Abstract
Basonuclin is a protein containing three pairs of C2H2 zinc fingers. The protein has been found in the basal (germinal) cell layer of stratified squamous epithelia, such as the epidermis, and in germ cells of the testis and ovary. We show here that the human protein has specific affinity for a segment of the promoter of the gene for rRNA. Basonuclin interacts with two separate parts of the promoter, each possessing dyad symmetry. The upstream part, but not the downstream part, is known to bind UBF1, a transcription factor for rDNA. Basonuclin is likely to be a cell-type-specific regulatory protein for rDNA transcription.
Keywords: dyad symmetry, gene selection with inclusion bodies, UBF1
Basonuclin is a cell-type-specific zinc finger protein discovered in cultured human epidermal cells (1). The primary structure of the protein, deduced from the nucleotide sequence of its cDNA, shows that it contains three pairs of C2H2 zinc fingers (Zfs), a nuclear localization signal, and a serine-rich region, called a serine stripe (1). The human gene contains five exons and is located on chromosome 15 (2).
During terminal differentiation of squamous epithelia, the mRNA and the protein disappear from the suprabasal cells, which also lose colony-forming ability and develop markers of terminal differentiation, such as involucrin. Basonuclin is also restricted to certain cells in the hair follicles of humans (3) and mice (4). In the cells of mouse testis, basonuclin is concentrated in the nuclei of germ cells, but during the formation of spermatozoa, which are inactive in transcription, the protein leaves the nucleus and becomes cytoplasmic (5, 6). Basonuclin is also concentrated in the nuclei of female germ cells (6).
In rapidly growing cultured human keratinocytes, the basonuclin gene is highly expressed. The protein is mostly located in the cell nuclei (3) but can be translocated to the cytoplasm by cultural conditions that poorly support cell growth. For example, human keratinocytes grown at low density in the absence of supporting 3T3 cells contain basonuclin mainly in the cytoplasm (7). After addition of 3T3 cells to the culture, the basonuclin of the keratinocytes again becomes mainly nuclear, and rapid cell growth is resumed.
It has been shown by mutagenesis that phosphorylation of serine residue 541 of basonuclin promotes cytoplasmic localization of the protein, whereas absence of phosphorylation of the same residue promotes nuclear localization. Thus, the subcellular localization of basonuclin is subject to regulation, mediated by phosphorylation (8).
These observations suggest that basonuclin may be a transcription factor in keratinocytes. Direct evidence for association of basonuclin with the ribosomal gene promoter is described below.
MATERIALS AND METHODS
Construction of a Human Genomic Library.
Human genomic DNA (10 μg; Roche Molecular Biochemicals) was digested with 20 units of Sau 3A overnight at 37°C. In parallel, pBluescript KS(+) DNA (4 μg; Stratagene) was digested with 50 units of BamHI and dephosphorylated with alkaline phosphatase. The genomic DNA and the pBluescript KS(+) were mixed and ligated with 10 units of the T4 enzyme overnight at 16°C.
Construction of Plasmids.
The plasmids pHUB2 and pET28a(+) have been described (8). A tightly controlled isopropyl β-d-thiogalactoside-inducible Escherichia coli system for expression of recombinant proteins was required to avoid toxicity of the Zfs. Accordingly, DNA of glutathione S-transferase (GST) was first put into pET28a(+), and then the DNA of pairs of Zfs of basonuclin was ligated to it. Both pGEX 5X-1, encoding GST, and pHUB19, encoding basonuclin Zfs, were amplified by PCR. Oligonucleotide primers used in the reaction were TATACCATGGGCCCTATACTAGGTTATTGG and CGAGGCAGATCGTCAGTC for GST, CGGAATTCGAGGCCAAAGTGAAGCCTG and GGGGTACCCCTTAGTCTTTGTCCCGGTTATTTC for Zfs 1+2 (340- to 425-aa residues of basonuclin), and CGGAATTCCTGGAGCACGTGGGTCAGC and GGGGTACCCCTTAACTCTCCAATGCTTCCTGG for Zfs 3+4 (703- to 786-aa residues).
Labeling of DNA.
DNAs used for gel-shift and DNase I protection assays were end-labeled with the Klenow fragment or polynucleotide kinase.
Preparation of Bacterial Extracts and Purification of Proteins.
Bacterial strain BL21 (DE3) harboring the appropriate plasmid was grown in LB medium and induced for 2.5–3 h at 37°C or for 7 h at 30°C to produce the recombinant proteins. The bacterial cells were collected by centrifugation for 10 min at 6,000 × g, resuspended in a zinc buffer (20 mM Hepes, pH 7.0/150 mM NaCl/0.1 mM ZnSO4/1 mM DTT), and lysed on ice by incubating with lysozyme to 1 mg/ml for 30 min with occasional stirring. To the lysate, a mixture of protease inhibitors (complete, EDTA-free, Roche Molecular Biochemicals) was added. After sonication for 1 min and addition of Triton X-100 to 1%, the lysate was separated into soluble and particulate fractions by centrifugation for 10 min at 14,000 × g. To obtain basonuclin inclusion bodies, the particulate fraction was resuspended in the zinc buffer containing the protease inhibitors and 50% (vol/vol) glycerol. To purify the recombinant basonuclin, inclusion bodies were solubilized in a loading buffer, separated by SDS/PAGE, and renatured with the addition of 0.1 mM ZnSO4 as described (9). To purify GST fusion proteins, the soluble fraction was applied to a glutathione Sepharose 4B column and processed according to the manufacturer’s protocol (Amersham Pharmacia). To the purified protein, 1 mM DTT, 0.1 mM ZnSO4, and 40% (wt/vol) glycerol were added.
Binding Conditions for Selection of the Target Gene.
The human genomic library (14 μg) and basonuclin inclusion bodies (25 μg) were mixed in 500 μl of a selection buffer (20 mM Hepes, pH 7.4/100 mM NaCl/10 mM MgSO4/1 mM DTT/0.1 mM ZnSO4/1 mg/ml tRNA/0.1 mg/ml BSA/0.1% Nonidet P-40) and incubated for 1 h in a rotating tube at room temperature. The plasmid-inclusion body complexes were collected by brief centrifugation, washed four times with the binding buffer, and suspended in 20 μl of TE buffer (10 mM Tris/1 mM EDTA, pH 8.0). The plasmids were removed from the inclusion bodies by heating for 5 min at 95°C and then kept on ice. All the plasmids were electroporated into XL-1 blue strain, propagated in it, and extracted. A portion of the extracted plasmids was used for the next round of the selection. This process was repeated six times. At each selection, 25 μg of inclusion bodies was used, but the amount of plasmid DNA was reduced to 14, 14, 1.0, 1.7, 1.3, and 1.0 μg for the six selections.
Gel-Shift Assay.
A 20-min incubation of 32P-labeled DNA was carried out at 25°C with GST–Zfs 1+2, GST–Zfs 3+4, or GST alone in a total volume of 10 μl containing a binding buffer [20 mM Hepes, pH 7.4/100 mM NaCl/10 mM MgSO4/1 mM DTT/0.1 mM ZnSO4/0.1 mM EDTA/1 unit/ml poly(dI-dC)]. Electrophoresis of the reaction was then carried out on a 4% acrylamide gel in 0.5× TBE buffer (90 mM Tris/64.6 mM boric acid/2.5 mM EDTA, pH 8.3) at 225 V for about 2.5 h at 20°C by using the Multiphor II chamber (Amersham Pharmacia). The gel was dried and exposed to Kodak BioMax MR film for autoradiography. The sequence of the double-stranded TATA-containing oligonucleotide used for a control experiment was GCA GAG CAT ATA AGG TGA GGT AGG A.
DNase I Footprinting.
An incubation was carried out with 32P-labeled DNA with basonuclin inclusion bodies, GST–Zfs 1+2, or GST alone, in the 20 μl binding buffer for 25 min at 25°C. The DNA was digested by adding a 20-μl solution containing 10–12.5 milliunits of DNase I (Roche Molecular Biochemicals) and incubating the mixture for 1 min at the same temperature. The digestion was terminated by adding 200 μl of stop solution [92% (vol/vol) ethanol/7 μg/ml of tRNA/7% (vol/vol) saturated ammonium acetate], and then the sample was prepared for denatured gel electrophoresis (10). The DNA sequencing reaction was carried out as described (11).
RESULTS
Isolation of Genomic DNA That Binds to Basonuclin.
Recombinant basonuclin produced in E. coli is insoluble and can be collected easily as inclusion bodies by centrifugation. Such a preparation contains small amounts of partially hydrolyzed basonuclin and bacterial proteins, but the most abundant protein is intact basonuclin (Fig. 1). We used such a preparation to isolate target genes to which basonuclin specifically binds. An aliquot of inclusion bodies was allowed to bind to plasmids of a human genomic library, and the unbound plasmids were removed by several washes with the binding buffer. The plasmids adhering to the inclusion bodies were released by heating, amplified in E. coli, and extracted; the selected library was then subjected to five more rounds of purification by binding to and elution from inclusion bodies. To assess the degree of purification after each cycle, the plasmids were linearized by digestion with KpnI, and the size of the plasmids in the digest was determined by agarose gel electrophoresis (Fig. 2). Up to the third cycle, a 3-kb band corresponding to the size of the plasmid was the only band seen. At the fourth cycle, a band of 5 kb appeared, and by the sixth cycle, this band was the only one detected. Three randomly picked colonies from the sixth cycle yielded a 5-kb plasmid containing a 2-kb insert. The three inserts seemed identical, because digestion by restriction enzymes gave the same fragment pattern for all. One of these clones, designated pTRG1, was analyzed extensively.
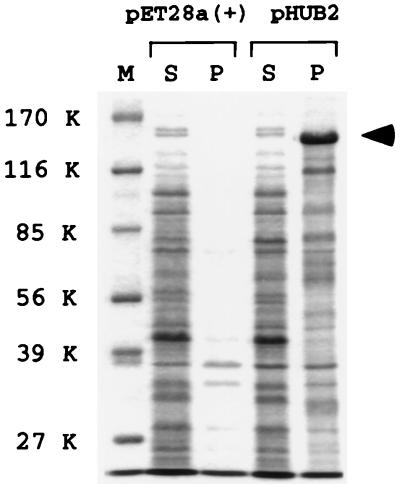
Figure 1.
Recombinant basonuclin recovered from inclusion bodies. E. coli expressing human basonuclin from plasmid pHUB2 were disrupted, and the lysate was divided into soluble and particulate fractions (S and P). Each fraction was subjected to SDS/PAGE, and the resolved proteins were stained with Coomassie blue. Human basonuclin, indicated by an arrowhead at 140 kDa (K), was the most abundant protein. This protein was absent from the particulate fraction of cells harboring the vector plasmid pET28a(+). Basonuclin was absent from both soluble fractions, which otherwise contained virtually identical proteins. M, standard markers.
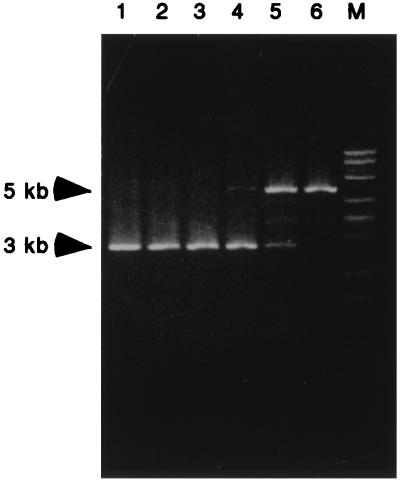
Figure 2.
A human genomic fragment with affinity for basonuclin. A genomic library was subjected to selection based on affinity binding to basonuclin inclusion bodies. After each cycle of binding and elution, the DNA was amplified, digested with KpnI, and analyzed by agarose electrophoresis and ethidium bromide staining. The number above each lane indicates the selection cycle. After the fifth cycle, a single band of 5 kilobases (kb) became dominant (pTRG1). The band of 3 kb, corresponding to the size of the vector, could no longer be detected after the sixth cycle. In lane M, a λDNA–BstEII digest is shown as standard marker.
Specific Binding of Basonuclin Zfs 1+2 to a 258-bp Segment of the Insert of pTRG1.
To identify the basonuclin-binding region, the 2-kb insert was cleaved with EcoRI and NotI into three fragments of 368, 779, and 819 bp. Each of these fragments was cloned into a plasmid; all three were mixed with a plasmid consisting of vector alone and subjected to selection with inclusion bodies. After one cycle, the abundance of the plasmid with the 779-bp fragment exceeded that of the other plasmids by a factor of 5 to 10. This result showed that the 779-bp fragment contained a sequence able to bind to basonuclin with high affinity (Fig. 3A).
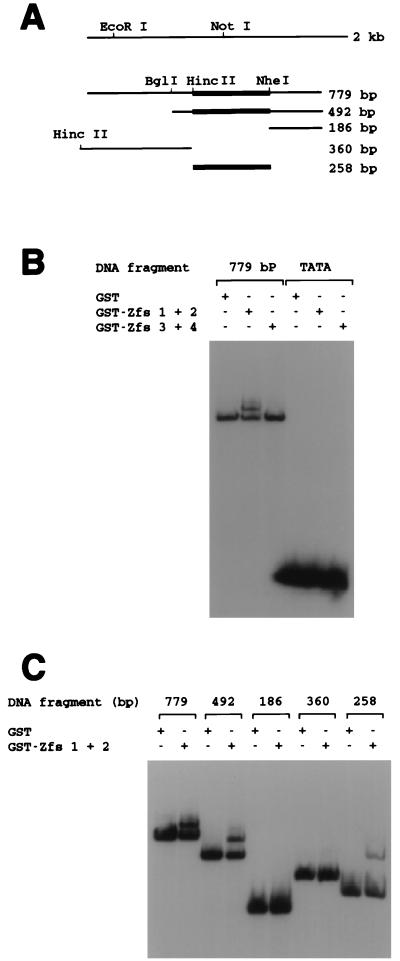
Figure 3.
Binding of the pTRG1 insert and its subfragments to Zfs of basonuclin. (A) Schematic of the subcloned human genomic DNA. The left end of the 360-bp fragment was created by digesting the vector sequence with HincII instead of EcoRI. (B) Gel-shift assay of binding between the 779-bp DNA fragment and Zfs coupled to GST. GST–Zfs 1+2 (40 ng), GST–Zfs 3+4 (60 ng), and GST itself (40 ng) were each incubated with the 32P-labeled 779-bp fragment (7.5 fmol), and the mobility of the DNA was tested by electrophoresis through a 4% nondenaturing polyacrylamide gel. A 32P-labeled 25-nucleotide oligomer containing TATA (17 fmol) served as a control. GST–Zfs 1+2 bound to the 779-bp fragment, but GST–Zfs 3+4 did not. No Zfs bound to the fragment containing TATA. (C) GST–Zfs 1+2 binding to subfragments. GST–Zfs 1+2 (40 ng) or GST itself (40 ng) was incubated with each labeled fragment (4,500 cpm; 779-, 492-, 186-, 360-, or 258-bp DNA), and the mobility of the DNA was assayed. A binding site for GST–Zfs 1+2 was present in the 799-, 492-, and 258-bp fragments, as indicated by thick lines in A, but not in the 186- or 360-bp fragments.
To study the binding sequence of the DNA by gel-shift or membrane-binding assay, we attempted to use basonuclin purified from inclusion bodies by SDS/PAGE and subjected to renaturation (9, 12). The basonuclin so obtained did not show specific binding to the 779-bp fragment of DNA in either assay, suggesting that the renaturation did not occur properly. As Zfs of other proteins may retain the natural binding activity of the intact protein (13–15), we examined, by gel-shift assay, the binding of the 779-bp fragment to two basonuclin Zfs, each fused to GST (GST–Zfs 1+2 and GST–Zfs 3+4). Purified GST–Zfs 1+2, when preincubated for 20 min with the 779-bp fragment in the presence of an excess of nonspecific oligonucleotide, retarded migration of the DNA fragment, whereas neither GST–Zfs 3+4 nor GST alone was able to do so (Fig. 3B). GST–Zfs 1+2 did not shift the mobility of an unrelated TATA-containing oligonucleotide of 25 nucleotides used as control. This result showed that GST–Zfs 1+2 selectively bound to the 779-bp DNA fragment. The inability of GST–Zfs 3+4 to bind to the DNA may indicate that these Zfs depend for binding on the presence of Zfs 1+2 or, like some other Zfs (14, 15), Zfs 3+4 do not interact with DNA.
To define more precisely the site to which human basonuclin binds, we digested the 779-bp fragment with HincII, BglI, or NheI and produced four overlapping fragments of 186, 258, 360, and 492 bp (Fig. 3A). As determined by gel-shift assay, the mobilities of the 186-bp and 360-bp fragments were not reduced by GST–Zfs 1+2, but the mobility of the 492-bp fragment was retarded (Fig. 3C). It could be concluded that basonuclin bound to that part of the 492-bp fragment corresponding to the HincII–NheI fragment of 258 bp. This conclusion was confirmed directly by showing that the mobility of the 258-bp fragment produced by HincII–NheI was retarded by GST–Zfs 1+2 (Fig. 3C).
Identification of the 258-bp Fragment as the rRNA Gene Promoter.
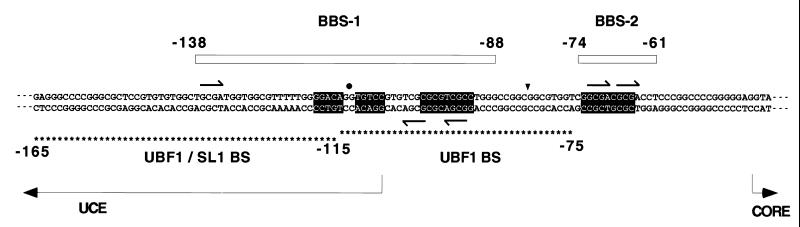
The entire 2-kb plasmid insert of the clone was sequenced, and a homology search of the GenBank database for the sequence was conducted by using the blast program of National Center for Biotechnology Information. This search revealed that the 2-kb insert contained the promoter region of the human rRNA gene (16, 17). The 258-bp sequence to which basonuclin binds contained all the cis-acting elements previously shown to be required for initiation of rRNA transcription: the upstream control element (UCE), the CORE, and the transcription start site (18, 19).
Footprinting to Identify the Binding Site for Basonuclin on the rRNA Gene Promoter.
Using the 258-bp fragment, we examined whether basonuclin inclusion bodies could protect the DNA against the action of DNase I. In the antisense or “noncoding” strand (20), the inclusion bodies protected two regions: the region between −138 and −88 [basonuclin-binding site 1 (BBS1)] and less strongly, the region between −74 and −61 (BBS2; Fig. 4A). In similar experiments, GST–Zfs 1+2 weakly protected BBS1, supporting the assumption that Zfs 1+2 were primarily responsible for the protection afforded by the inclusion-body preparation. As expected, basonuclin inclusion bodies protected BBS1 and BBS2 in the sense or “coding” strand as well (Fig. 4B).
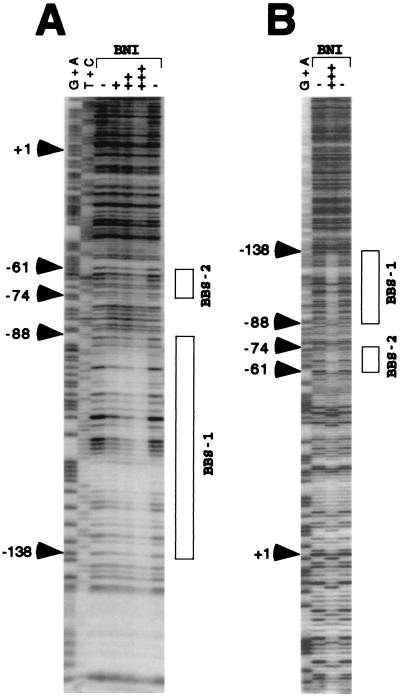
Figure 4.
Resistance to DNase I of rRNA promoter region bound to basonuclin inclusion bodies (BNI). (A) The noncoding strand (24 fmol) of the 258-bp DNA of the human rRNA promoter, to which a short stretch of vector sequence was added to label its 3′ end, was incubated with (+, 5 μg; ++, 10 μg; +++, 20 μg) or without (−) BNI and then digested with 10 milliunits of DNase I. BNI protected the BBS1 region and, less strongly, BBS2 (shown by boxes). The G + A and T + C sequence reactions were performed as described (11). (B) The coding strand (43 fmol) of the EcoRI–NheI fragment of the human rRNA promoter, labeled at the 3′ end, was incubated with (+++, 20 μg) or without (−) BNI and then digested with 12.5 milliunits of DNase I. Both BBS1 and BBS2 were protected by BNI. BBS2 was more strongly protected in this strand than in the noncoding strand.
In only the coding strand, an unexplained anomalous effect was observed in the region downstream of BBS2 and extending across the start site. This anomaly consisted of an apparently random distribution of single sites which, in the presence of basonuclin inclusion bodies, were protected from DNase, were made more sensitive to DNase, or were not affected (Fig. 4B).
The Basonuclin-Binding Sites Are Located in a Palindromic Sequence Containing Short Repeats.
The target of regulatory proteins often consists of a palindromic sequence permitting the proteins to bind at multiple sites (21–23). We examined BBS1 by aligning sequences of different regions within a single strand and the sequence of one strand against that of the other. We found that the region of BBS1 between −118 and −107 contains the dyad sequence GGACAggTGTCC, with an axis of symmetry between the two intervening guanines at −113 and −112 (Fig. 5). Extending the alignment revealed another dyad sequence, CGCGTCGCCX18GGCGACGCG, that spanned BBS1 and BBS2. The nonanucleotides of this dyad were protected against nuclease digestion, but most of the intervening X18 was not. Furthermore, five repeats of the tetranucleotide sequence, GCGA, were identified in both basonuclin-binding sites (Fig. 5).
Figure 5.
Location of binding sites of basonuclin relative to those of UBF1. The sequence shown is that of the promoter region of the human rRNA gene (18). The upper strand is the RNA-like (coding) strand. The CORE region is essential for transcription but is not protected from nuclease digestion by either UBF1 or basonuclin. BBS1 and BBS2 are indicated by boxes. BBS1 contains a dyad sequence (black background), whose center is indicated by a black dot. BBS2 contains a dyad sequence whose complement is located near the 3′ end of BBS1. The axis of symmetry, indicated by a triangle, is located between nucleotides −82 and −83. The intervening 18-bp spacer consisting of 15 nucleotides is not protected from DNase I. The UBF1-binding region is shown by asterisks below the sequence. The 3′ half is protected by UBF1 alone (upper row of asterisks), and the 5′ half is protected only when SL1 is present as well (lower row of asterisks). BBS1 is centered almost at the same point as the entire binding region of UBF1. BBS2 is not protected by UBF1. Arrows indicate the repeated sequence GCGA, which occurs in both strands of the dyad sequence.
Relation of Basonuclin-Binding Sites to Those of UBF.
The human rRNA gene promoter has been well characterized by linker scanning mutagenesis and divided into two cis-acting elements, UCE and CORE (refs. 18 and 24; Fig. 5). The trans-acting factor UBF1 (20) binds to the UCE (19, 25) and participates in the activation of transcription of the gene. Another factor, SL1, consisting of four different proteins, does not itself bind to UCE, but the complex UBF1/SL1 extends DNase protection in the 5′ direction. This complex is necessary for strong activation of transcription (19, 25).
The binding sequences for UBF1 and for basonuclin (BBS1 and BBS2) are compared in Fig. 5. In contrast to UBF1, the binding of basonuclin to the promoter requires no other factor. The regions of binding contain two elements possessing dyad symmetry. The combined binding regions of UBF1 alone (−75 to −115) and the UBF1/SL1 complex (−75 to −165) encompass the entire sequence of BBS1. However, UBF1 alone does not bind to the upstream half of the BBS1 dyad. It would therefore seem unlikely that UBF1 recognizes the dyad sequences. Also, UBF does not bind to BBS2, again indicating that UBF1 and basonuclin recognize different sequences within the rDNA promoter.
Other Genomic Fragments with Affinity for Basonuclin.
After five cycles of purification of genomic fragments by their affinity for basonuclin inclusion bodies, there remained, in addition to the principal band of rDNA, two faint bands corresponding to nucleotide sequences of less than 5 kb (Fig. 2). As these bands disappeared after an additional cycle of purification, the affinity of their DNAs for basonuclin was less than that of the rDNA promoter.
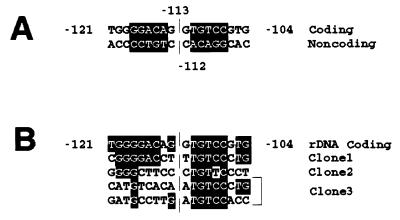
The two bands of the fifth cycle were pooled, ligated, and cloned. Three of these clones were isolated and sequenced (clones 1–3). They each contained an approximately 600-bp insert, of which part was found to possess homology to one of the following sequences in GenBank: band 1, AC003976 (chromosome 17); band 2, AC002065 (chromosome 7Q21); and band 3, U74497 (chromosome 4Q35). Neither the sequence of BBS2 nor the GCGA repeats were present in the nucleotide sequences of these clones. However, each clone contained all or nearly all of the pentanucleotide TGTCC, constituting the 3′ half of the dyad sequence of BBS1 (Fig. 6). Clone 1 also contained most of the 5′ half of the dyad of BBS1, and clones 2 and 3 had some similarities to BBS1 in this region.
Figure 6.
Homology between basonuclin-binding sites of human rRNA promoter and other genes. (A) BBS1. The dyad sequence, shown against a black background, with its axis of symmetry between positions −113 and −112. (B) The same sequence compared with homologous sequences of the three other selected DNA sequences (clones 1–3). Nucleotide identities are shown against a black background. In the case of clone 3, two sequences homologous to the sequence of rDNA were present.
DISCUSSION
Use of Inclusion Bodies for Gene Selection.
In contrast to fragments of basonuclin, which are soluble and can be purified, the complete protein in the form of inclusion bodies in E. coli was insoluble. When the protein was dissolved with ionic detergent, it could not be renatured to restore DNA-binding activity. However, we were able to overcome this difficulty by using the basonuclin inclusion bodies produced in E. coli as a natural solid-phase matrix for affinity purification of the target gene. The method is simple and should be applicable to any protein that forms inclusion bodies in E. coli or that can be made to do so by genetic engineering.
Nucleoplasmic Concentration of Basonuclin Suggests Other Interactions.
We have shown above that basonuclin has specific affinity for the human rRNA gene promoter, but no nucleolar concentration of basonuclin could be shown immunocytologically. This result might have been caused by masking of its immunological determinants. In interphase cells, the protein is very evident in the extranucleolar nucleoplasm, from which it could not be extracted with the nonionic detergent Triton X-100 (7). In mitotic cells, the protein is frequently localized in clusters of small particles bound to chromatin (3).
One possible explanation for nucleoplasmic basonuclin is that the protein may bind to some nucleoplasmic genes. For example, nonribosomal sequences of clones 1–3 bind to basonuclin inclusion bodies; this binding must be weaker than that of the ribosomal genes, because the latter were selected from all other genes by the inclusion-body purification. An alternative explanation for the presence of bound nucleoplasmic basonuclin might be that it makes the protein readily available to the nucleolar gene; in immunocytological studies, it has been noted that UBF1 could be detected in the nucleoplasm, though more faintly than in the nucleoli (26).
Significance of a Cell-Type-Specific Protein Interacting with the Gene for rDNA.
Basonuclin, like UBF1, is likely to be a transcription factor for the rRNA gene promoter. Similarities and differences between basonuclin and UBF1 can be summarized as follows. (i) Basonuclin binds to two dyad sequences, one completely within BBS1 and the other divided between BBS1 and BBS2. As UBF1 binds to the BBS1 region, it is unlikely that both proteins bind simultaneously, suggesting the possibility that the substitution of basonuclin for UBF1 would confer stronger or weaker transcriptional activation. UBF1 does not bind to BBS2. (ii) The two proteins UBF1 and basonuclin are not obviously homologous, but there is a similarity between their C-terminal sequences: in this region, basonuclin and the hyperacidic tail of UBF1 both contain numerous residues of serine, aspartic acid, and glutamic acid. This domain of UBF1 interacts with SL1, thereby activating DNA polymerase I (26–28). Possibly, the serine-stripe region of basonuclin also interacts with proteins. (iii) Basonuclin is a tissue- and cell-type-specific protein of squamous epithelium and testicular and ovarian germ cells, whereas UBF1 is a ubiquitous regulatory protein. In the epidermis, basonuclin is substantially confined to the basal cell layer, which contains most of the cells capable of proliferation. Basonuclin mRNA and protein disappear when keratinocytes become suprabasal and terminal differentiation proceeds; although these cells likely continue to accumulate ribosomes because they are enlarging, they probably do not require the same transcription control system as multiplying cells.
Basonuclin Binding and the rRNA Gene Promoter of the Mouse.
Although UBF1 transcription factors of mice and humans are virtually identical in amino acid sequence, the nucleotide sequences of the binding sites in the two species are only 36% identical. The substitution of the mouse UCE for the corresponding sequence of the human promoter does not permit the initiation of transcription from the human start site (20). Our examination of the mouse UCE region showed that it lacks the dyad symmetry characteristic of the human binding sites for basonuclin. This lack may mean that the basonuclins of the two species, like their UBFs, are not interchangeable. Evolutionary changes in the sequence of mouse basonuclin or accessory proteins may have compensated for the absence of dyad symmetry in the mouse ribosomal promoter.
Acknowledgments
This research was supported by a grant from the National Cancer Institute.
ABBREVIATIONS
- Zfs
zinc fingers
- GST
glutathione S-transferase
- kb
kilobase
- UCE
upstream control element
- BBS1 and BBS2
basonuclin-binding sites 1 and 2.
Note Added in Proof
From experiments beginning with a study of mitotic chromosomes, it has been independently concluded that basonuclin binds to the rDNA promoter (H. Tseng, J. A. Biegel, and R. S. Brown, unpublished work).
References
- 1.Tseng H, Green H. Proc Natl Acad Sci USA. 1992;89:10311–10315. doi: 10.1073/pnas.89.21.10311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Teumer J, Tseng H, Green H. Gene. 1997;188:1–7. doi: 10.1016/s0378-1119(96)00659-2. [DOI] [PubMed] [Google Scholar]
- 3.Tseng H, Green H. J Cell Biol. 1994;126:495–506. doi: 10.1083/jcb.126.2.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weiner L, Green H. Differentiation (Berlin) 1998;63:263–272. doi: 10.1046/j.1432-0436.1998.6350263.x. [DOI] [PubMed] [Google Scholar]
- 5.Yang Z-h, Gallicano G I, Yu Q-C, Fuchs E. J Cell Biol. 1997;137:657–669. doi: 10.1083/jcb.137.3.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mahoney M G, Tang W, Xiang M M, Moss S B, Gerton G L, Stanley J R, Tseng H. Biol Reprod. 1998;59:388–394. doi: 10.1095/biolreprod59.2.388. [DOI] [PubMed] [Google Scholar]
- 7.Iuchi, S., Easley, K., Matsuzaki, K., Weiner, L., O’Connor, N. & Green, H. (1999) Exp. Dermatol., in press. [DOI] [PubMed]
- 8.Iuchi S, Green H. Proc Natl Acad Sci USA. 1997;94:7948–7953. doi: 10.1073/pnas.94.15.7948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Iuchi S. J Biol Chem. 1993;268:23972–23980. [PubMed] [Google Scholar]
- 10.Brenowitz M, Senear D F, Kingston R E. In: DNase I Footprint Analysis of Protein–DNA Binding. Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Vol. 2. New York: Wiley; 1989. pp. 12.4.1–12.4.16. [Google Scholar]
- 11.Maxam A M, Gilbert W. Methods Enzymol. 1980;65:499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- 12.Vinson C R, LaMarco K L, Johnson P F, Landschulz W H, McKnight S L. Genes Dev. 1988;2:801–806. doi: 10.1101/gad.2.7.801. [DOI] [PubMed] [Google Scholar]
- 13.Fan C-M, Maniatis T. Genes Dev. 1990;4:29–42. doi: 10.1101/gad.4.1.29. [DOI] [PubMed] [Google Scholar]
- 14.Keller A D, Maniatis T. Mol Cell Biol. 1992;12:1940–1949. doi: 10.1128/mcb.12.5.1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zweidler-McKay P A, Grimes H L, Flubacher M M, Tsichlis P N. Mol Cell Biol. 1996;16:4024–4034. doi: 10.1128/mcb.16.8.4024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miesfield R, Arnheim N. Nucleic Acids Res. 1982;10:3933–3949. doi: 10.1093/nar/10.13.3933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Financsek I, Mizumoto K, Mishima Y, Muramatsu M. Proc Natl Acad Sci USA. 1982;79:3092–3096. doi: 10.1073/pnas.79.10.3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haltiner M M, Smale S T, Tjian R. Mol Cell Biol. 1986;6:227–235. doi: 10.1128/mcb.6.1.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bell S P, Learned R M, Jantzen H-M, Tjian R. Science. 1988;241:1192–1197. doi: 10.1126/science.3413483. [DOI] [PubMed] [Google Scholar]
- 20.Learned R M, Learned T K, Haltiner M M, Tjian R T. Cell. 1986;45:847–857. doi: 10.1016/0092-8674(86)90559-3. [DOI] [PubMed] [Google Scholar]
- 21.El-Deiry W S, Kern S E, Pietenpol J A, Kinzler K W, Vogelstein B. Nat Genet. 1992;1:45–49. doi: 10.1038/ng0492-45. [DOI] [PubMed] [Google Scholar]
- 22.Eiglmeier K, Honore N, Iuchi S, Lin E C C, Cole S T. Mol Microbiol. 1989;3:869–878. doi: 10.1111/j.1365-2958.1989.tb00236.x. [DOI] [PubMed] [Google Scholar]
- 23.Ptashne M. A Genetic Switch: Gene Control and Phage λ. Cambridge, MA; Blackwell Sci., Palo Alto, CA: Cell Press; 1986. [Google Scholar]
- 24.Jones M H, Learned R M, Tjian R. Proc Natl Acad Sci USA. 1988;85:669–673. doi: 10.1073/pnas.85.3.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bell S P, Jantzen H M, Tjian R. Genes Dev. 1990;4:943–954. doi: 10.1101/gad.4.6.943. [DOI] [PubMed] [Google Scholar]
- 26.Jantzen H-M, Admon A, Bell S P, Tjian R. Nature (London) 1990;344:830–836. doi: 10.1038/344830a0. [DOI] [PubMed] [Google Scholar]
- 27.Kihm A J, Hershey J C, Haystead T A J, Madsen C S, Owens G K. Proc Natl Acad Sci USA. 1998;95:14816–14820. doi: 10.1073/pnas.95.25.14816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tuan J A C, Zhai W, Comai L. Mol Cell Biol. 1999;19:2872–2879. doi: 10.1128/mcb.19.4.2872. [DOI] [PMC free article] [PubMed] [Google Scholar]