Abstract
Heritability estimates of birth weight have been inconsistent. Possible explanations are heritability changes during gestational age or the influence of covariates (e.g. chorionicity). The aim of this study was to model birth weights of twins across gestational age and to quantify the genetic and environmental components. We intended to reduce the common environmental variance to increase heritability and thereby the chance of identifying candidate genes influencing the genetic variance of birth weight. Perinatal data were obtained from 4232 live-born twin pairs from the East Flanders Prospective Twin Survey, Belgium. Heritability of birth weights across gestational ages was estimated using a non-linear multivariate Gaussian regression with covariates in the means model and in covariance structure. Maternal, twin-specific, and placental factors were considered as covariates. Heritability of birth weight decreased during gestation from 25 to 42 weeks. However, adjusting for covariates increased the heritability over this time period, with the highest heritability for first-born twins of multipara with separate placentas, who were staying alive (from 52% at 25 weeks to 30% at 42 weeks). Twin-specific factors revealed latent genetic components, whereas placental factors explained common and unique environmental factors. The number of placentas and site of the insertion of the umbilical cord masked the effect of chorionicity. Modeling genetic and environmental factors leads to a better estimate of their role in growth during gestation. For birth weight, mainly environmental factors were explained, resulting in an increase of the heritability and thereby the chance of finding genes influencing birth weight in linkage and association studies.
Keywords: Birth weight, Heritability, Twins, Gestational age, Growth curves, Variance.
Introduction
Birth weight is a complex trait in which both environmental and genetic factors are involved. Low birth weight is not only associated with higher neonatal morbidity and mortality (Kramer et al. 2005), but also with post-natal psychopathology (Wichers et al. 2002). It is even stated that birth weight is a causal risk factor for child problem behavior and that the effects may well extend into adulthood (van Os et al. 2001).
Heritability estimates for birth weight using twin pairs, based on structural equation modeling or study of the offspring of twins, have given inconsistent results ranging from 10 to 40% (Baird et al. 2001; Clausson et al. 2000; Hur et al. 2005; Luciano et al. 2004; Magnus et al. 2001; van Baal and Boomsma 1998; van Dommelen et al. 2004; Vlietinck et al. 1989; Whitfield et al. 2001). The relatively low heritability estimates may be explained by unequal sharing of nutrients or unequal placental blood supply to the twin pair (Machin 1996). Vlietinck et.al. (Vlietinck et al. 1989) revealed that chorionicity explained 12% of the variance of birth weight with DC twins having a significantly higher similarity than MC twins. Random environment explained 23%, additive genetic factors 22.5%, and maternal age 0.7% of the total variance. But, the largest source of variation on birth weight in this study was gestational age (42%). Until now however, no study has focused on the change of variance of birth weight during gestation. We have shown that this variance increases during gestation (Gielen et al. 2007). As a consequent, heritability of birth weight may change over gestational age or different covariates may explain different parts of the variance-covariance structure. For example, it is more likely that at 37 weeks of gestation, nutrient factors (environmental factor) will explain a larger part of the covariance matrix as compared to 13 weeks of gestation.
The advantage of twin studies, is that the total variance can be split up into genetic (A), shared or common environmental (C) and unique environmental (residual, E) components, enabling an accurate estimation of heritability (= genetic variance/total variance) (Boomsma et al. 2002). The degree of heritability determines the power to localize and identify the genes involved (Snieder et al. 2003). For linkage and association studies a high degree of heritability is preferable. Since heritability is the proportion of phenotypic variation of a trait that can be attributed to genetic variation (Boomsma et al. 2002), reducing environmental and residual components will give an increase of the heritability. The latter can be achieved by adjusting for covariates that explain part of the environmental factors in the analysis. Additionally, a covariate may also explain part of the genetic variance and as a result this may provide an indication for finding genes for this covariate, but of course the drawback is that with this covariate in the model the success of finding the latent gene (s) will be reduced. Consequently, it is mandatory to determine how each covariate influences the genetic, common environmental and residual factors of the variance. Covariates that influence birth weight of twins are gestational age, maternal factors (parity and maternal age), twin specific factors (zygosity, sex, birth order), and placental factors (chorionicity, fusion of placentas and insertion of the umbilical cord) (Gielen et al. 2007).
The aim of this twin study was to model birth weight across gestational ages and to quantify the genetic and environmental components in order to explain the effects of covariates on the variance of birth weight. We hypothesize that heritability changes during gestation and that covariates mainly explain the common environmental and residual factors of the variance. By entering these covariates in the means model and in the covariance matrix, heritability will increase and, as a consequence, the power to find genes by linkage and association studies will increase, although the effects of each covariate on the genetic and environmental component may be different.
Materials and methods
Subjects
The study sample consisted of live born twin pairs selected from the East Flanders Prospective Twin Survey (EFPTS), Belgium (Loos et al. 1998). Between July 1964 and the end of December 2002, the EFPTS registered 6315 twin pairs who met the World Health Organization criteria for live born infants (birth weight ≥ 500 grams or gestational age ≥ 22 weeks, if birth weight unknown). The vast majority consisted of twins of Caucasian origin. Methods of data collection have previously been described in detail (Gielen et al. 2006; Gielen et al. 2007; Loos et al. 2005). Briefly, a defined set of obstetric and perinatal data were recorded (including gestational age, mode of conception, maternal age, birth order, parity, sex and neonatal survival) and placentas were examined within 24 hours after delivery according to a standardized protocol. Based on the site of the cord insertion two groups were distinguished: (1) central insertion (central and eccentric insertions), and (2) peripheral insertion. Gestational age was reported by the obstetrician, based on the last menstruation or a first trimester ultrasound investigation, and was calculated as the number of completed weeks of pregnancy. Zygosity was determined through sequential analysis based on sex, fetal membranes, umbilical cord blood groups (ABO, Rh, CcDEe, MNSs, Duffy, Kell), placental alkaline phosphatase (Decorte et al. 1990; Vlietinck 1986), and, since 1982, DNA fingerprints. Zygosity and chorionicity were determined with an accuracy of 99%.
Twin pairs of whom one or both children were stillborn (205 pairs) or suffered from major congenital malformation (120 pairs) were excluded (Loos et al. 2001). Two MC pairs were excluded because they were recorded as being MC but at birth had two separate placentas. Twin pairs with missing values in one or more covariates for one or both twins (birth weight, gestational age, parity, chorionicity, mode of conception, unknown whether staying alive postnatally, maternal age, placental type and weight, site of insertion of the umbilical cord) were excluded (1756 pairs) (Gielen et al. 2007). Finally, 8464 twins (4232 pairs) with complete datasets were analyzed.
Statistical analysis
The birth weights of pregnancies of different gestational ages were analyzed using a non-linear multivariate Gaussian regression, MVN ((μ, Σ2) where μ is the means model and Σ2 is the covariance matrix.
Means model
Growth was best modeled by a logistic growth curve (non-linear multivariate Gaussian regression) in which the time of conception (gestational age minus two weeks) was chosen as time point zero (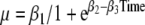 where β1 t/m β3 are the coefficients of the logistic growth curve and Time is time since conception; unadjusted model) (Neale and McArdle 2000). Furthermore, the two first weeks of pregnancy (the time between the last menstrual period and conception) were set to be zero. The logistic growth curve was modified to allow a change of growth rate (
where β1 t/m β3 are the coefficients of the logistic growth curve and Time is time since conception; unadjusted model) (Neale and McArdle 2000). Furthermore, the two first weeks of pregnancy (the time between the last menstrual period and conception) were set to be zero. The logistic growth curve was modified to allow a change of growth rate (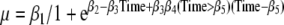 where β4 is the change in growth rate after the time point described by coefficient β5). The following covariates were considered: sex of the individual and of the co-twin, chorionicity, number of placentas, site of umbilical cord insertion, birth order, maternal age, mode of conception, parity, and neonatal death (Gielen et al. 2007). Because in 1964 all gestational ages were based on the last menstruation, while in 2002 almost all gestational ages were based on first trimester ultrasound and this might influence gestational age, we controlled for birth year. The covariates were consecutively added in the logistic growth curve,
where β4 is the change in growth rate after the time point described by coefficient β5). The following covariates were considered: sex of the individual and of the co-twin, chorionicity, number of placentas, site of umbilical cord insertion, birth order, maternal age, mode of conception, parity, and neonatal death (Gielen et al. 2007). Because in 1964 all gestational ages were based on the last menstruation, while in 2002 almost all gestational ages were based on first trimester ultrasound and this might influence gestational age, we controlled for birth year. The covariates were consecutively added in the logistic growth curve,
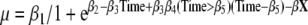 |
where X represents possible covariates and their interactions included in the model and β represents the corresponding coefficients. Finally, members of different groups were allowed to have different growth rates after a certain point in time as shown in the following equation
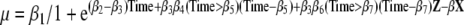 |
where Z is a binary covariate (indicator for belonging to group Z) and β6 is the additional change in growth rate after the time point described by coefficient β7 for members of group Z. (adjusted model). Examples of how to use the estimates can be found elsewhere (Gielen et al. 2007).
Covariance matrix
The twin model assumes that the total phenotypic variance can be decomposed into additive genetic effects (A), dominance genetic effects (D), effects of common environment (C) and effects of unique environment (E) (Neale and Maes 2002). The common environmental variance is assumed to be the same for both the MZ and DZ twin pairs and will account for the intra pair correlation between twin pair weights. MZ twins are assumed to share the same genetic variance (A and D), whereas DZ twins share half of the additive genetic variance and a quarter of the dominance genetic variance. In the case of a covariance matrix containing an additive genetic effect, common environment, and unique environment (ACE), the covariance matrix is respectively 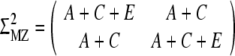 and
and 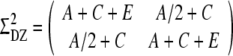 for the MZ and DZ pairs. Because former analyses showed that a model in which the variance allowed to change linearly during gestational age fitted best (Gielen et al. 2007), we allowed the variance and covariance to change over time (for example, A = eα1 + eα2Time, C = eδ1 + eδ2Time, and ln(E) = σ1 + σ2Time, where Time is gestational age, not time since conception).
for the MZ and DZ pairs. Because former analyses showed that a model in which the variance allowed to change linearly during gestational age fitted best (Gielen et al. 2007), we allowed the variance and covariance to change over time (for example, A = eα1 + eα2Time, C = eδ1 + eδ2Time, and ln(E) = σ1 + σ2Time, where Time is gestational age, not time since conception).
First, models containing the ACE and ADE components were fitted separately. Both models were fitted with only gestational age in the mean regression (unadjusted model) and with covariates in the mean regression (adjusted model). The best fitting unadjusted and adjusted models were tested with the (co-)variance being allowed to change during gestation. Next, the covariates of the mean regression of the adjusted model were entered as moderators in the covariance matrix the same way the mean regression of the adjusted model was built. This model is called the full model (for example, ln(E) = σ1 + σ2 Time + σ3X where σ3 is the coefficient of covariate X), which corresponds to the method described by Shaun Purcell (Purcell 2002). In this way, the covariance of the first-born twin was allowed to differ from the second born twin. The full model will show how heritability, A/(A + C + E), depends on the representative state of the covariates in the model.
For example, let us assume a model in which genetic, common and unique environmental components are defined as
A = e7.403 × Time, where the intercept α1 is set to zero
C = e6.688 × Time, where the intercept δ1 is set to zero
In(E) = 5.4054 + 0.1435 × Time + 0.4324 × SB + 0.724 × ND − 0.163 × MP + 0.2441 × OPM, where SB is the second born, ND is neonatal death, MP is multiparity, and OPM is one placental mass. The heritability for a first-born twin with a separate placenta, staying alive after one week of birth, of a multiparous mother, in case of two separate placentas would be at 25 weeks
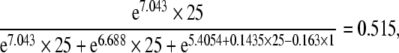 |
and at 40 weeks of gestation
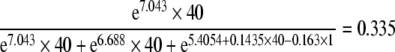 |
Model selection
Models were compared with the Akaike information criterion (AIC) (Akaike 1973; Lindsey and Jones 1998). Because the modeling process is exploratory, the inference criterion used for comparing the models under consideration is their ability to fit the observed data; i.e. models are compared directly through their minimized minus log likelihood. When the numbers of parameters in models differ, they are penalized by adding the number of estimated parameters. Smaller values indicate more preferable models. This criterion allows direct comparison among models, which are not required to be nested.
Effect of each separate covariate on the genetic and environmental component
To explain the effect of each covariate on the (co-)variance of birth weight at different gestational ages, we removed a significant covariate completely from the full model (out of the mean regression and out of the covariance matrix) and compared the (co-)variances, so that the influence of that specific covariate became apparent.
All analyses were conducted with R (Ihaka and Gentleman 1996) version 2.0.1 using the twin library (Lindsey 2001).
Results
Heritability decreases during gestation
For both the unadjusted model with only gestational age in the mean regression, and the adjusted model with covariates in the mean regression, the ACE model with additive genetic (A), common environmental (C), and unique environmental (E) variances fitted best. Allowing the covariance to change over time improved the fit (Table 1).
Table 1.
Model fit statistics of ACE and ADE models for the unadjusted model with only gestational age in the mean regression, the adjusted model with covariates in the mean regression, and the full model with covariates in the mean regression and in the covariance matrix. The best fitting model (lowest AIC) is presented in bold
| Unadjusted model | Adjusted model | |||||
|---|---|---|---|---|---|---|
| −Log likelihood | df | AIC | −Log likelihood | Df | AIC | |
| ACE | 61696.84 | 8457 | 61703.84 | 61350.49 | 8441 | 61373.49 |
| ADE | 61727.45 | 8457 | 61734.45 | 61380.6 | 8441 | 61403.6 |
| CE | 61711.88 | 8458 | 61717.88 | 61364.28 | 8442 | 61386.28 |
| AE | 61727.45 | 8458 | 61733.45 | 61380.58 | 8442 | 61402.58 |
| E | 62189.72 | 8459 | 62194.72 | 61826.93 | 8443 | 61847.93 |
| ACE; A and C allowed to change over time | 61688.44 | 8457 | 61695.44 | 61343.61 | 8441 | 61366.61 |
| Full model | ||||||
| ACE; A and C allowed to change over time and covariates in covariance matrix | 61297.26 | 8437 | 61324.26 | |||
A: Additive genetic effects, D: Dominance genetic effects, C: Effects of common environment, E: Effects of unique environment
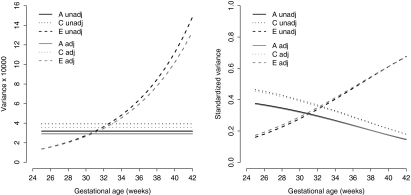
For both the unadjusted and adjusted ACE model, the common environmental variance was higher than the genetic variance. Although the variances of the adjusted model were lower, the proportions of the standardized variances of both models remained the same, and therefore heritability (standardized A-variance). Heritability decreased from 38% at 25 weeks to 15% at 42 weeks (Fig. 1). Allowing the covariance to change during gestation increased the genetic and common environmental variances from 25 to 42 weeks. The common environmental variance was only a little bit higher than the genetic variance, thereby increasing heritability at the end of gestation (from 36% at 25 weeks to 18% at 42 weeks). The model without an intercept for the genetic (α1) and common environmental variance (δ1) had a better fit, because it imposes these variance components to be zero at conception (Table 2).
Fig. 1.
Genetic (A), common environmental (C) and unique environmental (E) variance and standardized variance of birth weight of the unadjusted (unadj) model and the adjusted model (adj). E-variance was allowed to change over time, but the A- and C-variance not
Table 2.
Regression coefficients of the covariance matrix of the unadjusted model, the adjusted model, and the full model
| A | C | Log(E) | |||||
|---|---|---|---|---|---|---|---|
| Log(estimate) | SE | Log(estimate) | SE | estimate | SE | ||
| Unadjustedb | Intercept | 10.3700 | 0.1735 | 10.5800 | 0.1154 | 6.0055 | 0.3462 |
| Gestational age | – | – | 0.1405 | 0.0094 | |||
| Unadjusteda | Intercept | 0.0000 | – | 0.0000 | – | 6.3061 | 0.3468 |
| Gestational age | 6.8930 | 0.1604 | 6.942 | 0.1250 | 0.1317 | 0.0094 | |
| Adjustedb | Intercept | 10.2800 | 0.1857 | 10.4800 | 0.1222 | 6.1436 | 0.3495 |
| Gestational age | – | – | 0.1347 | 0.0094 | |||
| Adjusteda | Intercept | 0.0000 | – | 0.0000 | – | 6.4552 | 0.3489 |
| Gestational age | 6.7830 | 0.1737 | 6.8600 | 0.1297 | 0.1256 | 0.0094 | |
| Full model | Intercept | 0.0000 | – | 0.0000 | – | 5.4054 | 0.3963 |
| Gestational age | 7.0430 | 0.1421 | 6.6880 | 0.1571 | 0.1435 | 0.0103 | |
| Second born | 0.4324 | 0.0642 | |||||
| Neonatal death | 0.7240 | 0.1981 | |||||
| Multiparity | −0.1630 | 0.0491 | |||||
| One placental mass | 0.2441 | 0.0636 | |||||
Unadjusted:model with only gestational age in the mean regression
Adjusted:model with covariates in the mean regression
Full model:model with covariates in the mean regression and in the covariance matrix
The unadjusted and adjusted models are presented witha and withoutb A and C allowed changing during gestation
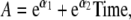
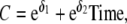 and
and 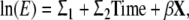 where Time is gestational age and X represent the covariates with the corresponding estimates β.
where Time is gestational age and X represent the covariates with the corresponding estimates β.
Effect of covariates in the covariance matrix on heritability
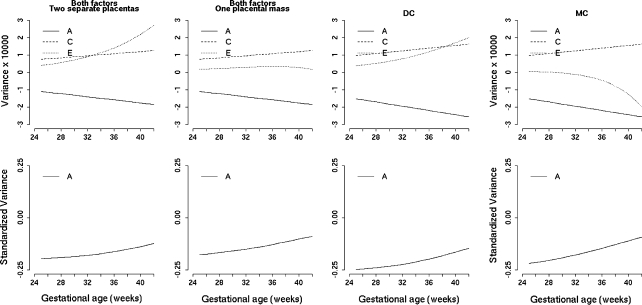
The full model with also covariates in the covariance matrix was the best fitting model (Table 1; for estimates of the means model see Table 3). The genetic variance became larger than the common environmental variance (Fig. 3; Table 2). Not all covariates of the mean regression explained part of the covariance matrix (Table 2 and Table 3). The genetic and environmental variance were only influenced by gestation, the unique environmental variance by parity, birth order, neonatal survival and number of placentas (estimates in Table 2, path diagram in Fig. 2). Although these dichotomous covariates were only present in the unique environmental variance, heritability will nevertheless change, because heritability is the proportion of the total variance that can be explained by genetic variation.
Table 3.
Regression coefficients of the full model with covariates in the mean regression and in the covariance matrix
| Estimate | SE | |
|---|---|---|
| Means model | ||
| Coefficients of the logistic growth curve | ||
| b1 = saturation level at which growth stops | 3688.0980 | 84.9100 |
| b2 = growth rate | 5.8061 | 0.1762 |
| b3 = location parameter | −0.1842 | 0.0068 |
| b4 = decrease in weight after inflection point | 0.9563 | 0.3017 |
| b5 = inflection pointa | 37.5934 | 0.3216 |
| b61 = decrease in weight for second bornb | 0.1054 | 0.0021 |
| b71 = inflection point for second borna | 30.7221 | 0.8399 |
| b62 = decrease in weight for neonatal deathb | 0.5733 | 0.1846 |
| b72 = inflection point for neonatal deatha | 29.0000 | 1.098 |
| b63= decrease in weight for MZ MCb | 1.1438 | 0.5059 |
| b73 = inflection point for MZ MCa | 37.7869 | 0.4227 |
| Covariates | ||
| Multiparity | 0.1165 | 0.0148 |
| Male | 0.1405 | 0.0118 |
| Birth year (form 0 [1964] to 38 [2002]) | 0.0018 | 0.0007 |
| One placental mass | −0.0525 | 0.0127 |
| Peripheral umbilical cord insertion | −0.0593 | 0.0227 |
| Maternal age | 0.0046 | 0.0014 |
| Interaction terms | ||
| Birth yeara Peripheral umbilical cord insertion | −0.0027 | 0.0009 |
| Multiparitya Second born | 0.0317 | 0.0131 |
aWeeks since conception
bAfter inflection point
MZ MC = monozygotic monochorionic. Inflection point = weeks of gestational age since conception (Time). Equation: 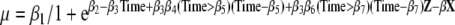
b1 t/m b7 coefficients, Z = second born, Neonatal death or MZ MC, X = covariates, and β = coefficient of covariates
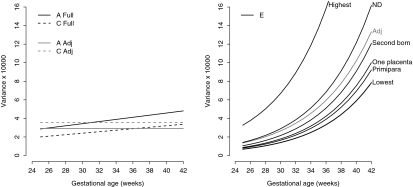
Fig. 3.
Genetic (A), environmental (C) and residual (E) variance of birth weight of the adjusted (Adj) model and the full model (Full). Legend: Lowest: E-variance for a first-born twin of a multipara with two separate placentas who is staying alive. Primipara: E-variance for a first-born twin of a primipara with two separate placentas who is staying alive. One placenta: E-variance for a first-born twin of a multipara with one placental mass who is staying alive. Second born: E-variance for a second born twin of a multipara with two separate placentas who is staying alive. Neonatal death (ND): E-variance a first-born twin of a multipara with two separate placentas who died neonatally. Highest: E-variance for a second born twin of a primipara with one placental mass who died neonatally
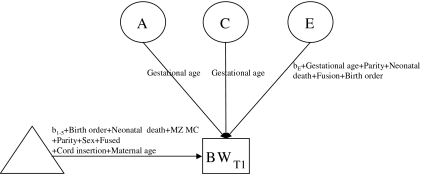
Fig. 2.
Partial path diagram of the full model shown for one twin only. For the estimates and interactions, see table 1 the adjusted model. BWT1 = Birth weight twin 1, Circle = latent variables, square = measured variables, triangle = definition variables in the means model, MZ MC = monozygotic monochorionic, A = additive genetic effects, C = effects of common environment, E = effects of unique environment
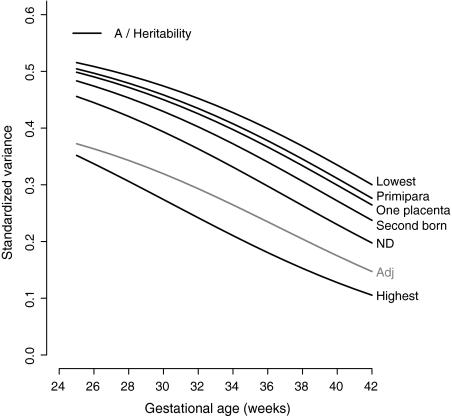
First-born twins of multipara with two separate placentas, who stay alive neonatally, have the lowest unique environmental variance (Table 2, Fig. 3: Lowest) and therefore the highest heritability (from 52% at 25 weeks to 30% at 42 weeks) (Fig. 4: Lowest). Allowing a dichotomous variable to be fixed to another representative state showed that the unique environmental variance remained lower than in the adjusted model, except for twins who died neonatally (Fig. 3). However, heritability was always higher in the unadjusted model than in the adjusted model (Fig. 4). When the covariates were chosen to represent second born twins of primipara with one placental mass and who died neonatally, the unique environmental variance became extremely high (Fig. 3: Highest) and heritability became lower than that of the adjusted model (Fig. 4: Highest).
Fig. 4.
Heritability (standardized A-variance) of birth weight of the adjusted (Adj) and full model (Full) of Fig. 3. Legend: Lowest: Standardized E-variance for a first-born twin of a multipara with two separate placentas who is staying alive. Primipara: Standardized E-variance for a first-born twin of a primipara with two separate placentas who is staying alive. One placenta: Standardized E-variance for a first-born twin of a multipara with one placental mass who is staying alive. Second born: Standardized E-variance for a second born twin of a multipara with two separate placentas who is staying alive. Neonatal death (ND): Standardized E-variance a first-born twin of a multipara with two separate placentas who died neonatally. Highest: Standardized E-variance for a second born twin of a primipara with one placental mass who died neonatally
Effect of each separate covariate on the genetic and environmental component
Comparing the variances of the full model with and without a specific covariate showed that maternal factors (age and parity) had minor influence on the genetic variance, but no influence on heritability. Removing sex, birth order and neonatal survival gave an increase in genetic variance. However, only for sex, heritability increased if removed from the model. Removing the placental factors from the full means model resulted in a decrease of the genetic variance and a decrease of heritability. Insertion of the umbilical cord (unique for each twin) explained mainly the unique environmental variance, whereas number of placentas also explained common environment.
Chorionicity or other placental factors
Since chorionicity, number of placentas and insertion of the umbilical cord are related (MC twins always have one placenta and have more often a peripheral cord insertion), as an additional step we built a model in which we replaced cord insertion and number of placentas with chorionicity (chorionicity model). Although the AIC of the chorionicity model was higher, it roughly explained as much of the genetic and unique environmental variances but not of the common environmental variance (Fig. 5). Removing of either the two placental factors or chorionicity resulted in a decrease of genetic variance and decrease of heritability. The variances of the DC twins were comparable to the variances of two separate placentas, whereas variances of the MC twins were comparable to the variances of one placental mass (Fig. 5).
Fig. 5.
Effect of either placental factors (site of insertion of umbilical cord and number of placentas), or chorionicity on the variance and heritability of birth weight The curves represent the differences between the full model and the model without the specific covariate. The presented curves are the curves for the first born twin of a primipara, who is staying alive
Discussion
The aim of this study was to model birth weight at different gestational ages and quantify the genetic and environmental components. To our knowledge we are the first to show that heritability of birth weight seems to decrease during gestation, in our case from 38% at 25 weeks to 15% at 42 weeks. Entering the covariates of the mean regression as moderators in the covariance matrix, explained more of the common environmental variance than of the genetic variance. However, the unique environmental variance depended upon the representative state of the covariates and was lowest for first-born twins of multipara with two separate placentas, who stayed alive neonatally. Therefore these twins have the highest heritability: from 52% at 25 weeks to 30% at 42 weeks.
Part of the explanation for a decrease in heritability during gestation could be the increasing demand for nutrients and oxygen in twin gestation, especially in the third trimester (Loos 2001). After about 29 weeks of gestation, intrauterine growth of twins deviates from growth of singletons, (Loos et al. 2005; Luke et al. 1993; Soucie et al. 2006), resulting in a lower birth weight and placental weight (per twin) (Gielen et al. 2006; Loos et al. 2005; Pinar et al. 1996). In addition, placental factors can be involved. In twin pregnancies, a lower placental weight, monochorionicity, one placental mass (one placenta or two fused), and a peripheral cord insertion result in a lower birth weight (this study) (Gielen et al. 2007; Loos et al. 2001). Also, vascular anastomoses in MC twins result in higher discordancy rates (Machin et al. 1996). These placental factors can partly determine the (unequal) blood supply to the fetus and therefore the amount of nutrients and oxygen for each fetus, resulting in a higher degree of discordancy (Machin 1996) and low heritability estimates in twins in late gestation. In this study, first-born twins of multipara with two separate placentas, who stay alive, were genetically the most homogeneous group. This group also had the highest birth weights and placental weights (Gielen et al. 2006; Gielen et al. 2007), suggesting that they may have minimal environmental restriction and therefore the highest heritability.
Modeling common and unique environmental factors can increase heritability and therefore optimize finding genes responsible for the genetic variance in combined linkage and association studies. By partitioning the association effects into between twin pair and within twin pair components, true from false associations can be distinguished in combined linkage and association studies (Fulker et al. 1999; Posthuma et al. 2004). As a rule, the power in statistical analysis will increase as the sample size increases, if a quantitative trait locus (QTL) explains a larger part of the variance it is easier to find, and if multivariate models are used (Peeters 2005). For example, if a QTL effect explains 0.05 of the variance (α = 0.05 β = 0.80), with genetic factors explaining 20%, and common and unique environmental factors each explaining 40% of the variance, 6422 sib-pairs are needed. If genetic factors explain a larger part of the variance e.g. 40%, and common and unique environmental factors each explaining 30% of the variance, only 3751 sib-pairs are needed. If in the latter case the QTL effect is 0.10 even less sib-pairs (n = 1080) are needed. Lack of statistical power may also be a reason for the inconsistent results of linkage studies to identify quantitative trait loci (QTLs) for birth weight. Only three genome wide linkage studies in different populations have identified QTLs on chromosome 6 (Arya et al. 2006), 7 (Fradin et al. 2006), and 11 (Lindsay et al. 2002) (LOD > 3) with relative small sample sizes (less then 413 sibships (Fradin et al. 2006), (Lindsay et al. 2002) and 840 individuals from families (Arya et al. 2006)) and only two covariates taken into account (sex and gestational age).
By building a means model and then entering the significant covariates in the covariance matrix, we controlled for environmental variables correlated with the genetic effects on the trait (gene environment correlation = rGE) (Purcell 2002). All the covariates with a unique environmental component, besides gestational age, must either explain differences between twins or describe gene environment interactions (Purcell 2002; van der Sluis et al. 2006). Parity is common for both twins, but because of the interaction with birth order in the mean regression, parity explains part of the unique environmental variance.
Covariates also contain useful information in order to identify genes, which may be associated with the trait. In this study, only sex contained latent genetic information. For all other covariates used in this study, either heritability did not change, or environmental factors were explained with heritability depending on the representative state of the covariate. Since sex explained part of heritability, entering sex in the means model will mask genes that act differently according to sex and it is better to have these genes in the model instead of sex.
In the past, Vlietinck et al (1989) showed an effect of chorionicity, although fusion of the placentas and site of insertion of the umbilical cord was not taken into account. Our analyses showed that only by replacing the placental factors cord insertion and number of placentas with chorionicity, the effect of chorionicity became visible, although the model fitted worse. Chorionicity certainly plays a role in growth (in mean regression), but mainly after 40 weeks of gestation if fusion of the placentas and insertion of the umbilical cord are taken into account (Gielen et al. 2007). Consequently, in order to achieve a higher heritability by explaining environmental factors, number of placentas and site of the insertion of the umbilical cord are preferable above chorionicity.
The strength of our study lies in the large sample size, accurate zygosity and chorionicity determination. This results in a decrease of the unique environmental variance and therefore a better estimation of heritability of birth weight. The large sample size further offers the opportunity to determine heritability at different gestational ages, although twins might not be representative for singletons due to differences in growth restriction. On the other hand, in studies without MZ and DZ twins, no difference can be made between genetic and common environmental factors, resulting in an overestimation of the genetic factors with, in case of birth weight, heritability estimates up to 80% (Arya et al. 2006; Clausson et al. 2000; Magnus et al. 1984; Nance et al. 1983).
Information on parental ethnicity was not always available. However, it has been shown that the genetic variance of birth weight does not differ between populations in which the majority (>95%) is Caucasian and Asian (Hur et al. 2005), suggesting that the genetic variance might be equal for populations. Since a vast majority of the inhabitants of East Flanders are Caucasians, we assume that our results are not biased by ethnicity. Finally, parental weight and height were not available. We assume however no biased results, since both traits are highly heritable (Carmichael and McGue 1995; Preece 1996; Silventoinen et al. 2001). Because the aim of linkage and association studies is to identify loci or alleles responsible for birth weight, adjusting for genetic factors will reduce the chance of finding them as stated above. But also, because we studied birth weight of twin pairs, rather than offspring of twins, the genes implicated in our study relate to fetal rather than maternal effects (Luciano et al. 2004). Therefore, maternal genes will be seen as common environment (Luciano et al. 2004).
In conclusion, modeling genetic and environmental factors gives a better insight in factors influencing growth during gestation, and therefore birth weight. Heritability of birth weight changed during gestation with a decrease from 25 to 42 weeks with highest heritability for twins with the least environmental restriction (first-born twins of multipara with two separate placentas who stayed alive neonatally). For this complex trait, modeling the covariance matrix with covariates, mainly environmental factors were explained, resulting in an increase of the heritability and subsequently the chance of finding genes by linkage and association studies.
Acknowledgements
Since its origin, the East Flanders Prospective Survey has been partly supported by grants from the Fund of Scientific Research, Flanders (Belgium), the Marguerite-Marie Delacroix Foundation, and by the Association for Scientific Research in Multiple Births (Belgium). We thank Ingeborg Berckmoes and Lut De Zeure for their excellent technical assistance.Grant Sources: This study was supported by grants from the National Fund for Scientific Research, Belgium (number 3.0269.97, G.0383.03) and the Dutch Diabetes Fund (2002.00.15). Patrick J. Lindsey was supported from the CARIM, GROW and CAPHRI Research Institutes of the Maastricht University (Netherlands).
Open Access This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Footnotes
Edited by David Allison.
References
- http://statgen.iop.kcl.ac.uk/gpc/qtllink.html
- Akaike H (1973) Information theory and an extension of the maximum likelihood principle. In: Petrov BNaC F (ed) Second International Symposium on Inference Theory, Budapest: Akadémiai Kiadó, pp 267–281
- Arya R, Demerath E, Jenkinson CP, Goring HH, Puppala S, Farook V, Fowler S, Schneider J, Granato R, Resendez RG et al (2006) A quantitative trait locus (QTL) on chromosome 6q influences birth weight in two independent family studies. Hum Mol Genet 15:1569–1579 [DOI] [PubMed]
- Baird J, Osmond C, MacGregor A, Snieder H, Hales CN, Phillips DI (2001) Testing the fetal origins hypothesis in twins: the Birmingham twin study. Diabetologia 44:33–39 [DOI] [PubMed]
- Boomsma D, Busjahn A, Peltonen L (2002) Classical twin studies and beyond. Nat Rev Genet 3:872–882 [DOI] [PubMed]
- Carmichael CM, McGue M (1995) A cross-sectional examination of height, weight, and body mass index in adult twins. J Gerontol A Biol Sci Med Sci 50:B237–B244 [DOI] [PubMed]
- Clausson B, Lichtenstein P, Cnattingius S (2000) Genetic influence on birthweight and gestational length determined by studies in offspring of twins. BJOG 107:375–381 [DOI] [PubMed]
- Decorte R, Cuppens H, Marynen P, Cassiman JJ (1990) Rapid detection of hypervariable regions by the polymerase chain reaction technique. DNA Cell Biol 9:461–469 [DOI] [PubMed]
- Fradin D, Heath S, Lepercq J, Lathrop M, Bougneres P (2006) Identification of distinct quantitative trait Loci affecting length or weight variability at birth in humans. J Clin Endocrinol Metab 91:4164–4170 [DOI] [PubMed]
- Fulker DW, Cherny SS, Sham PC, Hewitt JK (1999) Combined linkage and association sib-pair analysis for quantitative traits. Am J Hum Genet 64:259–267 [DOI] [PMC free article] [PubMed]
- Gielen M, Lindsey P, Derom C, Loos R JF, Derom R, Nijhuis J, Vlietinck R (2006) Curves of placental weights of live born twins. Twin Res Hum Genet 9:664–672 [DOI] [PubMed]
- Gielen M, Lindsey PJ, Derom C, Loos RJ, Derom R, Nijhuis J, Vlietinck R (2007) Twin birth weight standards. Neonatology 92:164–173 [DOI] [PubMed]
- Hur YM, Luciano M, Martin NG, Boomsma DI, Iacono WG, McGue M, Shin JS, Jun JK, Ooki S, van Beijsterveldt CE et al (2005) A comparison of twin birthweight data from Australia, the Netherlands, the United States, Japan, and South Korea: are genetic and environmental variations in birthweight similar in Caucasians and East Asians? Twin Res Hum Genet 8:638–648 [DOI] [PubMed]
- Ihaka R, Gentleman R (1996) R: a language for data analysis and graphics. J Comput Graph Stat 5:299–314 [DOI]
- Kramer MS, Barros FC, Demissie K, Liu S, Kiely J, Joseph KS (2005) Does reducing infant mortality depend on preventing low birthweight? An analysis of temporal trends in the Americas. Paediatr Perinat Epidemiol 19:445–451 [DOI] [PubMed]
- Lindsay RS, Kobes S, Knowler WC, Hanson RL (2002) Genome-wide linkage analysis assessing parent-of-origin effects in the inheritance of birth weight. Hum Genet 110:503–509 [DOI] [PubMed]
- Lindsey JK (2001) Nonlinear models in medical statistics. Oxford university press, Oxford
- Lindsey JK, Jones B (1998) Choosing among generalized linear models applied to medical data. Stat Med 17:59–68 [DOI] [PubMed]
- Loos R (2001) Fetal origins of cardiovascular and metabolic risk factors in young adults: a prospective twin study. Leuven: Katholieke Universiteit Leuven
- Loos R, Derom C, Vlietinck R, Derom R (1998) The East Flanders Prospective Twin Survey (Belgium): a population-based register. Twin Res 1:167–175 [DOI] [PubMed]
- Loos RJ, Derom C, Derom R, Vlietinck R (2001) Birthweight in liveborn twins: the influence of the umbilical cord insertion and fusion of placentas. BJOG 108:943–948 [DOI] [PubMed]
- Loos RJ, Derom C, Derom R, Vlietinck R (2005) Determinants of birthweight and intrauterine growth in liveborn twins. Paediatr Perinat Epidemiol 1(19 Suppl):15–22 [DOI] [PubMed]
- Luciano M, Wright MJ, Martin NG (2004) Exploring the etiology of the association between birthweight and IQ in an adolescent twin sample. Twin Res 7:62–71 [DOI] [PubMed]
- Luke B, Minogue J, Witter FR, Keith LG, Johnson TR (1993) The ideal twin pregnancy: patterns of weight gain, discordancy, and length of gestation. Am J Obstet Gynecol 169:588–597 [DOI] [PubMed]
- Machin G, Still K, Lalani T (1996) Correlations of placental vascular anatomy and clinical outcomes in 69 monochorionic twin pregnancies. Am J Med Genet 61:229–236 [DOI] [PubMed]
- Machin GA (1996) Some causes of genotypic and phenotypic discordance in monozygotic twin pairs. Am J Med Genet 61:216–228 [DOI] [PubMed]
- Magnus P, Berg K, Bjerkedal T, Nance WE (1984) Parental determinants of birth weight. Clin Genet 26:397–405 [DOI] [PubMed]
- Magnus P, Gjessing HK, Skrondal A, Skjaerven R (2001) Paternal contribution to birth weight. J Epidemiol Community Health 55:873–877 [DOI] [PMC free article] [PubMed]
- Nance WE, Kramer AA, Corey LA, Winter PM, Eaves LJ (1983) A causal analysis of birth weight in the offspring of monozygotic twins. Am J Hum Genet 35:1211–1223 [PMC free article] [PubMed]
- Neale MC, Maes HM (2002) Methodology for Genetic Studies of Twins and Families. Kluwer Academic Publishers B.V, Dordrecht, The Netherlands
- Neale MC, McArdle JJ (2000) Structured latent growth curves for twin data. Twin Res 3:165–177 [DOI] [PubMed]
- Peeters MW 2005 Genetic and environmental determination of somatic growth and muscular strength during adolescence: a multivariate and longitudianl analysis. Katholieke Universiteit Leuven, Leuven, pp 11–32
- Pinar H, Sung CJ, Oyer CE, Singer DB (1996) Reference values for singleton and twin placental weights. Pediatr Pathol Lab Med 16:901–907 [DOI] [PubMed]
- Posthuma D, de Geus EJ, Boomsma DI, Neale MC (2004) Combined linkage and association tests in mx. Behav Genet 34:179–196 [DOI] [PubMed]
- Preece MA (1996) The genetic contribution to stature. Horm Res 2(45 Suppl):56–58 [DOI] [PubMed]
- Purcell S (2002) Variance components models for gene-environment interaction in twin analysis. Twin Res 5:554–571 [DOI] [PubMed]
- Silventoinen K, Kaprio J, Lahelma E, Viken RJ, Rose RJ (2001) Sex differences in genetic and environmental factors contributing to body-height. Twin Res 4:25–29 [DOI] [PubMed]
- Snieder H, Harshfield GA, Treiber FA (2003) Heritability of blood pressure and hemodynamics in African- and European-American youth. Hypertension 41:1196–1201 [DOI] [PubMed]
- Soucie JE, Yang Q, Wen SW, Kee Fung KF, Walker M (2006) Neonatal mortality and morbidity rates in term twins with advancing gestational age. Am J Obstet Gynecol 195:172–177 [DOI] [PubMed]
- van Baal CG, Boomsma DI (1998) Etiology of individual differences in birth weight of twins as a function of maternal smoking during pregnancy. Twin Res 1:123–130 [DOI] [PubMed]
- van der Sluis S, Dolan CV, Neale MC, Boomsma DI, Posthuma D (2006) Detecting genotype-environment interaction in monozygotic twin data: comparing the Jinks and Fulker test and a new test based on Marginal Maximum Likelihood estimation. Twin Res Hum Genet 9:377–392 [DOI] [PubMed]
- van Dommelen P, de Gunst MC, van der Vaart AW, Boomsma DI (2004) Genetic study of the height and weight process during infancy. Twin Res 7:607–616 [DOI] [PubMed]
- van Os J, Wichers M, Danckaerts M, Van Gestel S, Derom C, Vlietinck R (2001) A prospective twin study of birth weight discordance and child problem behavior. Biol Psychiatry 50:593–599 [DOI] [PubMed]
- Vlietinck R (1986) Determination of the zygosity of twins. Katholieke Universiteit Leuven, Leuven, pp 1–123
- Vlietinck R, Derom R, Neale MC, Maes H, van Loon H, Derom C, Thiery M (1989) Genetic and environmental variation in the birth weight of twins. Behav Genet 19:151–161 [DOI] [PubMed]
- Whitfield JB, Treloar SA, Zhu G, Martin NG (2001) Genetic and non-genetic factors affecting birth-weight and adult Body Mass Index. Twin Res 4:365–370 [DOI] [PubMed]
- Wichers MC, Purcell S, Danckaerts M, Derom C, Derom R, Vlietinck R, Van Os J (2002) Prenatal life and post-natal psychopathology: evidence for negative gene-birth weight interaction. Psychol Med 32:1165–1174 [DOI] [PubMed]