fig. 8B). However, because 4-oxoretinol increases CRABPI and II levels, we hypothesize that 4-oxoretinol influences the fate of ES cells by altering their ability to respond to other retinoids that can bind to CRABPs. The fact that relatively high amounts of 4-oxoretinol and 4-oxoretinaldehyde, in comparison to RA, are detected in Xenopus oocytes and embryos (Blumberg et al., 1996) indicates a role for 4-oxoretinol in development.
Acknowledgments
The authors would like to thank Karl B. Ecklund for his editorial assistance in preparing this manuscript for publication, Marina Vivero and Martin Albert for some experiments, and members of the Gudas laboratory for useful discussions. F.D. thanks Dr. G. Massiot for encouragement. This research was funded by NIH grant R01CA43796 to LJG.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Achkar CC, Derguini F, Blumberg B, Langston A, Levin AA, Speck J, Evans RM, Bolado J, Jr, Nakanishi K, Buck J, Gudas LJ. 4-Oxoretinol, a new natural ligand and transactivator of the retinoic acid receptors. Proc Natl Acad Sci U S A. 1996;93:4879–84. doi: 10.1073/pnas.93.10.4879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Astrom A, Pettersson U, Chambon P, Voorhees JJ. Retinoic acid induction of human cellular retinoic acid-binding protein-II gene transcription is mediated by retinoic acid receptor-retinoid X receptor heterodimers bound to one far upstream retinoic acid-responsive element with 5-base pair spacing. J Biol Chem. 1994;269:22334–9. [PubMed] [Google Scholar]
- Astrom A, Pettersson U, Voorhees JJ. Structure of the human cellular retinoic acid-binding protein II gene. Early transcriptional regulation by retinoic acid. J Biol Chem. 1992;267:25251–5. [PubMed] [Google Scholar]
- Bhatt H, Brunet LJ, Stewart CL. Uterine expression of leukemia inhibitory factor coincides with the onset of blastocyst implantation. Proc Natl Acad Sci U S A. 1991;88:11408–12. doi: 10.1073/pnas.88.24.11408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumberg B, Bolado J, Jr, Derguini F, Craig AG, Moreno TA, Chakravarti D, Heyman RA, Buck J, Evans RM. Novel retinoic acid receptor ligands in Xenopus embryos. Proc Natl Acad Sci U S A. 1996;93:4873–8. doi: 10.1073/pnas.93.10.4873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehm MF, Gawinowicz MA, Foucault A, Derguini F, Nakanishi K. Photoaffinity labeling studies of bacteriorhodopsin with [15-3H]-3-diazo-4-keto-all-trans-retinal. J Am Chem Soc. 1990;112:7779–7782. [Google Scholar]
- Boylan JF, Gudas LJ. Overexpression of the cellular retinoic acid binding protein-I (CRABP-I) results in a reduction in differentiation-specific gene expression in F9 teratocarcinoma cells. J Cell Biol. 1991;112:965–79. doi: 10.1083/jcb.112.5.965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boylan JF, Gudas LJ. The level of CRABP-I expression influences the amounts and types of all-trans-retinoic acid metabolites in F9 teratocarcinoma stem cells. J Biol Chem. 1992;267:21486–91. [PubMed] [Google Scholar]
- Bucco RA, Melner MH, Gordon DS, Leers-Sucheta S, Ong DE. Inducible expression of cellular retinoic acid-binding protein II in rat ovary: gonadotropin regulation during luteal development. Endocrinology. 1995;136:2730–40. doi: 10.1210/endo.136.6.7750498. [DOI] [PubMed] [Google Scholar]
- Bucco RA, Zheng WL, Wardlaw SA, Davis JT, Sierra-Rivera E, Osteen KG, Melner MH, Kakkad BP, Ong DE. Regulation and localization of cellular retinol-binding protein, retinol-binding protein, cellular retinoic acid-binding protein (CRABP), and CRABP II in the uterus of the pseudopregnant rat. Endocrinology. 1996;137:3111–22. doi: 10.1210/endo.137.7.8770937. [DOI] [PubMed] [Google Scholar]
- Chen AC, Gudas LJ. An analysis of retinoic acid-induced gene expression and metabolism in AB1 embryonic stem cells. J Biol Chem. 1996;271:14971–80. doi: 10.1074/jbc.271.25.14971. [DOI] [PubMed] [Google Scholar]
- Chen AC, Guo X, Derguini F, Gudas LJ. Human breast cancer cells and normal mammary epithelial cells: retinol metabolism and growth inhibition by the retinol metabolite 4-oxoretinol. Cancer Res. 1997;57:4642–51. [PubMed] [Google Scholar]
- Cornic M, Delva L, Castaigne S, Lefebvre P, Balitrand N, Degos L, Chomienne C. In vitro all-trans retinoic acid (ATRA) sensitivity and cellular retinoic acid binding protein (CRABP) levels in relapse leukemic cells after remission induction by ATRA in acute promyelocytic leukemia. Leukemia. 1994;8(Suppl 2):S16–9. [PubMed] [Google Scholar]
- Delva L, Bastie JN, Rochette-Egly C, Kraiba R, Balitrand N, Despouy G, Chambon P, Chomienne C. Physical and functional interactions between cellular retinoic acid binding protein II and the retinoic acid-dependent nuclear complex. Mol Cell Biol. 1999;19:7158–67. doi: 10.1128/mcb.19.10.7158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong D, Ruuska SE, Levinthal DJ, Noy N. Distinct roles for cellular retinoic acid-binding proteins I and II in regulating signaling by retinoic acid. J Biol Chem. 1999;274:23695–8. doi: 10.1074/jbc.274.34.23695. [DOI] [PubMed] [Google Scholar]
- Durand B, Saunders M, Leroy P, Leid M, Chambon P. All-trans and 9-cis retinoic acid induction of CRABPII transcription is mediated by RAR-RXR heterodimers bound to DR1 and DR2 repeated motifs. Cell. 1992;71:73–85. doi: 10.1016/0092-8674(92)90267-g. [DOI] [PubMed] [Google Scholar]
- Faria TN, Rivi R, Derguini F, Pandolfi PP, Gudas LJ. 4-Oxoretinol, a metabolite of retinol in the human promyelocytic leukemia cell line NB4, induces cell growth arrest and granulocytic differentiation. Cancer Res. 1998;58:2007–13. [PubMed] [Google Scholar]
- Fiorella PD, Giguere V, Napoli JL. Expression of cellular retinoic acid-binding protein (type II) in Escherichia coli. Characterization and comparison to cellular retinoic acid-binding protein (type I) J Biol Chem. 1993;268:21545–52. [PubMed] [Google Scholar]
- Fogh K, Voorhees JJ, Astrom A. Expression, purification, and binding properties of human cellular retinoic acid-binding protein type I and type II. Arch Biochem Biophys. 1993;300:751–5. doi: 10.1006/abbi.1993.1104. [DOI] [PubMed] [Google Scholar]
- Giguere V. Retinoic acid receptors and cellular retinoid binding proteins: complex interplay in retinoid signaling. Endocr Rev. 1994;15:61–79. doi: 10.1210/edrv-15-1-61. [DOI] [PubMed] [Google Scholar]
- Grippo JF, Gudas LJ. The effect of dibutyryl cyclic AMP and butyrate on F9 teratocarcinoma cellular retinoic acid-binding protein activity. J Biol Chem. 1987;262:4492–500. [PubMed] [Google Scholar]
- Guo X, Gudas LJ. Metabolism of all-trans-retinol in normal human cell strains and squamous cell carcinoma (SCC) lines from the oral cavity and skin: reduced esterification of retinol in SCC lines. Cancer Res. 1998;58:166–76. [PubMed] [Google Scholar]
- Lane MA, Chen AC, Roman SD, Derguini F, Gudas LJ. Removal of LIF (leukemia inhibitory factor) results in increased vitamin A (retinol) metabolism to 4-oxoretinol in embryonic stem cells. Proc Natl Acad Sci U S A. 1999;96:13524–9. doi: 10.1073/pnas.96.23.13524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langston AW, Gudas LJ. Identification of a retinoic acid responsive enhancer 3′ of the murine homeobox gene Hox-1.6. Mech Dev. 1992;38:217–27. doi: 10.1016/0925-4773(92)90055-o. [DOI] [PubMed] [Google Scholar]
- Li R, Faria TN, Boehm M, Nabel EG, Gudas LJ. Retinoic acid causes cell growth arrest and an increase in p27 in F9 wild type but not in F9 retinoic acid receptor beta2 knockout cells. Exp Cell Res. 2004;294:290–300. doi: 10.1016/j.yexcr.2003.11.014. [DOI] [PubMed] [Google Scholar]
- Li XH, Ong DE. Cellular retinoic acid-binding protein II gene expression is directly induced by estrogen, but not retinoic acid, in rat uterus. J Biol Chem. 2003;278:35819–25. doi: 10.1074/jbc.M302551200. [DOI] [PubMed] [Google Scholar]
- MacGregor TM, Copeland NG, Jenkins NA, Giguere V. The murine gene for cellular retinoic acid-binding protein type II. Genomic organization, chromosomal localization, and post-transcriptional regulation by retinoic acid. J Biol Chem. 1992;267:7777–83. [PubMed] [Google Scholar]
- Martinez-Ceballos E, Chambon P, Gudas LJ. Differences in gene expression between wild type and Hoxa1 knockout embryonic stem cells after retinoic acid treatment or leukemia inhibitory factor (LIF) removal. J Biol Chem. 2005;280:16484–98. doi: 10.1074/jbc.M414397200. [DOI] [PubMed] [Google Scholar]
- McClean SW, Ruddel ME, Gross EG, DeGiovanna JJ, Peck GL. Liquid-chromatographic assay for retinol (vitamin A) and retinol analogs in therapeutic trials. Clin Chem. 1982;28:693–6. [PubMed] [Google Scholar]
- Means AL, Gudas LJ. The roles of retinoids in vertebrate development. Annu Rev Biochem. 1995;64:201–33. doi: 10.1146/annurev.bi.64.070195.001221. [DOI] [PubMed] [Google Scholar]
- Means AL, Gudas LJ. The CRABP I gene contains two separable, redundant regulatory regions active in neural tissues in transgenic mouse embryos. Developmental Dynamics. 1997;209:59–69. doi: 10.1002/(SICI)1097-0177(199705)209:1<59::AID-AJA6>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- Means AL, Thompson JR, Gudas LJ. Transcriptional regulation of the cellular retinoic acid binding protein I gene in F9 teratocarcinoma cells. Cell Growth Differ. 2000;11:71–82. [PubMed] [Google Scholar]
- Murray P, Edgar D. The regulation of embryonic stem cell differentiation by leukaemia inhibitory factor (LIF) Differentiation. 2001;68:227–34. doi: 10.1046/j.1432-0436.2001.680410.x. [DOI] [PubMed] [Google Scholar]
- Napoli JL, Posch KP, Fiorella PD, Boerman MH. Physiological occurrence, biosynthesis and metabolism of retinoic acid: evidence for roles of cellular retinol-binding protein (CRBP) and cellular retinoic acid-binding protein (CRABP) in the pathway of retinoic acid homeostasis. Biomed Pharmacother. 1991;45:131–43. doi: 10.1016/0753-3322(91)90101-x. [DOI] [PubMed] [Google Scholar]
- Nichols J, Davidson D, Taga T, Yoshida K, Chambers I, Smith A. Complementary tissue-specific expression of LIF and LIF-receptor mRNAs in early mouse embryogenesis. Mech Dev. 1996;57:123–31. doi: 10.1016/0925-4773(96)00531-x. [DOI] [PubMed] [Google Scholar]
- Noy N. Retinoid-binding proteins: mediators of retinoid action. Biochem J 348 Pt. 2000;3:481–95. [PMC free article] [PubMed] [Google Scholar]
- Ong DE, Newcomer ME, Chytil F. Cellular retinoid-binding proteins. In: Sporn MB, et al., editors. The Retinoids: Biology, Chemistry, and Medicine. Raven Press; New York: 1994. pp. 283–318. [Google Scholar]
- Park SW, Li G, Lin YP, Barrero MJ, Ge K, Roeder RG, Wei LN. Thyroid hormone-induced juxtaposition of regulatory elements/factors and chromatin remodeling of Crabp1 dependent on MED1/TRAP220. Mol Cell. 2005;19:643–53. doi: 10.1016/j.molcel.2005.08.008. [DOI] [PubMed] [Google Scholar]
- Penniston KL, Tanumihardjo SA. Elevated serum concentrations of beta-glucuronide metabolites and 4-oxoretinol in lactating sows after treatment with vitamin A: a model for evaluating supplementation in lactating women. Am J Clin Nutr. 2005;81:851–8. doi: 10.1093/ajcn/81.4.851. [DOI] [PubMed] [Google Scholar]
- Rathjen PD, Nichols J, Toth S, Edwards DR, Heath JK, Smith AG. Developmentally programmed induction of differentiation inhibiting activity and the control of stem cell populations. Genes Dev. 1990;4:2308–18. doi: 10.1101/gad.4.12b.2308. [DOI] [PubMed] [Google Scholar]
- Ruff SJ, Ong DE. Cellular retinoic acid binding protein is associated with mitochondria. FEBS Lett. 2000;487:282–6. doi: 10.1016/s0014-5793(00)02366-8. [DOI] [PubMed] [Google Scholar]
- Sauter CN, McDermid RL, Weinberg AL, Greco TL, Xu X, Murdoch FE, Fritsch MK. Differentiation of murine embryonic stem cells induces progesterone receptor gene expression. Exp Cell Res. 2005;311:251–64. doi: 10.1016/j.yexcr.2005.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sessler RJ, Noy N. A ligand-activated nuclear localization signal in cellular retinoic acid binding protein-II. Mol Cell. 2005;18:343–53. doi: 10.1016/j.molcel.2005.03.026. [DOI] [PubMed] [Google Scholar]
- Stewart CL, Kaspar P, Brunet LJ, Bhatt H, Gadi I, Kontgen F, Abbondanzo SJ. Blastocyst implantation depends on maternal expression of leukaemia inhibitory factor. Nature. 1992;359:76–9. doi: 10.1038/359076a0. [DOI] [PubMed] [Google Scholar]
- Tighe AP, Gudas LJ. Retinoic acid inhibits leukemia inhibitory factor signaling pathways in mouse embryonic stem cells. J Cell Physiol. 2004;198:223–9. doi: 10.1002/jcp.10424. [DOI] [PubMed] [Google Scholar]
- Vasios GW, Gold JD, Petkovich M, Chambon P, Gudas LJ. A retinoic acid-responsive element is present in the 5′ flanking region of the laminin B1 gene. Proc Natl Acad Sci. 1989;86:9099–9103. doi: 10.1073/pnas.86.23.9099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White JA, Beckett-Jones B, Guo YD, Dilworth FJ, Bonasoro J, Jones G, Petkovich M. cDNA cloning of human retinoic acid-metabolizing enzyme (hP450RAI) identifies a novel family of cytochromes P450. J Biol Chem. 1997;272:18538–41. doi: 10.1074/jbc.272.30.18538. [DOI] [PubMed] [Google Scholar]
- Zheng WL, Bucco RA, Schmitt MC, Wardlaw SA, Ong DE. Localization of cellular retinoic acid-binding protein (CRABP) II and CRABP in developing rat testis. Endocrinology. 1996;137:5028–35. doi: 10.1210/endo.137.11.8895377. [DOI] [PubMed] [Google Scholar]