Abstract
Basic helix–loop–helix (bHLH) DNA-binding proteins have been demonstrated to regulate tissue-specific transcription within multiple cell lineages. The Id family of helix–loop–helix proteins does not possess a basic DNA-binding domain and functions as a negative regulator of bHLH proteins. Overexpression of Id proteins within a variety of cell types has been shown to inhibit their ability to differentiate under appropriate conditions. We demonstrate that ectopic expression of Id-1 leads to activation of telomerase activity and immortalization of primary human keratinocytes. These immortalized cells have a decreased capacity to differentiate as well as activate phosphorylation of the retinoblastoma protein. Additionally, these cells acquire an impaired p53-mediated DNA-damage response as a late event in immortalization. We conclude that bHLH proteins play a pivotal role in regulating normal keratinocyte growth and differentiation, which can be disrupted by the immortalizing functions of Id-1 through activation of telomerase activity and inactivation of the retinoblastoma protein.
Keywords: telomerase, differentiation, senescence, transcription
The differentiation program of multiple cell types has been shown to be dependent on the activity of basic helix–loop–helix (bHLH) transcription factors (reviewed in refs. 1 and 2). These proteins share a common sequence motif of a stretch of basic amino acids responsible for site-specific DNA binding adjacent to a helix–loop–helix dimerization domain. The Id family of helix-loop-helix proteins, which does not possess a basic DNA-binding domain, functions as a negative regulator of bHLH proteins through the formation of inactive heterodimers with intact bHLH transcription factors (3, 4). The family of Id proteins has been demonstrated to bind the ubiquitously expressed E proteins or cell lineage-restricted bHLH transcription factors leading to inhibition of lineage-specific gene expression and differentiation (3, 5–7). A growing body of evidence has implicated Id proteins as playing a critical role in promoting G1/S cell cycle transitions. Id gene expression is elevated in undifferentiated cells and tumor cells, supporting the notion of their role as inhibitors of differentiation and growth-promoting factors (8, 9). Overexpression of Id proteins within myoblasts, myeloid precursor cells, mammary epithelium, and preadipose cells inhibits their ability to differentiate under appropriate conditions (6, 7, 10, 11). To determine whether Id proteins might play a role in regulating human keratinocyte differentiation, we evaluated the effects of deregulated expression of Id proteins in these cells. We determined that Id-1, Id-2, and Id-3 all extend the normal in vitro lifespan of primary human keratinocytes. Furthermore, we determined that deregulated expression of Id-1 alone could immortalize primary human keratinocytes and activate telomerase activity and retinoblastoma protein phosphorylation in these cells.
MATERIALS AND METHODS
Cell Culture.
Monolayer cultures of primary human foreskin keratinocytes (HFKs) were prepared from a pool of neonatal foreskins obtained from routine circumcisions by using a modified version of the protocol of Rheinwald and Green (19). Keratinocytes were isolated by using dispase incubation of foreskin tissue to allow for dermal–epidermal separation. Epidermal specimens were trypsinized and plated on standard tissue culture dishes. Cells were maintained in serum-free keratinocyte growth medium (KGM; GIBCO/BRL). Differentiation was induced by allowing for keratinocyte growth in high-calcium-containing medium (2.0 mM calcium) consisting of DMEM supplemented with 10% FBS, penicillin, streptomycin, and l-glutamine as described previously (13). Confluency of cultures under growth and differentiating conditions was regulated strictly to ensure that differentiation effects were attributable to the supplemented medium provided as opposed to confluency of cells. Cells were transfected 4–6 weeks after initial preparation from foreskins at 70% density by using 10 μl of lipofectamine (GIBCO/BRL) lipofection reagent and 8 μg of plasmid DNA. Human Id-1, Id-2, and Id-3 cDNAs cloned into a pCMV-Neo vector were used in the transfections (14). Cells were selected for plasmid expression with neomycin and grown in KGM (GIBCO/BRL). Cells were passaged continuously in 10-cm tissue culture dishes until 80% confluent and then replated at 1:3 cell density. Lifespan of cells was determined for pools of transfectants. Cells that did not reach 80% confluency within 2 weeks of passaging were determined to have senesced. For retroviral infection, human Id-1 was cloned into the recombinant retroviral vector LXSN and packaging cell lines (PA317) were maintained in DMEM supplemented with 10% FBS, penicillin, and streptomycin. Recombinant retroviruses were prepared by using standard methods, and infections of HFKs were performed as described previously (15).
Protein Analysis.
Cell lysates of human keratinocytes were prepared as described previously (16), and Western blotting was performed according to standard procedures. Primary antibodies against Id-1 (Santa Cruz Biotechnology; SC-734), retinoblastoma protein (pRb) (PharMingen 14001A), proliferating cell nuclear antigen (Santa Cruz Biotechnology; SC-56), and p53 (Oncogene Science, Ab-2) were obtained commercially. Monoclonal involucrin antibodies were a gift from J. Kvedar (Massachusetts General Hospital, Charlestown, MA) (17).
Telomerase Assays.
Telomeric repeat amplification protocol (TRAP) assays were performed by using the TRAPeze telomerase detection kit (Oncor).
Reverse Transcription–PCR.
Total cellular RNA was isolated by using Trizol reagent (Life Technologies, Gaithersburg, MD) and treated with RNase-free DNase (Boehringer Mannheim). RT was carried out on 5 μg of total RNA by using Moloney murine leukemia virus reverse transcriptase (Promega) and following the manufacturer’s protocol. PCR assays were carried out on 1/10th the RT reaction in 50 μl of total volume by using the recommended buffers and Taq polymerase concentration (Perkin–Elmer). PCR parameters consisted of denaturing at 95°C for 45 s, annealing at 68°C for 45 s, and extension at 72°C for 60 s for 10 cycles followed by 20 cycles by using an annealing temperature of 60°C. Final primer concentrations were 1 μM each. Human telomerase reverse transcriptase (hTERT) primers HT-1 and HT-5 were used to amplify a 377-bp fragment as described previously (18). Human Id-1 primers were 5′-CACCCTCAACGGCGAGATC and 5′-CCACAGAGCACGTAATTCCTC.
Cell Cycle Analysis.
Cells were prepared for fluorescence-activated cell sorter analysis as follows: 106 cells were trypsinized and washed three times in cold PBS. Cells then were fixed in ethanol for 30 min at 4°C, washed in PBS, and resuspended in 6.9 × 10−5 M propidium iodide (Sigma) containing RNase A. Cells were incubated at 37°C for 30 min and then analyzed in a cell sorter.
Cytogenetics.
Cultured cells were arrested in mitosis with 0.1 μg/ml colcemid. Arrested cells were spun down and resuspended in 75 μM KCl for 10 min. Cells then were spun down and resuspended in fresh 3:1 methanol/acetic acid overnight. The next day, the cells were rinsed twice in fixative and then resuspended in 1 ml of methanol/acetic acid. Cells were dropped onto cold, wet slides and allowed to air dry. Cell preps were reviewed under phase-contrast microscopy to ensure adequate spread. Slides then were incubated in 30 μg/ml aqueous solution of quinacrine mustard for 7 min, washed one time in water, mounted in Tris-maleate buffer, pH 5.6, and viewed by fluorescence microscopy. Photographs of metaphase spreads were taken for karyotype analysis.
RESULTS
Id-1 Immortalizes Primary Human Keratinocytes.
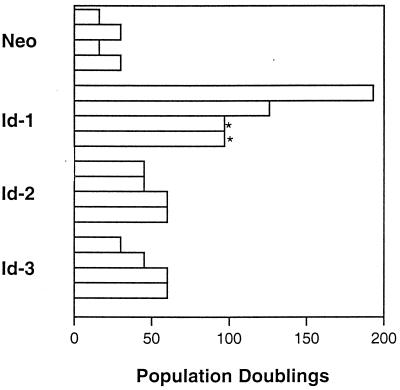
We investigated the effects of ectopic expression of Id proteins in primary human keratinocytes to determine whether Id genes might play a role in regulating human keratinocyte differentiation. Because only Id-1, Id-2, and Id-3 are expressed in human keratinocytes (data not shown), we undertook investigations of these particular Id family members. Id genes were introduced into primary human keratinocytes by either plasmid transfection or retroviral infection, and cells were grown in selection medium and, subsequently, as pools of selected colonies. Cellular lifespan and number of population doublings was recorded for pools of transfected cells (Fig. 1), with initiation of measurement of lifespan (after G418 selection) approximately 4–6 weeks (20–30 doublings) after preparation of primary cells. Similar results were obtained in four separate experiments and demonstrate a clear extension of keratinocyte lifespan in all Id-overexpressing pools of cells vs. vector controls. None of the Id-transfectant pools demonstrated an obvious crisis stage at any point during their passage. Keratinocyte clones that achieved an extended lifespan of >70 population doublings without undergoing crisis were considered immortal (19, 20). Using such criteria, all pools of Id-1-overexpressing cells possessed an unlimited lifespan in cell culture [two clones were contaminated after 6 months of passage (Fig. 1, asterisk) whereas two additional clones were stored as frozen cell lines after 8 months and 1 year, respectively] and therefore were determined to be immortalized. These experiments were repeated by using an Id-1 retroviral expression construct, and similar results were obtained (data not shown). Given these results, we investigated whether human keratinocytes that expressed Id-1 ectopically were able to respond to differentiation stimuli.
Figure 1.
Population doublings of Id-transfected primary human keratinocytes. Population doublings were determined for four separate transfections. All cells were passaged continuously since the day of transfection (after 20–30 doublings). Two of the Id-1 cell lines were contaminated after 6 months of passage (∗). Two additional cell lines were frozen down after 8 months and 1 year of passage.
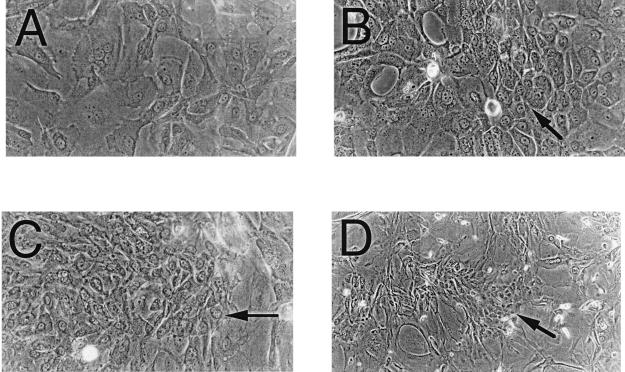
Id-overexpressing cells were induced to differentiate utilizing an in vitro HFK differentiation system described previously (16) at 4 weeks after transfection. Differentiation was monitored by marker protein expression and cellular morphology. Although all Id-overexpressing cells did contain small amounts of the differentiation marker protein involucrin without a differentiating stimulus (the cells differentiate spontaneously to a certain extent over time), the level of expression did not increase under a differentiation stimulus to the extent that was seen in control cells. Furthermore, Id-overexpressing cells grown in differentiation medium showed many focal regions of poorly differentiated cells within regions of more differentiated cells, which was not seen in control cells, suggesting that clonal populations of Id transfectants that were resistant to differentiating stimuli may have arisen (Fig. 2).
Figure 2.
Photomicrographs of Id-transfected HFKs under differentiating stimuli. Phase-contrast photomicrographs demonstrate focal regions of smaller, phenotypically less differentiated cells (arrows) in Id-1- (B), Id-2- (C), and Id-3- (D) transfected HFKs vs. vector control-transfected cells (A), which show a typical differentiated phenotype of large, flattened cells with increased cytoplasmic-to-nuclear ratios.
Id-1 Activates Phosphorylation of the Retinoblastoma Protein and Inactivates the p53 DNA-Damage Response.
Having determined that Id-1 was able to immortalize primary human keratinocytes, we wished to determine potential mechanisms by which Id-1 might prevent cellular senescence in cultures of primary human keratinocytes. Given the model systems of the DNA tumor viruses whose oncoproteins are able to affect the p53 or pRb pathways to induce cellular immortalization, we investigated whether these pathways also might be affected by Id-1 immortalization.
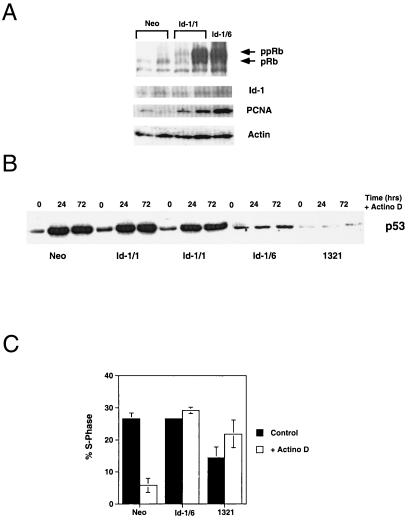
To test the integrity of the pRb tumor-suppressor pathway and the cyclin D-cdk4/6-p16 pathway, we evaluated the expression of the various phosphorylated forms of the retinoblastoma protein in Id-1 transfectants. We have found that phosphorylation of the retinoblastoma protein and its expression level are elevated in Id-1-overexpressing keratinocytes (Fig. 3A) at both 1 month (Id-1/1) and 6 months (Id-1/6) after transfection with Id-1. This pRb expression level and phosphorylation correlated well with the levels of Id and proliferating cell nuclear antigen expression in the cells examined.
Figure 3.
(A) Immunoblot of pRb, proliferating cell nuclear antigen, and Id-1 expression in Id-1 transfectants. Neo, vector-transfected HFKs 1 month after transfection; Id-1/1, Id-1-transfected HFKs 1 month after transfection; Id-1/6, Id-1-transfected HFKs 6 months after transfection. Actin panel is used as a loading control. (B) Immunoblot of p53 expression in response to DNA damage by actinomycin D for 24 and 72 hr. Neo cells are vector-transfected HFKs 1 month after transfection, Id-1/1 cells are Id-1-transfected HFKs 1 month after transfection, Id-1/6 cells are Id-1-transfected HFKs 6 months after transfection, and 1321 cells are HPV 16 E6/E7-immortalized HFKs. (C) Fluorescence-activated cell sorter analysis of Id-1-immortalized HFK response to DNA damage. Cells were exposed to 2.5 nM actinomycin D for 48 hr, and cell cycle profiles were determined by propidium iodide staining. Neo cells are vector-transfected HFKs, Id-1/6 are Id-1-transfected HFKs 6 months after transfection, and 1321 cells are HPV 16 E6/E7 immortalized HFKs.
To assay for p53 function, DNA damage was induced by treating Id-1-transfected keratinocytes with 2.5 nM actinomycin D, and p53 expression was determined by immunoblotting (Fig. 3B). Normal HFKs and a human keratinocyte cell line immortalized by human papillomavirus HPV-16 E6/E7 (1321) (21) were used as positive and negative controls, respectively. These experiments demonstrate a normal p53 DNA-damage response in early Id-1 transfectants (Id-1/1). However, complete abrogation of p53 up-regulation in response to DNA damage is seen in Id-1-immortalized keratinocytes (Id-1/6). Consistent with this finding, fluorescence-activated cell-sorting analysis of these cells revealed a G1 growth arrest in response to DNA damage in normal human keratinocytes that was absent in Id-1-immortalized (Id-1/6) and 1321 cells (Fig. 3C). These data support the idea that abrogation of the p53 pathway in Id-1-immortalized cells is a late effect of Id-1 expression and not a direct result of Id-1 regulation of p53.
Id-1 Activates Telomerase Activity and hTERT Expression in Primary Human Keratinocytes.
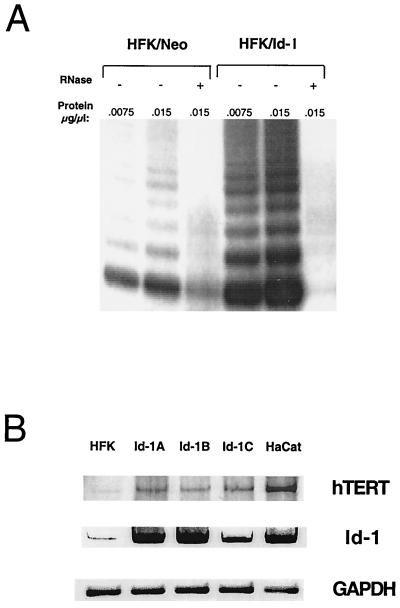
Recent studies have demonstrated that telomerase functions as a potential regulator of senescence in human cells and that TERT expression levels correlate with telomerase activity and cellular immortalization (22). Furthermore, overexpression of hTERT has been demonstrated to extend the normal lifespan of primary human cells (18). We determined telomerase activity in matched sets of vector-transfected human keratinocytes and Id-1-transfected human keratinocytes to determine whether Id-1 might function in regulating telomerase activity as a means of preventing cellular senescence. Consistent with earlier studies (23, 24), we detected small amounts of telomerase activity in vector control-transfected human keratinocytes; however, in the Id-1-transfected cells that were examined at 1 month after transfection, we detected significantly increased telomerase activity (Fig. 4A). Furthermore, we were able to demonstrate elevated expression of hTERT expression in these cells at early and late passages (Fig. 4B).
Figure 4.
(A) Telomerase activity in Id-1-expressing human keratinocytes. Normal human keratinocytes were transfected with either vector alone (HFK-Neo) or with a vector expressing Id-1 (HFK-Id). Cells were selected for expression of neomycin resistance and were analyzed for telomerase activity by the TRAP assay at 1 month after transfection. (B) hTERT expression in Id-1-transfected human keratinocytes. hTERT and Id-1expression was evaluated in normal human keratinocytes (HFK) and Id-1-transfected keratinocytes at 1 month (Id-1A, Id-1B) and 1 year (Id-1C) after transfection by using RT-PCR analysis. The spontaneously immortalized human keratinocyte cell lines, HaCaT, were used as a positive control for hTERT expression. Glyceraldehyde-3-phosphate dehydrogenase expression is shown as an internal control.
DISCUSSION
The above data demonstrate that constitutive expression of Id-1 results in the activation of cellular telomerase activity, inactivation of the retinoblastoma protein (through phosphorylation), and immortalization of primary human keratinocytes. Although the precise frequency of these immortalization events has not yet been determined, we have been able to show that these events occurred several times in each pool of selected cells because chromosomal analyses of cells after 6–12 months of passage revealed several distinct clonal populations within pools of immortalized cells. The populations possessed numerous chromosomal abnormalities including chromosomal translocations, ring chromosomes, aneuploidy, and polyploidy.
Normal somatic cells possess a finite lifespan that is limited by replicative senescence (25–27). Upon senescence, cells exit the cell cycle and arrest in G1, are unable to respond to mitogenic stimuli by reentering S-phase, but remain viable and metabolically active (28). Until recently, the only known genes capable of reactivating DNA synthesis in senescent cells were DNA tumor virus oncogenes including the SV40 large tumor antigen and the human papillomavirus E6/E7 oncoproteins. These genes have been shown previously to inactivate the tumor-suppressor proteins p53 and pRb through various mechanisms. Id family members also have been implicated in regulating the process of cellular senescence. It has been shown that Id-1 and Id-2 are required for G1 progression in human diploid fibroblasts and are repressed in senescent human fibroblasts (29). Furthermore, Id-1 has been shown to cooperate with a pRb-binding mutant of SV40 T antigen to reactivate DNA synthesis in senescent human fibroblasts (30). Our data demonstrate that, as with the viral oncoproteins, Id-1 is able to abrogate functional activity of the retinoblastoma protein through activated phosphorylation. Furthermore, much like the papillomavirus E6 protein, Id-1 can activate telomerase activity in primary human keratinocytes (31). Additionally, Id-1 is able to abrogate p53 function but only as a late effect of its expression, much like the late abrogation of p53 activity in primary human tumors.
The experiments presented here are among the first demonstrations of a normal cellular gene functioning alone to induce primary human cell immortalization. Because the Id-1-transfected HFKs were passaged continuously without an obvious crisis or senescence, it is likely that HLH proteins play a specific role in regulating these processes. Recently, it was reported that the bHLH-ZIP transcription factor, c-myc, is also able to activate telomerase activity in primary human cells (32). This further supports a potential role for the involvement of the helix–loop–helix family of transcriptional regulatory proteins in mediating cell-fate decisions of growth and senescence. Although Id proteins typically are thought of as being inhibitory to differentiation, our data suggest that they may have a more general effect on regulating the cellular “mitotic clock” by transcriptionally regulating genes involved in cellular growth control and/or senescence and reactivating telomerase activity. Recent data have suggested that this “mitotic clock” also might be reset to some degree by maintaining active telomerase function alone in primary human cells (18). Furthermore, it has been demonstrated that activation of telomerase activity and inactivation of the pRb/p16 pathway are sufficient for immortalization of primary human keratinocytes (20). Because both of these functions can be achieved with constitutive expression of Id-1 alone in primary human keratinocytes, it is likely that Id-1 is a critical regulator of senescence in these cells. Aberrant expression of Id proteins may provide a potent replicative stimulus in primary human cells, which allows for circumventing normal cellular growth controls and may lead indirectly to the up-regulation of telomerase function and pRb inactivation. Alternatively, bHLH proteins may directly regulate the transcription of telomerase subunits and pRb kinase complexes. Because Id-1 can perform both steps necessary for keratinocyte immortalization, we suggest that Id-1 may be a potential target for intervention in primary cutaneous malignancies.
Acknowledgments
We are grateful to Jeannette-MyLe Le and Ramana Tantravahi for their help in cytogenetic analyses, Agata Smogorzewska for technical assistance with the telomerase assays, and Robert Benezra and Maurice Markus for their careful review of this manuscript and helpful discussions. This work was supported by Grants T32 AR07098–21 and K08 AR01975–01A1 (R.M.A.) and CA66980 (K.M.) from the National Institutes of Health and by a pilot and feasibility grant from the Harvard Skin Disease Research Center (P30 AR42689; T. Kupper, principal investigator). K.M. is supported by a Junior Faculty Research Award (JFRA-597) from the American Cancer Society. J.H. was a Fellow of the Biomedical Sciences Exchange Program between North America and Europe and was supported by a fellowship from the Deutscher Akademischer Austauschdienst.
ABBREVIATIONS
- bHLH
basic helix–loop–helix
- HFK
human foreskin keratinocyte
- RT
reverse transcription
- hTERT
human telomerase reverse transcriptase
- pRb
retinoblastoma protein
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
References
- 1.Olson E N, Klein W H. Genes Dev. 1994;8:1–8. doi: 10.1101/gad.8.1.1. [DOI] [PubMed] [Google Scholar]
- 2.Weintraub H. Cell. 1993;75:1241–1244. doi: 10.1016/0092-8674(93)90610-3. [DOI] [PubMed] [Google Scholar]
- 3.Benezra R, Davis R L, Lockshon D, Turner D L, Weintraub H. Cell. 1990;61:49–59. doi: 10.1016/0092-8674(90)90214-y. [DOI] [PubMed] [Google Scholar]
- 4.Sun X H, Copeland N A, Jenkins N A, Baltimore D. Mol Cell Biol. 1991;11:5603–5611. doi: 10.1128/mcb.11.11.5603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wilson R B, Kiledjian M, Shen C P, Benezra R, Zwollo P, Dymecki S M, Desiderio S V, Kadesch T. Mol Cell Biol. 1991;11:6185–6191. doi: 10.1128/mcb.11.12.6185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jen Y, Weintraub H, Benezra R. Genes Dev. 1992;6:1466–1479. doi: 10.1101/gad.6.8.1466. [DOI] [PubMed] [Google Scholar]
- 7.Kreider B, Benezra R, Roverea G, Kadesch T. Science. 1992;255:1700–1702. doi: 10.1126/science.1372755. [DOI] [PubMed] [Google Scholar]
- 8.Ellmeier W, Aguzzi A, Kleiner E, Kurzbauer R, Weith A. EMBO J. 1992;11:2563–2571. doi: 10.1002/j.1460-2075.1992.tb05321.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhu W, Dahmen J, Bulfone A, Rigolet M, Hernandez M C, Kuo W L, Puelles L, Rubenstein J L, Israel M A. Brain Res Mol Brain Res. 1995;30:312–326. doi: 10.1016/0169-328x(95)00017-m. [DOI] [PubMed] [Google Scholar]
- 10.Moldes M, Lasnier F, Feve B, Pairault J, Djian P. Mol Cell Biol. 1997;17:1796–1804. doi: 10.1128/mcb.17.4.1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Desprez P Y, Hara E, Bissell M J, Campisi J. Mol Cell Biol. 1995;1555:3398–3404. doi: 10.1128/mcb.15.6.3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rheinwald J G, Beckett M A. Cancer Res. 1981;41:1657–1663. [PubMed] [Google Scholar]
- 13.Schlegel R, Phelps W C, Zhang Y-L, Barbosa M. EMBO J. 1988;7:3181–3187. doi: 10.1002/j.1460-2075.1988.tb03185.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lasorella A, Iavarone A, Israel M A. Mol Cell Biol. 1996;16:2570–2578. doi: 10.1128/mcb.16.6.2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Halbert C L, Demers W, Galloway D A. J Virol. 1991;65:473–478. doi: 10.1128/jvi.65.1.473-478.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jones D L, Alani R M, Munger K. Genes Dev. 1997;11:2101–2111. doi: 10.1101/gad.11.16.2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baden H P, Kvedar J C. J Invest Dermatol. 1993;101:72S–74S. doi: 10.1111/1523-1747.ep12362869. [DOI] [PubMed] [Google Scholar]
- 18.Bodnar A, Ouellette M, Frolkis M, Holt S, Chiu C-P, Morin G, Harley C, Shay J, Lichtsteiner S, Wright W. Science. 1998;279:349–352. doi: 10.1126/science.279.5349.349. [DOI] [PubMed] [Google Scholar]
- 19.Rheinwald J G, Green H. Cell. 1975;6:331–343. doi: 10.1016/s0092-8674(75)80001-8. [DOI] [PubMed] [Google Scholar]
- 20.Kiyono T, Foster S A, Koop J I, McDougall J K, Galloway D A, Klingelhutz A J. Nature (London) 1998;396:84–88. doi: 10.1038/23962. [DOI] [PubMed] [Google Scholar]
- 21.Munger K, Phelps W C, Bubb V, Howley P M, Schlegel R. J Virol. 1989;63:4417–4421. doi: 10.1128/jvi.63.10.4417-4421.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meyerson M, Counter C M, Eaton E N, Ellisen L W, Steiner P, Caddle S D, Ziaugra L, Beijersbergen R L, Davidoff M J, Liu Q, et al. Cell. 1997;90:785–795. doi: 10.1016/s0092-8674(00)80538-3. [DOI] [PubMed] [Google Scholar]
- 23.Yasumoto S, Kunimura C, Kikuchi K, Tahara H, Ohji H, Yamomoto H, Ide T, Utakoji T. Oncogene. 1996;13:433–439. [PubMed] [Google Scholar]
- 24.Harle-Bachor C, Boukamp P. Proc Natl Acad Sci USA. 1996;93:6476–6481. doi: 10.1073/pnas.93.13.6476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smith J R, Pereira-Smith O M. Science. 1996;273:63–67. doi: 10.1126/science.273.5271.63. [DOI] [PubMed] [Google Scholar]
- 26.Goldstein S. Science. 1990;249:1129–1133. doi: 10.1126/science.2204114. [DOI] [PubMed] [Google Scholar]
- 27.Hayflick L. Exp Cell Res. 1965;37:614–636. doi: 10.1016/0014-4827(65)90211-9. [DOI] [PubMed] [Google Scholar]
- 28.Campisi J. Cell. 1996;84:497–500. doi: 10.1016/s0092-8674(00)81023-5. [DOI] [PubMed] [Google Scholar]
- 29.Hara E, Yamaguchi T, Nojima H, Ide T, Campisi J, Okayama H, Oda K. J Biol Chem. 1994;269:2139–2145. [PubMed] [Google Scholar]
- 30.Hara E, Uzman J A, Dimri G P, Nehlin J O, Testori A, Campisi J. Dev Genet. 1996;18:161–172. doi: 10.1002/(SICI)1520-6408(1996)18:2<161::AID-DVG9>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 31.Klingelhutz A J, Foster S A, McDougall J K. Nature (London) 1996;380:79–82. doi: 10.1038/380079a0. [DOI] [PubMed] [Google Scholar]
- 32.Wang J, Xie L Y, Allan S, Beach D, Hannon G J. Genes Dev. 1998;12:1769–1774. doi: 10.1101/gad.12.12.1769. [DOI] [PMC free article] [PubMed] [Google Scholar]