Abstract
Bread wheat quality is mainly correlated with high molecular weight glutenin subunits (HMW-GS) of endosperm. The number of HMW-GS alleles with good processing quality is limited in bread wheat cultivars, while there are plenty of HMW-GS alleles in wheat-related grasses to exploit. We report here on the cloning and characterization of HMW-GS alleles from the decaploid Agropyron elongatum. Eleven novel HMW-GS alleles were cloned from the grass. Of them, five are x-type and six y-type glutenin subunit genes. Three alleles Aex4, Aey7, and Aey9 showed high similarity with another three alleles from the diploid Lophopyrum elongatum, which provided direct evidence for the Ee genome origination of A. elongatum. It was noted that C-terminal regions of three alleles of the y-type genes Aey8, Aey9, and Aey10 showed more similarity with x-type genes than with other y-type genes. This demonstrates that there is a kind of intermediate state that appeared in the divergence between x- and y-type genes in the HMW-GS evolution. One x-type subunit, Aex4, with an additional cysteine residue, was speculated to be correlated with the good processing quality of wheat introgression lines. Aey4 was deduced to be a chimeric gene from the recombination between another two genes. How the HMW-GS genes of A. elongatum may contribute to the improvement of wheat processing quality are discussed.
Introduction
High molecular weight glutenin subunits (HMW-GS) are conserved endosperm storage proteins in the seeds of wheat and wheat-related species (Lawrence and Shepherd 1981; Shewry et al. 1995, 2003a). They explain up to 70% of the variation in bread making performance among European wheat cultivars (Branlard and Dardevet 1985; Payne et al. 1987, 1988), despite they only accounting for up to about 12% of the total protein in the endosperm of common wheat (Halford et al. 1992).
Due to the importance of HMW-GS to the improvement of wheat processing quality, genes encoding these subunits have been cloned from wheat and wheat-related species (Forde et al. 1985; Sugiyama et al. 1985; Thompson et al. 1985; Halford et al. 1987; Anderson and Greene 1989; Anderson et al. 1989; Halford et al. 1992; Reddy and Appels 1993; De Bustos et al. 2001; Wan et al. 2002; Liu et al. 2003; Wang et al. 2004; Guo et al. 2005; Wang et al. 2006; Yan et al. 2006). It has been confirmed that the HMW-GS genes are located on the long arms of the homoeologous group 1 chromosomes of hexaploid bread wheat at loci designated as Glu-1 (Lawrence and Shepherd 1980; Payne et al. 1980; Lawrence and Shepherd 1981; Payne et al. 1982). Each locus consists of two tightly linked genes which encode two types of subunits, the greater one termed x-type and the smaller one y-type (Harberd et al. 1986). Complete amino acid sequences of these subunits include three distinct domains: two highly conserved N- and C-terminal domains and a central repetitive domain. The central repetitive domains of both x- and y-type subunits comprising of hexapeptide and nonapeptide motifs while x-type subunits also contain tripeptide motifs.
The sequences of known HMW-GS genes shed light on how the different allelic genes have evolved and diverged. The similarity in structures of different HMW glutenin subunits indicates that they probably evolved from the same ancestor (Shewry and Tatham 1990; Shewry et al. 1995). Analyses through aligning N- and C-terminal sequences of some known HMW-GS confirmed that x- and y-type subunits represent two different subclasses, which indicated that the first step in the evolutionary process of HMW subunits was the duplication of a single ancestral gene into two closely linked copies (Wan et al. 2002; Shewry et al. 2003b). These copies diverged to be distinguishable (x- and y-type) before the speciation of wheat and wheat-related species (Shewry et al. 2003b).
Decaploid Agropyron elongatum (syn. Lophopyrum elongatum = Thinopyrum ponticum, 2n = 10x = 70) has many excellent characteristics such as high content of seed protein and high resistance to stress (Xia et al. 2003). So, it is an important resource for improving cultivated wheat (Triticum aestivum L). A great deal of hybrid cultivars with good processing quality were derived from sexual hybridization between A.elongatum and T. aestivum, e.g., Xiaoyan no. 6 (1, 14 + 15, 2 + 12) (Zhou et al. 1995) and Xiaoyan no. 54 (1, 14 + 15, 2 + 12); moreover, some somatic hybrid introgression lines with good processing quality were obtained from somatic hybrids (Xia et al. 2003; Liu et al. 2006) between T. aestivum cv. Jinan 177 (7 + 9, 2 + 12) and A. elongatum (most of the HMW-GS of A. elongatum are not clear before this report). A series of novel HMW-GS correlated with good bread-making quality were present in these hybrid progenies (Zhao et al. 2003; Feng et al. 2004a; Liu et al. 2006). It is necessary to investigate the contribution of HMW-GS of A. elongatum to those of hybrids.
In the work reported in this paper, we cloned and sequenced the open reading frames (ORFs) encoding HMW-GS from the decaploid A. elongatum. These results could enable us to compare the primary structure of HMW-GS from this wheat related polyploid with the published HMW-GS from wheat and other wheat-related grasses. In addition, the result will also assist us to understand the evolutionary process of HMW-GS genes in this decaploid grass and even the process of allopolyploidization of many Triticeae species.
Materials and methods
Cloning and sequencing of HMW glutenin gene ORFs
Seeds of A. elongatum stored in our laboratory were grown for 20 days at room temperature. Genomic DNA was extracted from a single seedling by the CTAB method according to Murray and Thompson (1980). HMW subunit genes contain no introns; so, genomic DNA is suitable as a template for PCR amplification of the entire coding region. In order to amplify the complete ORFs of HMW-GS genes of A. elongatum via genomic PCR, we designed a pair of degenerate primers according to published DNA sequences of HMW-GS alleles of wheat and wheat-related grasses. The sequences of the two primers were P1 (5′-ATGGCTAAGCGGC/TTA/GGTCCTCTTTG-3′) and P2 (5′-CTATCACTGGCTA/GGCCGACAATGCG-3′), respectively. Genomic PCR was carried out using the LA Taq polymerase (TaKaRa Biotechnology) with GC buffer for GC-rich template. The parameters for the reaction were: one cycle at 95°C for 5 min, followed by 30 cycles of 94°C for 40 s, 68°C for 4 min, and a final extension step at 72°C for 7 min. PCR products were separated in 1.0% agarose gels. All of the amplicon were recovered from the agarose gels and cloned into pUCm-T vector, then transferred into Escherichia coli DH10B competent cells. By restriction enzyme digestion mapping and terminal DNA sequencing, we found a series of new inserts that are not published before. To determine the complete DNA sequences of selected inserts, a series of subclones were prepared for each insert using the nested deletion method of Sambrook et al. (1989). Sequencing was performed commercially (Invitrogen). Sequence analyses were performed with the help of MEGA (Version 3.1, Kumar et al. 2004) and programs from the NCBI and EBI networks.
Bacterial expression of cloned ORFs
For bacterial expression of the mature proteins of HMW-GS from A. elongatum, two sets of PCR primers were designed for amplifying mutant ORFs without signal peptides and introducing appropriate restriction enzyme sites for the mutant ORFs to facilitate subsequent cloning experiments. The sequence of forward primer is PF: 5′-ACCCATATGGAAGGTGAGGCCTCT-3′, while the sequences of two reverse primers are PR1: 5′-CTAGAATTCCTATCACTGGCTGGCCGA (for Aex1, Aex4, Aey7, and Aey9) and PR2: 5′-CTAGAATTCCTATCACTGGC TAGCCGA (for Aey2). Introduced restriction site is Nde I for forward primer and EcoR I for both reverse primers, respectively. Mutant ORFs of these five alleles were cloned into the expression vector pET-24a (Novagen). The constructs were transferred into E. coli DE3 competent cells (Promega) for inducing bacterial expression. Inducement of bacterial expression of these five alleles was carried out according to Sambrook et al. (1989). Expressed proteins were extracted by dissolving cells in SDS-PAGE sample buffer (Wan et al. 2002).
Results
HMW-GS genes in A. elongatum
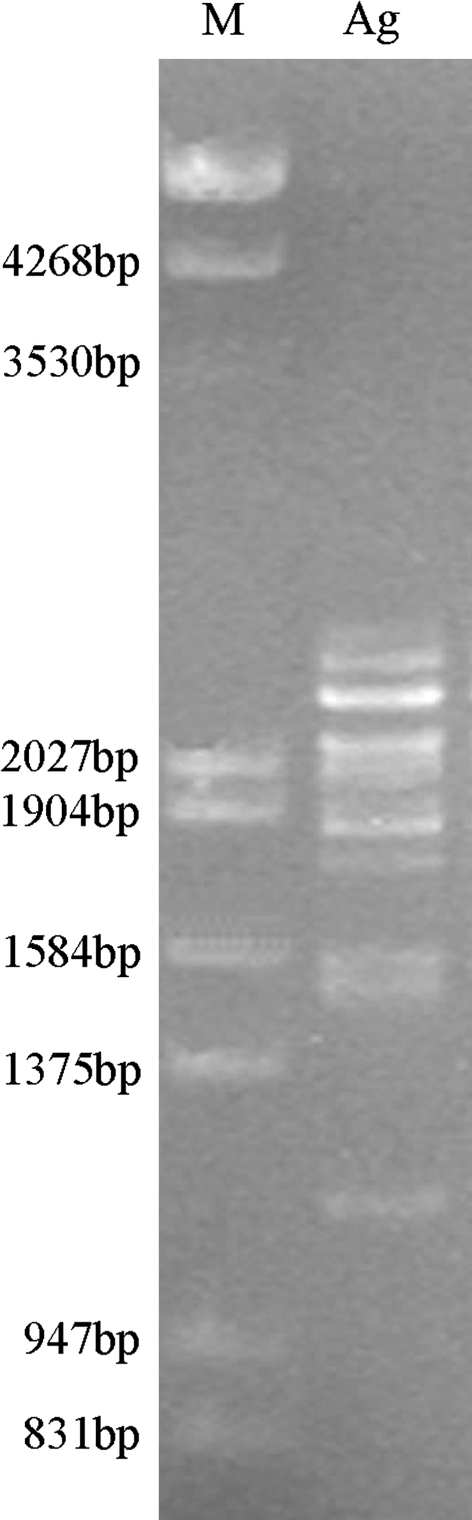
The amplicon of A. elongatum genomic DNA included over ten bands in all (Fig. 1). We reclaimed and cloned all the amplicon together. After restriction enzyme digestion mapping and terminal DNA sequencing, we confirmed that 15 different inserts were obtained; five for x-type subunits and ten for y-type subunits. These inserts were designated as Aex1–Aex5 (x-type) and Aey1–Aey10 (y-type) according to their type and length. The length of these sequences shows a very large range, with the largest, Aex1, containing 2,424 base pairs while the smallest, Aey10, comprised only 1,150 base pairs. The latter is one of the smallest known HMW-GS genes identified to date. The five x-type sequences were larger than most of the y-type genes except that Aex3 (2,184 bp), Aex4 (2,082 bp), and Aex5 (2,004 bp) were smaller than the largest y-type allele Aey1 (2,219 bp). Of the 15 HMW-GS alleles, Aey2, Aey6, Aey8, and Aey9 were the same as those published by Feng et al. (2004b, c) in our lab, while the other 11 were found for the first time, which included five x-type and six y-type ones.
Fig. 1.
PCR amplification of HMW-GS coding sequences from genomic DNA of A. elongatum. M lambda DNA digested by EcoR I + Hind III, Ag amplicon of A. elongatum
Expression of the HMW-GS genes in bacterial cells
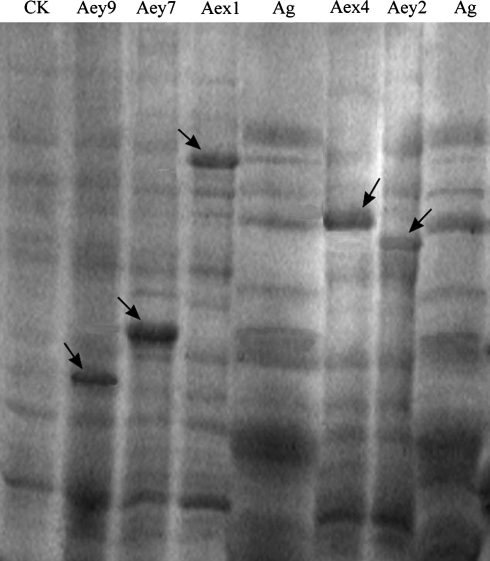
For validating the correspondence of the HMW-GS genes we cloned with the HMW-GS proteins in the seeds of A. elongatum, we expressed five alleles with intact ORFs in bacterial cells. The signal peptide sequences were removed from the ORFs by PCR mutagenesis. After cloning the modified ORFs into the pET-24a vector, we chose five expression constructs, pET–Aex1, pET–Aex4, pET–Aey2, pET–Aey7, and pET–Aey9 to express the mature proteins in bacterial cells. In SDS-PAGE analysis of proteins extracted from induced bacterial cells, the electrophoretic mobility of proteins directed by pET–Aex1, pET–Aex4, and pET–Aey7 was similar to three subunits extracted from seeds of A. elongatum (Fig. 2). However, we have not found proteins from the seeds that showed similar migration with the proteins directed by pET–Aey2 and pET–Aey9 in bacterial cells (Fig. 2).
Fig. 2.
Expression of the modified ORFs of five alleles Aex1, Aex4, Aey2, Aey7, and Aey9 in E. coli and SDS-PAGE analysis of expressed products. The modified ORFs were prepared by removing the signal peptide sequence from each of the sequences by mutagenesis. Protein extracts were prepared by dissolving cells directly in SDS-PAGE sample buffer. The glutenin proteins synthesized in E. coli directed by Aex1, Aex4, and Aey7 under IPTG induction showed identical electrophoretic mobility to those from seeds of A. elongatum (shown by arrows). No proteins from seeds of A. elongatum displayed similar mobility with those directed by Aey2 and Aey9 in bacteria (shown by arrows). CK proteins extracted from bacteria harboring pET–Aex1 without IPTG induction for control, Ag proteins extracted from seeds of A. elongatum
Derived amino acid sequences of HMW-GS genes
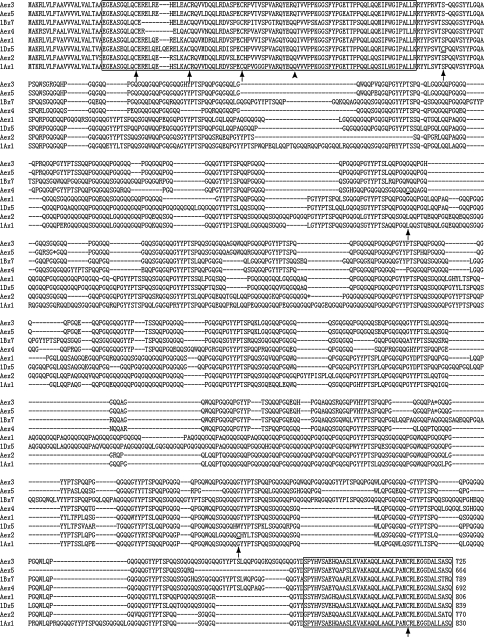
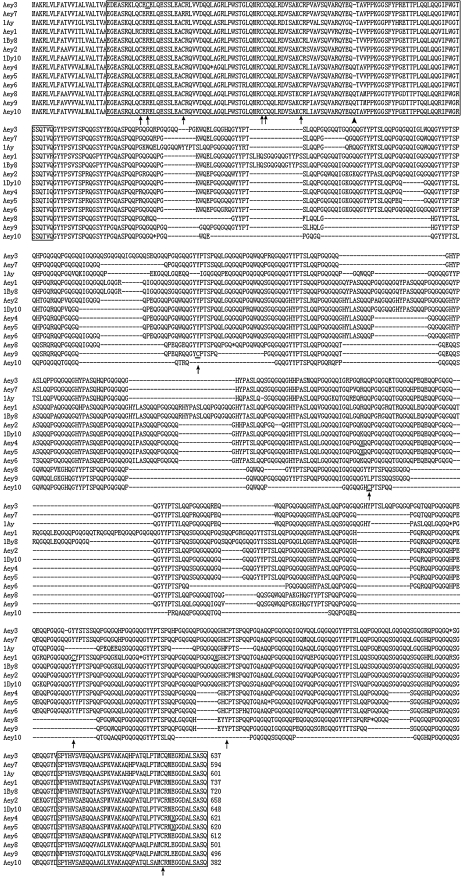
Analysis of the amino acid sequences deduced by the DNA sequences showed that 11 HMW-GS possessed a typical primary structure shared by other published HMW-GS (Figs. 3, 4). Each subunit consists of a signal peptide of 21 amino acid residues, a conserved N-terminal region, a central repetitive domain, and a conserved C-terminal region (Figs. 3, 4). The N-terminal regions of x-type subunits possess 86 amino acid residues except that Aex4 contains only 81 amino acid residues, while that of y-type subunits include 104 amino acid residues except that Aey4, Aey8, Aey9, and Aey10 have 105 amino acid residues. N-terminal regions of these four y-type subunits contain an extra glutamine (Q) residue compared to other y-type subunits with only 104 residues (Fig. 4). This glutamine residue is also present in all the known x-type subunits. Conserved C-terminal regions of all the 11 subunits comprise 42 amino acid residues. Central repetitive region of all the 11 subunits consists of hexapeptide and nonapeptide motifs; however, the five x-type subunits also contain tripeptides (GQQ), which is the typical character of x-type HMW glutenin subunits. Difference between these subunits and those from wheat is mainly due to single residue substitution and insertion or deletion of repeat motifs in central repetitive region (Figs. 3, 4).
Fig. 3.
Comparison of primary structure of five x-type subunits from A. elongatum with that of three representative x-type subunits from common wheat. The N- and C-terminal regions were boxed. The tailed arrows indicated the cysteine residues and the additional cysteine residues of 1Dx5, Aex2, and Aex4 were underlined. The glutamine (Q) residues conserved in N-terminal domain of x-type subunits but absent from most y-type subunits were shown by non-tailed arrows. The in-frame stop codon was represented by asterisk. The Genbank accession numbers of these sequences were displayed in Table 1
Fig. 4.
Comparison of primary structure of all the ten y-type subunits from A. elongatum and that of three representative y-type subunits from common wheat. The N- and C-terminal regions were boxed. The tailed arrows indicated the cysteine residues and additional cysteine residues of Aey1, Aey3, and Aey10 were underlined. The sequences of Aey1, Aey4, and Aey5 were rectified to diminish the influence of frame shift. The revised amino acid residues were substituted by underlined X. The in-frame stop codon was represented by asterisk. The extra glutamine (Q) residues in N-terminal domain of Aey4, Aey8, Aey9, and Aey10 were shown by non-tailed arrows. The Genbank accession numbers of these sequences were displayed in Table 1
All the conserved cysteine residues that present in published HMW-GS of wheat and wheat-related grasses were observed in the deduced amino acid sequences of the 11 alleles except that the conserved cysteine (TGC) mutated to arginine (CGC) at position 43 of the N-terminal region of Aey7 (Fig. 4). Additional cysteine residues were observed at position 659, 261, 581, and 33 of the deduced amino acid sequences of Aex2, Aex4, Aey1, and Aey3, respectively. The cysteine residues situated on the bottom of repetitive regions of most y-type subunits were absent from Aey10, while an extra cysteine residue appeared at its position 257. The detailed properties of the 11 subunits and some representative subunits of wheat are summarized in Table 1.
Table 1.
A summary of properties of the primary structure of HMW-GS from A. elongatum in comparison with some HMW-GS of common wheat
| Subunit | Signal peptide | N-terminal region | Repetitive region | C-terminal region | Total | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Name | Accession number | Size | Size | Cys | Size | Cys | Size | Cys | Size | Cys |
| 1Ax1 | X61009 | 21 | 86 | 3 | 681 | 0 | 42 | 1 | 830 | 4 |
| 1Bx7 | X13927 | 21 | 81 | 3 | 645 | 0 | 42 | 1 | 789 | 4 |
| 1Dx5 | X12928 | 21 | 89 | 3 | 687 | 1b | 42 | 1 | 839 | 5 |
| Aex1 | DQ478575 | 21 | 86 | 3 | 657 | 0 | 42 | 1 | 806 | 4 |
| Aex2 | DQ478576 | 21 | 86 | 3 | 623 | 1b | 42 | 1 | 772 | 5 |
| Aex3 | DQ478574 | 21 | 86 | 3 | 577 | 0 | 42 | 1 | 726 | 4 |
| Aex4 | DQ534448 | 21 | 81 | 3 | 548 | 1b | 42 | 1 | 692 | 5 |
| Aex5 | EF190195 | 21 | 86 | 3 | 517 | 0 | 42 | 1 | 666 | 4 |
| 1Ay | X03042 | 21 | 104 | 5 | 420 | 0 | 42 | 1 | 587 | 6 |
| 1By8 | AY245797 | 21 | 104 | 5 | 553 | 1 | 42 | 1 | 720 | 7 |
| 1Dy10 | X12929 | 21 | 104 | 5 | 481 | 1 | 42 | 1 | 648 | 7 |
| Aey1 | AY899822 | 21 | 104 | 5 | 571 | 2b | 42 | 1 | 738 | 8 |
| Aey2a | AY263343 | 21 | 104 | 5 | 491 | 1 | 42 | 1 | 658 | 7 |
| Aey3 | EF190196 | 21 | 104 | 6c | 472 | 1 | 42 | 1 | 639 | 8 |
| Aey4 | EF190197 | 21 | 105 | 5 | 454 | 1 | 42 | 1 | 622 | 7 |
| Aey5 | EF190198 | 21 | 104 | 5 | 454 | 1 | 42 | 1 | 621 | 7 |
| Aey6a | AY263344 | 21 | 104 | 5 | 445 | 1 | 42 | 1 | 612 | 7 |
| Aey7 | DQ078273 | 21 | 104 | 4d | 427 | 1 | 42 | 1 | 594 | 6 |
| Aey8a | AY319518 | 21 | 105 | 5 | 335 | 0 | 42 | 1 | 503 | 6 |
| Aey9a | AY264065 | 21 | 105 | 5 | 328 | 1 | 42 | 1 | 496 | 7 |
| Aey10 | DQ078274 | 21 | 105 | 5 | 215 | 1 | 42 | 1 | 383 | 7 |
aAey2, Aey6, Aey8, and Aey9 were cloned by Feng et al. (2004b)
b1Dx5, Aex2, Aex4, and Aey1 contain extra cysteine residues in central repetitive domain
cAey3 contains an additional cysteine in N-terminal domain
dThe N-terminal domain of Aey7 has one cysteine residue less than other y-type subunits
Out of the 11 novel alleles, only three (Aex1, Aex4, and Aey7) have intact open reading frames; five (Aex2, Aex3, Aex5, Aey3, and Aey10) contain in-frame stop codon; the remainder three (Aey1, Aey4, and Aey5) show frame shift mutation resulting from single nucleotide insertion or deletion. The analysis of amino acid sequences derived from Aey1, Aey4, and Aey5 is based on the rectified sequences. It is interesting that except N-terminal region, repetitive region and C-terminal region of Aey4 and Aey5 were nearly identical, with only three SNPs. The N-terminal region of Aey4 is the same as Aey10 with only a SNP, and both of them contain an additional glutamine residue than most y-type subunits (Fig. 4).
Evolutionary relationship
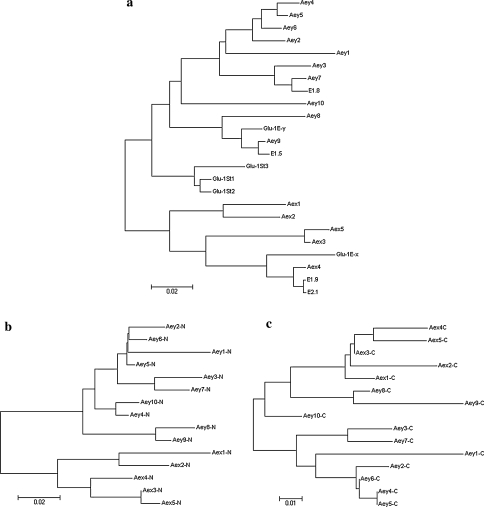
To investigate evolutionary relationships among the subunits characterized in this study and the published HMW glutenin subunits cloned from Lophopyrum elongatum (Ee) and Pseudoroegneria stipifolia (St), phylogenetic trees were drawn from the alignment of these alleles based on both full length sequences and the two conserved terminal sequences (Fig. 5). Alignment according to full length sequences indicated that the phylogenetic tree was divided into two halves, comprising the alleles of y-type genes at the top and x-type genes at the bottom. The ten y-type alleles of A. elongatum have been further divided into five clades. Of them, Aey2, Aey4, Aey5, and Aey6 showed close relationship, while Aey3 with Aey7 and Aey8 with Aey9 clustered together, respectively; moreover, Aey1 and Aey10 were located in two independent branches. The five x-type sequences were subdivided into three clades. Aex1 with Aex2 and Aex3 with Aex5 stayed together, respectively, while Aex4 was far from the other four sequences. It was noted from the phylogenetic tree that the published y-type HMW-GS alleles E1.5 and Glu-1E-y from the diploid L. elongatum were more similar to Aey9 of A. elongatum; another y-type allele E1.8 was closer with Aey7 of A. elongatum, while two x-type alleles E2.1 and E1.9 from the diploid L. elongatum showed higher similarity to Aex4 than any other alleles of A. elongatum. Sequence alignment also indicated that the sequences of Aex4, Aey7, and Aey9 displayed homology to those of E2.1, E1.8 (Wang et al. 2006) and E1.5 (Wang et al. 2004) from the diploid L. elongatum (Ee), respectively (data not shown). HMW-GS sequences from the diploid P. stipifolia were very smaller in length than most alleles from A. elongatum and all of them did not exhibit homology to those from A. elongatum (Fig. 5a).
Fig. 5.
Phylogenetic analysis of HMW-GS from A. elongatum and some other wheat-related grass. a Neighbor-Joining tree of full length sequences of Glu-1 genes from A. elongatum and some other wheat-related grass. b Neighbor-Joining tree of N-terminal regions of HMW-GS from A. elongatum. c Neighbor-Joining tree of C-terminal regions of HMW-GS from A. elongatum. This work was done under the help of MEGA program (Version 3.1)
As observed for the tree based on full length sequences, phylogenetic trees based on the N- and C-terminal region sequences can be divided into two halves. The relationship of all 15 sequences from A. elongatum reflected by trees of terminal sequences was similar to those in the full length sequences. However, there was one obvious difference in the position of Aey4: the N-terminal region of Aey4 showed a higher degree of relatedness to that of Aey10 (Fig. 5b), while the full length sequence and C terminal region of this gene were closer to Aey5 (Fig. 5a, c). Another difference was that the C-terminal sequences of Aey8, Aey9, and Aey10 showed more similarity with x-type genes than with the other seven y-type genes (Fig. 5c).
Discussion
Previous research indicates that each genome of wheat and its wild-related grasses, all contain a locus consisting of two tightly linked HMW-GS genes. Hence, we deduce that there may be ten pairs of tightly linked HMW-GS alleles in cross-pollinated decaploid A. elongatum. We have cloned 15 HMW-GS alleles from a seedling of A. elongatum in all, including ten y-type and five x-type alleles. The reason why we have not obtained the other x-type alleles may be that the degenerate primer pairs we used did not match those sequences very well and/or that the x-type alleles were less polymorphic or heterozygous than the y-type ones.
Five HMW-GS alleles from A. elongatum were successfully expressed in E. coli and three of the proteins directed by Aex1, Aex4, and Aey7 have the same mobility with those from the seeds; thus, they were the coding genes for the three subunits. However, we have not found proteins from the seeds that showed similar migration with those directed by Aey2 and Aey9 in bacteria, which may be due to silencing of the two alleles in the seeds. The reason for this is not clear, but it has been reported that inactivation of promoter leads to the silencing of 1Ay of bread wheat (Halford et al. 1989).
Decaploid A. elongatum is an allopolyploid, but there is still controversy about the composition of its genome. Based on the results of cytogenetics, biochemistry, RAPD, and ISH, Zhang et al. (1996) speculated that the genome composition of decaploid A. elongatum was StStStStEeEeEbEbExEx, and the St genome might come from Pseudoroegneria while the E genome derived from the diploid Thinopyrum elongatum (L. elongatum, EeEe) and Thinopyrum Bessarabicum (EbEb). Sequence alignment of HMW-GS genes from the decaploid A. elongatum with that of the diploid L. elongatum and P. stipifolia indicated that Aex4, Aey7, and Aey9 from A. elongatum showed very high similarity with three alleles from the diploid L. elongatum (Fig. 5), which confirmed that the diploid L. elongatum was the donor of the Ee genome of A. elongatum. Because all the 15 alleles from A. elongatum showed no high similarity with those of P. stipifolia, we concluded that the diploid P. stipifolia might not be the ancestor of the decaploid A. elongatum, while the St genome of A. elongatum might come from other species of the Pseudoroegneria genera.
The C-terminal region of three y-type alleles Aey8, Aey9, and Aey10 showed higher similarity with the five x-type alleles than other y-type alleles (Fig. 5). Therefore, the structure of these three y-type subunits was not as typical as that of other y-type ones. Their structures lied between x- and y-type but inclined to y-type subunits. The extra glutamine residues in N-terminal of Aey8, Aey9, and Aey10 also presented in x-type subunits, while this residue was absent from most y-type subunits. Thus, we speculated that this glutamine residue might be deleted after the divergence of x- and y-type subunits, and Aey8, Aey9, and Aey10 were older than those subunits that did not contain this residue. Furthermore, we concluded that there was a kind of intermediate state in the divergence between x- and y-type subunits.
The strange structure of Aey4 indicated that it might be a chimeric gene originating from recombination between Aey5 and Aey10. Chimeric HMW-GS gene has also been reported in Aegilops searsii (Sun et al. 2006). Through which way were these chimeric genes created was not known, but Wang et al. (2002) and Arguello et al. (2006) have referred that some new chimeric genes observed in Drosophila originated through retroposition and illegitimate recombination. Therefore, the mechanism of origination of chimeric HMW-GS gene might be the same as that found in Drosophila.
The greater part of HMW-GS genes from A. elongatum possess in-frame stop codon or frame shift mutation which result in their inability to express normal proteins in seeds. The reason for the appearance of so many mutations in HMW-GS genes may be due to the special structure of these subunits and their biological function. The amino acid sequences of HMW-GS include many glutamine (Gln) residues whose codons, CAA or CAG, can easily convert to stop codon TAA or TAG, respectively, in the process of evolution. The in-frame stop codons in these genes that we cloned are all due to this conversion. Such circs have also been reported in silent 1Ay and 1Ax from bread wheat (Forde et al. 1985; De Bustos et al. 2000) and 1Dx from Aegilops cylindrica (Wan et al. 2002). They were also present in gliadin and LMW subunit pseudogenes (Rafalski 1986; Anderson and Greene 1997). As seed storage proteins, the biological function of HMW glutenin subunits is to provide carbon, nitrogen, and energy sources for seed germination and seedling growth. The mutation or silence of such genes is not lethal for the plant, so the selection pressure on these genes is much lower than on other functional genes in evolution; this may be another reason for these genes to accumulate more mutation.
Four of the 15 subunits Aex2, Aex4, Aey1, and Aey3 contain extra cysteine residues in their amino acid sequences, even though only one subunit, Aex4, has intact ORFs. It has been shown that in all the known HMW-GS of wheat, only the 1Dx5 subunit possesses an additional cysteine residue in its structure and this cysteine exerts a positive influence on dough properties (Lafiandra et al. 1993; Gupta and MacRitchie 1994). Because of the presence of relatively more cysteine residues in glutenin subunits of A. elongatum than those of wheat, we predicted that this grass could provide HMW-GS genes for wheat quality improvement, e.g., the extra cysteine residue may confer on Aex4 potential value in improving the processing properties of wheat. Moreover, asymmetric somatic hybrid lines between common wheat and A. elongatum have been shown to possess a series of novel HMW-GS with good processing quality (Xia et al. 2003; Feng et al. 2004a; Liu et al. 2006). It has been proved that the presence of some novel HMW-GS in the hybrid lines was correlated with HMW-GS sequences from the donor A. elongatum. For example, Aex4 and Aey1 were found to be introgressed into some high quality hybrid lines from A. elongatum, while another novel hybrid allele H1Dy12 may be the outcome of recombination between Aey2 and another HMW-GS allele of Jinan177 (Liu et al. 2007). Therefore, HMW-GS genes of A. elongatum will contribute to the improvement of wheat processing quality and they deserve further investigation.
Acknowledgments
This work was supported by Projects of National 863 High Technology Research and Development (Project no. 2006AA100102), National Basic Research Program of China (Project no. 2006CB100100), and Key and Doctor Position of ministry of education.
Open Access This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
References
- Anderson OD, Greene FC (1989) The characterization and comparative analysis of high-molecular-weight glutenin genes from genomes A and B of a hexaploid wheat. Theor Appl Genet 77:689–700 [DOI] [PubMed]
- Anderson OD, Greene FC (1997) The α-gliadin gene family. II. DNA and protein sequence variation, subfamily structure, and origins of pseudogenes. Theor Appl Genet 95:59–65 [DOI]
- Anderson OD, Greene FC, Yip RE, Halford NG, Shewry PR, Malpica-Romero J-M (1989) Nucleotide sequences of the two high-molecular-weight glutenin genes from the D-genome of a hexaploid bread wheat, Triticum aestivum L. cv Cheyenne. Nucleic Acids Res 17:461–462 [DOI] [PMC free article] [PubMed]
- Arguello JR, Chen Y, Yang S, Wang W, Long M (2006) Origination of an X-linked testes chimeric gene by illegitimate recombination in Drosophila. PLoS Genet 2(5):e77 [DOI] [PMC free article] [PubMed]
- Branlard G, Dardevet M (1985) Diversity of grain protein and bread wheat quality. II. Correlation between high molecular weight subunits of glutenin and flour quality characteristics. J Cereal Sci 3:345–354
- De Bustos A, Rubio P, Jouve N (2000) Molecular characterization of the inactive allele of the gene Glu-A1 and the development of a set of AS-PCR markers for HMW glutenins of wheat. Theor Appl Genet 100:1085–1094 [DOI]
- De Bustos A, Rubio P, Jouve N (2001) Characterisation of two gene subunits on the 1R chromosome of rye as orthologs of each of the Glu-1 genes of hexaploid wheat. Theor Appl Genet 103:733–742 [DOI]
- Feng DS, Xia GM, Zhao SY, Chen FG (2004a) Two quality-associated HMW glutenin subunits in a somatic hybrid line between Triticum aestivum and Agropyron elongatum. Theor Appl Genet 110:136–144 [DOI] [PMC free article] [PubMed]
- Feng DS, Chen FG, Zhao SY, Xia GM (2004b) High-molecular-weight glutenin subunit genes in decaploid Agropyron elongatum. Acta Bot Sin 46:489–496
- Feng DS, Chen FG, Zhao SY, Xia GM (2004c) Study on a novel HMW glutenin subunit coding region from Agropyron elongatum (in Chinese with English Abstract). Acta Bot Boreal Occident Sin 24:237–242
- Forde J, Malpica J-M, Halford NG, Shewry PR, Anderson OD, Greene FC, Miflin BJ (1985) The nucleotide sequence of a HMW glutenin subunit gene located on chromosome 1A of wheat (Triticum aestivum L.). Nucleic Acids Res 13:6817–6832 [DOI] [PMC free article] [PubMed]
- Guo ZF, Yan ZH, Wang JR, Wei YM, Zheng YL (2005) Characterization of HMW prolamines and their coding sequences from Crithopsis delileana. Hereditas 142:56–64 [DOI] [PubMed]
- Gupta RB, MacRitchie F (1994) Allelic variation at glutenin subunit and gliadin loci, Glu-1, Glu-3 and Gli-1, of common wheats. II. Biochemical basis of the allelic effects on dough properties. J Cereal Sci 19:19–29 [DOI]
- Halford NG, Forde J, Anderson OD, Greene FC, Shewry PR (1987) The nucleotide and deduced amino acid sequences of an HMW glutenin subunit gene from chromosome 1B of bread wheat (Triticum aestivum L.) and comparison with those of genes from chromosomes 1A and 1D. Theor Appl Genet 75:117–126 [DOI]
- Halford NG, Forde J, Shewry PR, Kreis M (1989) Functional analysis of the upstream regions of a silent and an expressed member of a family of wheat seed protein genes in transgenic tobacco. Plant Sci 62:207–216 [DOI]
- Halford NG, Field JM, Blair H, Urwin P, Moore K, Robert L, Thompson R, Flavell RB, Tatham AS, Shewry PR (1992) Analysis of HMW glutenin subunits encoded by chromosome 1A of bread wheat (Triticum aestivum L.) indicates quantitative effects on grain quality. Theor Appl Genet 83:373–378 [DOI] [PubMed]
- Harberd NP, Bartels D, Thompson RD (1986) DNA restriction-fragment variation in the gene family encoding high molecular weight (HMW) glutenin subunits of wheat. Biochem Genet 24:579–596 [DOI] [PubMed]
- Kumar S, Tamura K, Nei M (2004) MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform 5:150–163 [DOI] [PubMed]
- Lafiandra D, D’Ovidio R, Porceddu E, Margiotta B, Colaprico G (1993) New data supporting high Mr glutenin subunit 5 as the determinant of quality differences among the pairs 5 + 10 vs. 2 + 12. J Cereal Sci 18:197–205 [DOI]
- Lawrence GJ, Shepherd KW (1980) Variation in glutenin protein subunits of wheat. Aust J Biol Sci 33:221–233 [DOI]
- Lawrence GJ, Shepherd KW (1981) Chromosomal location of genes controlling seed proteins in species related to wheat. Theor Appl Genet 59:25–31 [DOI] [PubMed]
- Liu Z, Yan Z, Wang Y, Liu K, Zheng Y, Wang D (2003) Analysis of HMW glutenin subunits and their coding sequences in two diploid Aegilops species. Theor Appl Genet 106:1368–1378 [DOI] [PubMed]
- Liu H, Shi L, Zhao JS, Xia GM (2006) Genetic characteristic of high molecular weight glutenin subunits in somatic hybrid wheat lines––potential application to wheat breeding. J Agric Food Chem 54:5007–5013 [DOI] [PubMed]
- Liu S, Zhao S, Chen F, Xia G (2007) Generation of novel high quality HMW-GS genes in two introgression lines of Triticum aestivum/Agropyron elongatum. BMC Evol Biol 7:76 [DOI] [PMC free article] [PubMed]
- Murray MG, Thompson WF (1980) Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res 8:4321–4325 [DOI] [PMC free article] [PubMed]
- Payne PI, Law CN, Mudd EE (1980) Control by homoeologous group 1 chromosomes of the high-molecular-weight subunits of glutenin, a major protein of wheat endosperm. Theor Appl Genet 58:113–120 [DOI] [PubMed]
- Payne PI, Holt LM, Worland AJ, Law CN (1982) Structural and genetical studies on the high-molecular-weight subunits of wheat glutenin. Part 3. Telocentric mapping of the subunit genes on the long arms of the homoeologous group 1 chromosomes. Theor Appl Genet 63:129–138 [DOI] [PubMed]
- Payne PI, Nightingale MA, Krattiger AF, Holt LM (1987) The relationship between HMW glutenin subunit composition and the breadmaking quality of British grown wheat varieties. J Sci Food Agric 40:51–65 [DOI]
- Payne PI, Holt LM, Krattiger AF, Carrillo JM (1988) Relationship between seed quality characteristics and HMW glutenin subunit composition determined using wheats grown in Spain. J Cereal Sci 7:229–235
- Rafalski JA (1986) Structure of wheat gamma-gliadin genes. Gene 43:221–229 [DOI] [PubMed]
- Reddy P, Appels R (1993) Analysis of a genomic DNA segment carrying the wheat high-molecular-weight (HMW) glutenin Bx17 subunit and its use as an RFLP marker. Theor Appl Genet 85:616–624 [DOI] [PubMed]
- Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory, New York
- Shewry PR, Tatham AS (1990) The prolamin storage proteins of cereal seeds: structure and evolution. Biochem J 267:1–12 [DOI] [PMC free article] [PubMed]
- Shewry PR, Napier JA, Tatham AS (1995) Seed storage proteins: structure and biosynthesis. Plant Cell 7:945–956 [DOI] [PMC free article] [PubMed]
- Shewry PR, Halford NG, Tatham AS, Popineau Y, Lafiandra D, Belton PS (2003a) The high molecular weight subunits of wheat glutenin and their role in determining wheat processing properties. Adv Food Nutr Res 45:219–302 [DOI] [PubMed]
- Shewry PR, Halford NG, Lafiandra D (2003b) Genetics of wheat gluten proteins. Adv Genet 49:111–184 [DOI] [PubMed]
- Sugiyama T, Rafalski A, Peterson D, Söll D (1985) A wheat HMW glutenin subunit gene reveals a highly repeated structure. Nucleic Acids Res 13:8729–8737 [DOI] [PMC free article] [PubMed]
- Sun X, Hu SL, Liu X, Qian WQ, Hao ST, Zhang AM, Wang DW (2006) Characterization of the HMW glutenin subunits from Aegilops searsii and identification of a novel variant HMW glutenin subunit. Theor Appl Genet 113:631–641 [DOI] [PubMed]
- Thompson RD, Bartels D, Harberd NP (1985) Nucleotide sequence of a gene from chromosome 1D of wheat encoding a HMW-glutenin subunit. Nucleic Acids Res 13:6833–6846 [DOI] [PMC free article] [PubMed]
- Wang W, Brunet FG, Nevo E, Long M (2002) Origin of Sphinx, a young chimeric RNA gene in Drosophila melanogaster. Proc Natl Acad Sci USA 99:4448–4453 [DOI] [PMC free article] [PubMed]
- Wang JR, Yan ZH, Wei YM, Zheng YL (2004) A novel high-molecular-weight glutenin subunit gene Ee1.5 from Elytrigia elongata (Host) Nevski. J Cereal Sci 40:289–294 [DOI]
- Wang JR, Yan ZH, Wei YM, Zheng YL (2006) Characterization of high molecular weight glutenin subunit genes from Elytrigia elongata. Plant Breed 125:89–95 [DOI]
- Wan Y, Wang D, Shewry PR, Halford NG (2002) Isolation and characterization of five novel high molecular weight subunit of glutenin genes from Triticum timopheevi and Aegilops cylindrica. Theor Appl Genet 104:828–839 [DOI] [PubMed]
- Xia GM, Xiang FN, Zhou AF, Wang H, Chen HM (2003) Asymmetric somatic hybridization between wheat (Triticum aestivum L.) and Agropyron elongatum (Host) Nevishi. Theor Appl Genet 107:299–305 [DOI] [PubMed]
- Yan ZH, Wei YM, Wang JR, Liu DC, Dai SF, Zheng YL (2006) Characterization of two HMW glutenin subunit genes from Taenitherum Nevski. Genetica 127:267–276 [DOI] [PubMed]
- Zhang XY, Dong YS, Wang RRC (1996) Characterization of genomes and chromosomes in partial amphiploids of the hybrid Triticum aestivum × Thinopyrum ponticum by in situ hybridization, isozyme analysis and RAPD. Genome 39:1062–1071 [DOI] [PubMed]
- Zhao TJ, Quan TY, Xia GM, Chen HM (2003) Glutenin and SDS sedimentation analysis of the F5 somatic hybrids between Triticum aestivum L. and Agropyron elongatum (in Chinese with English Abstract). J Shandong Univ (Nat Sci) 38(3):112–116
- Zhou HP, Li B, Li ZS (1995) The study of breeding blue-grain gene translocation of wheat (in Chinese with English Abstract). Acta Bot Boreal Occident Sin 15:125–128