Abstract
The Bcl-2 family of proteins regulates apoptosis, the cell death program triggered by activation of certain proteases (caspases). An attractive model for how Bcl-2 and its closest relatives prevent caspase activation is that they bind to and inactivate an adaptor protein required for procaspase processing. That model has been supported by reports that mammalian prosurvival Bcl-2 relatives bind the adaptor Apaf-1, which activates procaspase-9. However, the in vivo association studies reported here with both overexpressed and endogenous Apaf-1 challenge this notion. Apaf-1 could be immunoprecipitated together with procaspase-9, and the Apaf-1 caspase-recruitment domain was necessary and sufficient for their interaction. Apaf-1 did not bind, however, to any of the six known mammalian prosurvival family members (Bcl-2, Bcl-xL, Bcl-w, A1, Mcl-1, or Boo), or their viral homologs adenovirus E1B 19K and Epstein–Barr virus BHRF-1. Endogenous Apaf-1 also failed to coimmunoprecipitate with endogenous Bcl-2 or Bcl-xL, or with two proapoptotic relatives (Bax and Bim). Moreover, apoptotic stimuli did not induce Apaf-1 to bind to these family members. Thus, the prosurvival Bcl-2 homologs do not appear to act by sequestering Apaf-1 and probably instead constrain its activity indirectly.
Keywords: cell death, CED-4, CED-9, Bcl-xL, caspase-9
Programmed cell death is critical for the physiologic removal of unwanted cells during development, for tissue homeostasis, and in host defense mechanisms (1). Because deregulation of this program also underlies many pathological states, including cancer, autoimmunity, and degenerative diseases (2, 3), clarifying the biochemical mechanisms of apoptosis is a major goal of current biological research.
The pathway to apoptosis has been conserved during evolution. Genetic studies in the nematode Caenorhabditis elegans have identified three regulators required for the activation and execution of apoptosis (CED-3, CED-4, EGL-1) and one critical inhibitor (CED-9) (4, 5). CED-3, a cysteine protease of the caspase family (6), serves as the executioner by cleaving vital cellular substrates. Premature CED-3 activation is precluded by its synthesis as a zymogen, which requires the adaptor protein CED-4 for autocatalytic processing (7). CED-4 activity normally is held in check by CED-9, but CED-9 in turn can be neutralized by its distant relative EGL-1, which directly associates with CED-9 (5).
The mammalian apoptotic program, albeit more complex, is closely analogous. The eight or so mammalian caspases that mediate apoptosis include both those that initiate the program, such as procaspase-9, and those the initiator caspases then process, such as procaspase-3, which cleave downstream targets (6). Activation of procaspase-9 is promoted by Apaf-1, the only well-characterized mammalian homolog of CED-4, in the presence of the cofactors cytochrome c and dATP (8–11). The mammalian relatives of CED-9 include the oncoprotein Bcl-2, the first inhibitor of apoptosis to be identified (12), and its close relatives, such as Bcl-xL, whereas the counterparts of EGL-1 comprise a number of more distant Bcl-2 relatives, such as Bax or Bim, that can bind to the prosurvival molecules and ablate their function (reviewed in ref. 13).
A central unresolved issue in cell death research is how the prosurvival Bcl-2 family members prevent caspase activation. The death programs impaired in cells from mice lacking either Apaf-1 (14, 15) or caspase-9 (16, 17) suggest that both are essential components of the pathway controlled by the Bcl-2 family (see Discussion). If so, the critical issue becomes how these family members keep Apaf-1 in a latent form. Two major types of models remain under debate. The indirect model argues that Bcl-2, which resides on the endoplasmic reticulum, nuclear envelope, and mitochondrial outer membrane, acts to maintain organelle function or integrity (18, 19). For instance, Bcl-2 may prevent the escape of the Apaf-1 cofactor cytochrome c from the intermembrane space of the mitochondrion (10, 20). Other evidence instead favors direct interaction between the prosurvival molecules and the adaptor protein. Studies of C. elegans genes in heterologous overexpression systems have suggested that CED-9, CED-4, and CED-3 exist in a multiprotein complex, dubbed the apoptosome (21), in which CED-9 inhibits apoptosis by binding to CED-4, thereby preventing it from activating CED-3 (22–25). Upon induction of apoptosis, the binding of EGL-1 to CED-9 is envisaged to displace CED-4, allowing it to activate CED-3 (5, 7, 26).
Recently, evidence for analogous mammalian apoptosomes has been reported by two laboratories (27–30). Coimmunoprecipitation studies with epitope-tagged molecules from transiently transfected cells led them to conclude that Bcl-xL (and later also Boo) bound to Apaf-1. However, the similar in vivo association studies reported here for Bcl-xL or its relatives, Apaf-1 and procaspase-9, challenge that conclusion. We find that Apaf-1 can interact with procaspase-9 but we have been unable to detect association between Apaf-1 and any of the six known mammalian prosurvival family members or two viral homologs. Moreover, a new mAb to Apaf-1 has allowed us to show that endogenous Apaf-1 does not interact detectably with either overexpressed or endogenous Bcl-2 or Bcl-xL. Apaf-1 also failed to associate with two proapoptotic family members. Finally, we show that apoptotic stimuli did not induce association of Bcl-2 relatives with Apaf-1. These results suggest that Bcl-2 and its prosurvival homologs do not function by sequestering Apaf-1.
MATERIALS AND METHODS
Expression Constructs.
The expression vector pEF PGKpuro and derivatives expressing wild-type (wt) or FLAG-tagged human Bcl-xL or human Bcl-2, mouse FLAG Bcl-w, mouse FLAG A1, FLAG E1B19K and FLAG BHRF-1 have been described (31–33). Mcl-1 cDNA was amplified by PCR from a mouse thymoma library and cloned into pEF FLAGA PGKpuro (31) to generate pEF Mcl-1FLAG puro. The pcDNA3 mouse Boo FLAG construct was a gift from C. Vincenz, University of Michigan, Ann Arbor (30). Human caspase-9 cDNA, a gift from S. Kumar (Hanson Centre, Adelaide, Australia), and an inactivating C287A mutant generated by PCR using splice overlap extension (34) were subcloned into pEF PGKpuro. C-terminal hemagglutinin (HA)-tagged wt or truncation mutants of human Apaf-1 were generated by PCR using Apaf-1 plasmids isolated from a human HeLa cDNA library (Stratagene) or provided by X.D. Wang, Southwestern Medical Center, Dallas (8). They were cloned into derivatives of pcDNA3 (Invitrogen) that we derived to introduce a C-terminal HA tag. Point mutants of Apaf-1 (K149R and DD232/233AA) were generated by splice overlap extension (34), and pcDNA3 FLAGApaf-1 (27) was a gift from V. Dixit, Genentech, San Francisco.
All PCRs were performed by using proof-reading Pfu Turbo DNA polymerase (Stratagene), and the sequences of all constructs were verified by automated sequencing (ABI Perkin–Elmer). Details of the oligonucleotides are available on request.
Cell Culture, Transfection, Immunoprecipitation, and Western Blot Analysis.
The 293T, HeLa, and Jurkat cell lines were maintained as described (31–33, 35). Cell death was induced by culturing cells in 1 nm to 1 μM staurosporine (Sigma), 5–500 ng/ml doxorubicin (Sigma), or 100 J/m2 UV irradiation.
Liposome-mediated DNA transfection of 293T human embryonal kidney cells was performed essentially as described (32). Briefly, 106 cells were plated in a 10-cm dish and 24 h later were transfected by incubation overnight with 2.5 μg of total plasmid DNA and 15 μl of Lipofectamine (GIBCO/BRL) in 5 ml of high-glucose DMEM lacking antibiotics. This step was followed by addition of 5 ml of DMEM supplemented with 20% FCS. The cells were harvested 18 h after transfection, washed twice in cold PBS, then suspended in lysis buffer (20 mM Tris⋅HCl pH 7.4/135 mM NaCl/1% Triton X-100/10% glycerol, supplemented with 0.5 μg/ml Pefabloc/1 μg/ml each of leupeptin, aprotinin, soybean trypsin inhibitor, and pepstatin/50 mM NaF/5 mM Na3VO4). Protein concentration was determined by the Micro BCA (bicinchoninic acid) Protein Assay Kit (Pierce) according to the manufacturer’s protocol.
For immunoprecipitation, the lysates (1 mg protein per reaction) were precleared by incubation at 4°C for 1.5 h with 20 μl of 50% (vol/vol) Protein G Sepharose 4 Fast Flow beads (Amersham Pharmacia), followed by centrifugation at 13 K for 5 min at 4°C. The supernatant was mixed with excess (>1 μg) antibody at 4°C for 1.5 h, and then with 20 μl of 50% (vol/vol) Protein G Sepharose beads for 1.5 h at 4°C. Then the beads were washed 3–6 times with 0.5 ml of lysis buffer, and the bound proteins were eluted by boiling in 25 μl of loading buffer (0.5M Tris⋅HCl, pH 6.8/4% SDS/20% glycerol/0.005% bromophenol blue) containing 50 mM DTT. The eluate was fractionated by SDS/PAGE (Novex) and transferred to nitrocellulose membranes (Amersham Pharmacia).
Immunoblotting was carried out in PBS containing 5% skim milk powder and 0.05% Tween 20. Antibodies included mouse monoclonal anti-FLAG M2 (Sigma), mouse monoclonal anti-HA 16B12 (HA.11; Babco), rat monoclonal anti-HA 3F10 (Boehringer Mannheim), rabbit polyclonal anti-Bcl-x (PharMingen), mouse monoclonal anti-Bcl-x 7B2.5 (gift of C. Thompson, University of Chicago), mouse monoclonal anti-human Bcl-2–100 (36), hamster monoclonal anti-human Bcl-2 6C8 (37), mouse monoclonal anti-Bax B-9 (Santa Cruz Biotechnology), and rat monoclonal anti-human Apaf-1 2E12 (G. Hausmann, L. A. O’Reilly, D.C.S.H., A. Strasser, and J.M.A., unpublished results). Secondary horseradish peroxidase-conjugated goat antibodies to mouse, rat, and rabbit Ig were from Southern Biotechnology Associates. Immunoreactive proteins were detected by enhanced chemiluminescence (ECL, Amersham Pharmacia), with exposure times of < 5 min.
RESULTS
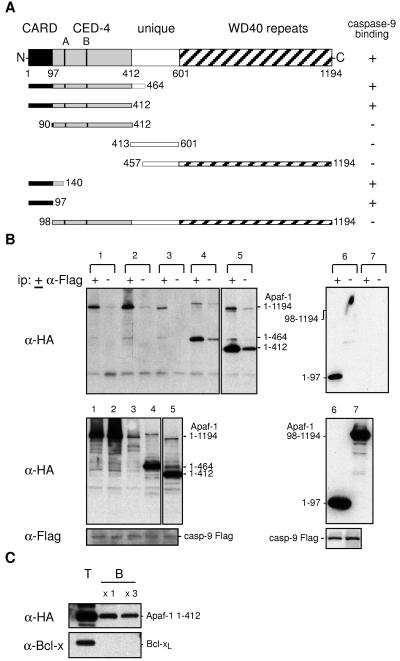
Apaf-1 (8) has several distinctive domains (Fig. 1A). The N-terminal CARD (caspase recruitment domain) motif is followed by a region having extensive homology with C. elegans CED-4, including the Walker A and B boxes needed for ATP binding and a unique region without homology to any known protein, whereas the C-terminal half is comprised of 12 or 13 repeats of the WD40 motif, which often is involved in protein association (8, 11). To explore the role of these domains in Apaf-1 function, we constructed cDNAs that encode eight Apaf-1 deletion mutants (Fig. 1A), as well as two full-length proteins having a point mutation in the A box (K149R) or the B box (DD232/233AA) that should block ATP binding (38).
Figure 1.
Apaf-1 binds through its CARD to procaspase-9. (A) The wt and truncated human Apaf-1 proteins used in this study, all with a C-terminal HA tag, and their binding to procaspase-9. (B) The Apaf-1 CARD (residues 1–97) is necessary and sufficient for binding to procaspase-9. 293T cells were transiently cotransfected with a vector expressing mutant procaspase-9 Flag and ones expressing HA-tagged wt or mutant Apaf-1: wt (lane 1), Apaf-1 K149R (A box mutant) (lane 2), Apaf-1 KDD232/233AA (B box mutant) (lane 3), Apaf-1 (1–464) (lane 4), Apaf-1 (1–412) (lane 5), Apaf-1 (1–97) (lane 6), and Apaf-1 (98–1194) (lane 7). Lysates (1 mg protein) were incubated in the presence (+) or absence (−) of monoclonal anti-FLAG M2, and the resulting immunoprecipitates were fractionated by SDS/PAGE and blotted with monoclonal anti-HA 3F10 antibody (Upper). Expression of procaspase-9 and Apaf-1 was confirmed by blots of lysates (Lower). The larger bands in lanes 4 and 5 represent aggregated Apaf-1 mutants. These data are representative of three experiments. Results with other Apaf-1 derivatives are summarized (A). (C) Apaf-1, particularly its N-terminal region, binds to Sepharose beads. Lysates (0.5 mg protein) from 293T cells transfected with vectors expressing Apaf-1 1–412 HA or Bcl-xL were mixed with 50% (vol/vol) Sepharose CL4B beads (Sigma) at 4°C for 1.5 h, and the beads then were washed once or three times with lysis buffer. Equivalent amounts of total (T) and Sepharose-bound (B) lysate then were Western-blotted. The data are representative of two experiments. Full-length Apaf-1 yielded lower background binding (see text).
The CARD Region of Apaf-1 Allowed Its Association with Procaspase-9.
To identify the regions of Apaf-1 critical for interaction with procaspase-9, we transiently expressed a catalytically inactive mutant of procaspase-9 (C287A) together with the wt or mutant forms of Apaf-1 in the human embryonal kidney cell line 293T, and detected the proteins by the C-terminal Flag epitope on procaspase-9 or the C-terminal HA tag on Apaf-1 and its derivatives. Immunoblots of lysates confirmed that each protein was readily detectable (Fig. 1B, Lower). To reveal any association between them, procaspase-9 was immunoprecipitated from the lysates with anti-Flag antibody. The immune complexes, captured on Protein G Sepharose beads, were fractionated by electrophoresis and blotted with anti-HA antibody to reveal any coprecipitated Apaf-1.
As reported by others (27, 28), full-length Apaf-1 bound to procaspase-9 (Fig. 1B, Upper, lysate 1). Many, but not all, of the Apaf-1 mutants also coimmunoprecipitated with procaspase-9, as summarized in Fig. 1A. The interaction was not prevented by mutation of the Walker A box or B box (lysates 2 and 3), nor by deletion of the unique or WD40 regions (lysates 4 and 5). On the other hand, the CARD was absolutely required. Indeed, that domain (residues 1–97) alone sufficed for interaction with procaspase-9 (lysate 6), and the remainder of Apaf-1 was unable to bind (lysate 7). These data suggest that, at least when coexpressed at high levels, the CARD region of Apaf-1 allows it to complex with procaspase-9 and the remainder of the molecule appears dispensable for the association.
An unexpected technical problem noted during these studies and apparent in Fig. 1B is that some control immunoprecipitates prepared without the anti-Flag antibody (lanes marked −) contained significant levels of Apaf-1. This background, particularly evident with the N-terminal polypeptides containing only the CARD and CED-4 homology regions (lysates 4 and 5), suggested that Apaf-1 could bind nonspecifically to the Sepharose beads. Indeed, a direct test (Fig. 1C) showed that a significant proportion of the truncated Apaf-1 (1–412) bound to these beads in the absence of antibody and remained bound even after several washes. Neither Bcl-xL (Fig. 1C) nor procaspase-9 (not shown) bound significantly to the beads. In view of the “stickiness” of Apaf-1, our protocols routinely included preclearance of the lysates with Protein G Sepharose beads, several wash steps and controls without the immunoprecipitating antibody.
No Interaction Between Overexpressed Bcl-xL and Apaf-1.
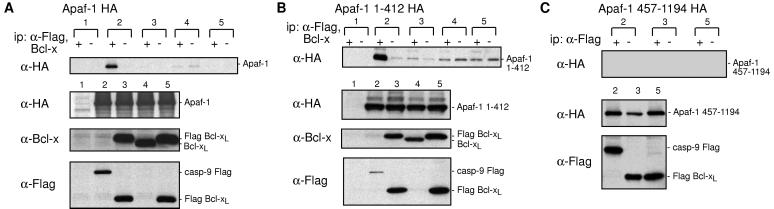
In the apoptosome model, Apaf-1 in healthy cells is sequestered in an inactive form by prosurvival Bcl-2 family members. In view of previous reports (27, 28), we first sought to detect association of Apaf-1 with Bcl-xL, by using procaspase-9 as a positive control. Full-length Apaf-1HA was transiently expressed in 293T cells together with either Flag-tagged procaspase-9 or Bcl-xL, either the native protein or derivatives with Flag at either end. Immunoprecipitates were prepared with anti-Bcl-x or anti-Flag antibodies and blotted with anti-HA antibody (Fig. 2A, Upper). As expected, the procaspase-9 immunoprecipitate (lysate 2) reproducibly contained substantial Apaf-1. In contrast, no evidence was found for specific interaction between Apaf-1 and either native Bcl-xL or its tagged derivatives (the trace of Apaf-1 in certain precipitates was also evident in the control lacking antibody). Inability to detect association could not be ascribed to poor expression of Bcl-xL, because its level was in fact higher than that of procaspase-9, as judged by analysis of the lysates with anti-Flag antibody (Fig. 2A, Lower). Moreover, both procaspase-9 and Bcl-xL were efficiently immunoprecipitated by the antibodies (data not shown). In case the C-terminal HA tag on Apaf-1 had prevented the putative Apaf-1:Bcl-xL interaction, we also tested Apaf-1 with an N-terminal HA tag, or an N- or C- terminal Flag tag, but again we found no evidence for interaction (data not shown).
Figure 2.
Overexpressed Bcl-xL and Apaf-1 do not interact. Tests with full-length Apaf-1 (A), its N-terminal moiety (B), or its C-terminal moiety (C). 293T cells were transiently transfected with vectors encoding (A) Apaf-1 HA, (B) Apaf-1 (1–412) HA, or (C) Apaf-1 (457–1194) HA, together with either a vector control (lane 1) or vectors encoding mutant caspase-9 FLAG (lane 2), FLAG Bcl-xL (lane 3), Bcl-xL (lane 4), or Bcl-xL FLAG (lane 5). Proteins were immunoprecipitated in the presence (+) or absence (−) of anti-FLAG M2 or polyclonal anti-Bcl-x, fractionated by SDS/PAGE and then blots were analyzed with anti-HA 3F10 (Upper). Expression of the constructs was documented by Western blots on lysates (Lower). Data shown are representative of three experiments.
Because the WD40 region of Apaf-1 may negatively regulate its action (8, 39, 40), we next tested whether removal of that domain would allow interaction with Bcl-xL. An HA-tagged N-terminal portion of Apaf-1 (residues 1–412), including the CARD and CED-4 homology domain, was transiently coexpressed in 293T cells with the Bcl-xL derivatives. Under conditions where Apaf-1(1–412) clearly interacted with procaspase-9 (lysate 2 in Fig. 2B), it did not interact specifically with either native Bcl-xL (lysate 4) or the tagged versions (lysates 3 and 5). Similarly, the C-terminal moiety of Apaf-1 containing the WD40 repeats (residues 457-1194) also failed to interact with the Bcl-xL (Fig. 2C).
Thus, in contrast to recent reports (27, 28), our immunoprecipitation experiments have not revealed any evidence of specific interaction between overexpressed Bcl-xL and Apaf-1, under conditions where association between Apaf-1 and procaspase-9 was readily demonstrable. It is highly unlikely that the putative Apaf-1:Bcl-xL complex was disrupted by the immunoprecipitating antibody, because the diverse antibodies tested included a polyclonal antiserum to Bcl-x (Fig. 2) and a monoclonal anti-Bcl-x antibody (data not shown), as well as the anti-Flag antibody, interacting with a tag at either end of Bcl-xL (Fig. 2). Moreover, we tested lysates prepared with other detergents (1% NP-40 or 1% digitonin) or in the absence of detergent (data not shown). Hence, the lack of interaction is unlikely to reflect epitope masking, antibody interference, or lysis conditions. Nor is it peculiar to the cell line used, because cotransfected COS cells yielded identical results (data not shown). Some of our experiments also included protocols that closely replicated those reported to reveal such complexes (27, 28), but no specific complexes were found.
Endogenous Apaf-1 Did Not Interact with Any Prosurvival Bcl-2 Family Member.
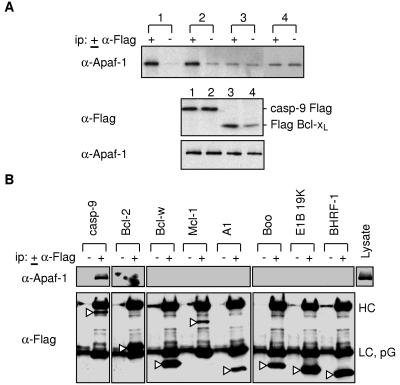
Because all of the studies described above had been performed with epitope-tagged Apaf-1, it remained possible that the tag had inhibited interaction with Bcl-xL (although not with procaspase-9). The recent generation of mAbs for Apaf-1 by L. O’Reilly (G. Hausmann, L. A. O’Reilly, D.C.S.H., A. Strasser, and J.M.A., unpublished results) enabled us to look for interactions with endogenous Apaf-1. The 2E12 mAb used here is specific for human Apaf-1 and recognizes its CARD. Lysates of the human 293T cells transfected with vectors for Flag-tagged procaspase-9 or Bcl-xL were precipitated with anti-Flag antibody and the immune complexes were analyzed with the 2E12 antibody (Fig. 3A). Endogenous Apaf-1 coprecipitated with procaspase-9 (lysates 1 and 2) but not detectably with Bcl-xL (lysates 3 and 4). Irrespective of whether Bcl-xL bore an N- or C-terminal Flag tag, the levels of Apaf-1 in those anti-Flag immunoprecipitates were no greater than in the control without antibody (Fig. 3A, compare + and − lanes).
Figure 3.
Endogenous Apaf-1 does not interact with Bcl-xL or the other prosurvival homologs. (A) Tests with Bcl-xL and procaspase-9. Lysates from 293T cells transfected with wt procaspase-9 FLAG (lane 1), procaspase-9 C287A FLAG (lane 2), FLAG Bcl-xL (lane 3), or Bcl-xL FLAG (lane 4) were immunoprecipitated by using monoclonal anti-FLAG M2, fractionated by SDS/PAGE and then analyzed by Western blotting with monoclonal anti-Apaf-1 2E12. (B) Tests with other prosurvival Bcl-2 homologs. Lysates from 293T cells transfected with constructs expressing Flag-tagged mutant procaspase-9, Bcl-2, Bcl-w, Mcl-1, A1, Boo, E1B19K, or BHRF-1 were immunoprecipitated with anti-FLAG M2, and the blot was probed with anti-Apaf-1 2E12 (Upper). Probing of the same blot (Lower) with anti-Flag M2 confirmed expression and immunoprecipitation of each of the transfected Bcl-2 homologs (open arrows). HC, heavy chain; LC, light chain; PG, protein G. Data shown are representative of four experiments.
If there prove to be several mammalian CED-4 homologs, Apaf-1 might preferentially interact with another Bcl-2 prosurvival relative. To explore this possibility, we transiently expressed in 293T cells each of the five other known mammalian prosurvival family members (Bcl-2, Bcl-w, Mcl-1, A1, and Boo), as well as two viral functional homologs (adenovirus E1B 19K and Epstein–Barr virus BHRF-1). Western blots confirmed adequate expression and precipitation of each of these Flag-tagged proteins (arrowed bands in Fig. 3B, Lower). However, none of their immunoprecipitates contained endogenous Apaf-1, although it clearly interacted with transfected procaspase-9 (Fig. 3B, Upper).
Endogenous Apaf-1 Did Not Associate with Endogenous Bcl-2 or Bcl-xL.
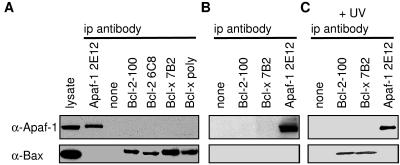
To avoid any possible artefacts associated with overexpression, we next tried to detect interaction of endogenous Apaf-1 with endogenous Bcl-2 or Bcl-xL in two human lines containing substantial Apaf-1: HeLa epithelial cells and Jurkat lymphoid cells. When Apaf-1 was immunoprecipitated from the HeLa cell lysates with mAb 2E12, neither Bcl-2 nor Bcl-xL was detected (data not shown). In the converse experiment, both prosurvival proteins were precipitated with each of two antibodies and none of the immunoprecipitates contained Apaf-1 (Fig. 4A, Upper). Lysates from Jurkat cells gave the same result. Importantly, endogenous Bax did coprecipitate with the prosurvival molecules under these conditions (Fig. 4A, Lower). The formation of Bcl-2/Bax heterodimers appears to involve a conformational change in Bax induced by detergents in the cell lysates (41). In case detergents had interfered with interaction of Bcl-2 (or Bcl-xL) with Apaf-1, we repeated all these experiments with lysates made in the absence of detergents but still observed no association of endogenous Apaf-1 with either prosurvival protein (Fig. 4B).
Figure 4.
Endogenous Apaf-1 does not interact with endogenous Bcl-2 or Bcl-xL in HeLa cells. Lysates prepared in the presence of Tritan X-100 from healthy cells (A), or without detergent from healthy cells (B), or those dying from UV treatment (C) cells were immunoprecipitated with anti-Apaf-1 2E12, or the indicated anti-Bcl-2 or anti-Bcl-x antibodies. The blots were probed with anti-Apaf-1 2E12 (Upper) or anti-Bax B-9 (Lower). Viability of the healthy cells was 98% and remained 90% for the dying cells at this time point (6 h after exposure to UV irradiation). Data shown are representative of three experiments.
Apoptotic Stimuli Did Not Induce Apaf-1 to Associate with Bcl-2 Family Members.
An alternative hypothesis for control of Apaf-1 would be that Bcl-2 prosurvival relatives sequester Apaf-1 after a death stimulus, thereby serving as a “sink” to prevent activated Apaf-1 from initiating the caspase cascade. To test this possibility, we subjected HeLa cells to a cytotoxic dose of UV radiation and immunoprecipitated Bcl-2 and Bcl-xL from lysates made without detergent. As reported with another cytotoxic stimulus (42), Bax underwent a change that allowed it to associate with the prosurvival proteins, but no Apaf-1 was discernible in the immune complexes (Fig. 4C). Treatment with staurosporine or doxorubicin yielded the same result (data not shown).
Yet another model would be that Apaf-1 binds to the proapoptotic members of the Bcl-2 family. For instance, the change in Bax might allow it to trigger Apaf-1 activation. However, immunoprecipitates of Apaf-1 did not contain Bax (Fig. 4 A and B), even after induction of cell death (Fig. 4C). Similarly, in both healthy Jurkat cells and those induced to die, Apaf-1 was not found in association with Bax or with Bim, a representative of the second major class of proapoptotic Bcl-2 family members (data not shown).
DISCUSSION
Recently the strong biochemical evidence that Apaf-1 is the adaptor protein controlling procaspase-9 activation (9, 11, 39) has been firmly supported genetically by the similar phenotypes of mice lacking Apaf-1 or caspase-9 (14–17). Our immunoprecipitation data confirm that Apaf-1 can interact with procaspase-9 and show that the Apaf-1 N-terminal CARD is necessary and sufficient for the association (Fig. 1) when either protein is overexpressed. We currently are testing whether these proteins interact in healthy cells when both are at physiological levels. Presumably, the binding of dATP to Apaf-1 is not required for its association with procaspase-9, because Apaf-1 with mutated A or B boxes bound as well as the wt protein (Fig. 1B). Regions of Apaf-1 other than the CARD appeared not to contribute to the interaction. Their roles remain less well defined, although the CED-4 region may be required for oligomerization (7, 11), whereas the WD40 repeat region may play a negative regulatory role (8, 39, 40).
Biochemical studies on the key apoptotic players from C. elegans have suggested that both CED-9 and CED-3 interact with CED-4 in healthy cells. The apoptosome model (21) suggests that CED-9 holds CED-4 in an inactive form until a death stimulus induces expression of EGL-1, which binds to CED-9, thereby releasing CED-4 to activate proteolytic activity of CED-3 (5, 26). The central issue addressed in this paper is whether, in an analogous manner, the mammalian prosurvival Bcl-2 family members function by binding to and neutralizing Apaf-1. In experiments equivalent in design to those described here, two laboratories have reported that overexpressed Apaf-1 binds to Bcl-xL (27, 28) and the newly described Boo/Diva (29, 30), whereas one experiment has suggested binding to Bcl-2 (43).
Our coimmunoprecipitation results are in conflict with those reports. We have been unable to detect any interaction of Apaf-1 with Bcl-xL or any other prosurvival Bcl-2 family member (Figs. 2–4). As well as testing full-length Apaf-1, we tested its N-terminal and C-terminal moieties. We excluded trivial technical explanations such as the presence or location of epitope tags, the cell line, and the lysis and immunoprecipitation conditions. Importantly, in case the epitope tag on Apaf-1 had inhibited interaction with Bcl-xL, we also looked for interaction with endogenous Apaf-1 by using a new Apaf-1-specific mAb, but found no evidence for interaction with transfected Bcl-2, Bcl-xL, Bcl-w, A1, Mcl-1, Boo, or their viral counterparts adenovirus E1B19K and Epstein–Barr virus BHRF-1 (Fig. 3). Moreover, we found no association of endogenous Apaf-1 with endogenous Bcl-2 or Bcl-xL, nor with Bax or Bim, representatives of the two classes of proapoptotic family members (Fig. 4). Importantly, no detectable association with Bcl-2 or Bcl-xL was induced by several cytotoxic stimuli (Fig. 4 and data not shown).
A plausible sequestration model would seem to require that a great proportion, if not all, of the adaptor resides in a tight complex with the prosurvival family members. No evidence for the existence of such complexes with Apaf-1 was found here, and it is unclear whether the Apaf-1 complexes reported by others (27–30, 43) would account for more than a few percent of the Apaf-1 molecules. Our current studies on the localization of Apaf-1 also argue against a sequestration model. Both confocal microscopy and subcellular fractionation indicate that neither overexpressed nor endogenous Apaf-1 co-localize with Bcl-2 or Bcl-xL (G. Hausmann, L. A. O’Reilly, D.C.S.H., A. Strasser, and J.M.A., unpublished results). Thus, we have found no support for the hypothesis that prosurvival Bcl-2 homologs physically sequester Apaf-1. Our observation that Apaf-1 can bind nonspecifically to Sepharose beads, particularly after removal of its WD40 repeat region (Fig. 1C), may in part explain the apparent interaction between Apaf-1 and Bcl-2-like proteins reported previously. The evidence for interaction of the homologous C. elegans proteins appears stronger, but as yet it all derives from overexpression in heterologous cell types (yeast or mammalian cells). It will be important to learn whether endogenous CED-9 and CED-4 interact in the worm.
Is it plausible that Apaf-1 is not the bona fide functional homolog of CED-4, and that a true, as-yet-undiscovered, mammalian counterpart is regulated by the Bcl-2 family? Certainly Apaf-1 has domains not present in CED-4, so its regulation may well differ from that of CED-4. Indeed, C. elegans CED-4 does not appear to require cytochrome c to activate CED-3 (38). On the other hand, the phenotype of mice deficient in Apaf-1 argues strongly that Apaf-1 is an essential, nonredundant component of the pathway regulated by Bcl-2, because cells from Apaf-1−/− mice are refractory to several of the death stimuli inhibitable by Bcl-2 and behave normally toward the signals from “death receptors” such as CD95 (14, 15), a pathway largely independent of Bcl-2 control (35). Nevertheless, another mammalian CED-4 homolog might participate. Two molecules with trace homology to CED-4 have been described very recently (44, 45), but their roles remain to be established.
Our results strongly suggest that the regulation of Apaf-1 by Bcl-2 is indirect rather than direct. More specifically, Bcl-2 and its prosurvival homologs may prevent the release of apoptogenic factors from mitochondria (18, 19) and, in view of their distribution in the cell, presumably also from the nucleus and endoplasmic reticulum. Cytochrome c is thus far the strongest candidate; it usually is released from mitochondria of cells undergoing apoptosis and serves as an Apaf-1 cofactor (10, 20, 46–48), probably by forming a multimeric complex with Apaf-1 and procaspase-9 (11). On the other hand, cytochrome c release is unlikely to be the sole and critical step regulated by Bcl-2 and its prosurvival relatives because Bcl-2 (and Bcl-xL) retains antiapoptotic activity in cells microinjected with cytochrome c (49, 50), cytochrome c loss from mitochondria can be reversed (51), and none was released during killing by a Bcl-xL peptide antagonist (52). Cytochrome c release may, in fact, serve mainly to amplify the apoptosis process (53), and Bcl-2 may control a crucial, as-yet-unknown, upstream initiating step.
Acknowledgments
We thank S. Novakovic, J. Beaumont, and S. Ahmad for expert technical assistance and Drs. J. Trapani and S. Kumar for comments on the manuscript. The anti-Apaf-1 antibodies were generated by Drs. L. O’Reilly and A. Strasser. We thank Drs. A. Strasser, L. O’Reilly, H. Puthalakath, L. O’Connor, and G. Hausmann for advice and discussions, and V. Dixit, S. Kumar, C. Thompson, C. Vincenz, X.D. Wang, and T. Willson for generous gifts of cell lines, antibodies, cDNA libraries, and expression vectors. K.M. was a recipient of a fellowship from the Japan Science and Technology Corporation; D.C.S.H. is a Special Fellow of the Leukemia Society of America. This work was supported by the National Health and Medical Research Council (Canberra, Reg. Key 973002) and U.S. National Cancer Institute Grant CA80188.
ABBREVIATIONS
- wt
wild type
- HA
hemagglutinin
- CARD
caspase recruitment domain
References
- 1.Vaux D L, Korsmeyer S J. Cell. 1999;96:245–254. doi: 10.1016/s0092-8674(00)80564-4. [DOI] [PubMed] [Google Scholar]
- 2.Thompson C B. Science. 1995;267:1456–1462. doi: 10.1126/science.7878464. [DOI] [PubMed] [Google Scholar]
- 3.Strasser A, Huang D C S, Vaux D L. Biochim Biophys Acta. 1997;1333:F151–F178. doi: 10.1016/s0304-419x(97)00019-x. [DOI] [PubMed] [Google Scholar]
- 4.Ellis R E, Yuan J, Horvitz H R. Annu Rev Cell Biol. 1991;7:663–698. doi: 10.1146/annurev.cb.07.110191.003311. [DOI] [PubMed] [Google Scholar]
- 5.Conradt B, Horvitz H R. Cell. 1998;93:519–529. doi: 10.1016/s0092-8674(00)81182-4. [DOI] [PubMed] [Google Scholar]
- 6.Thornberry N A, Lazebnik Y. Science. 1998;281:1312–1316. doi: 10.1126/science.281.5381.1312. [DOI] [PubMed] [Google Scholar]
- 7.Yang X, Chang H Y, Baltimore D. Science. 1998;281:1355–1357. doi: 10.1126/science.281.5381.1355. [DOI] [PubMed] [Google Scholar]
- 8.Zou H, Henzel W J, Liu X, Lutschg A, Wang X. Cell. 1997;90:405–413. doi: 10.1016/s0092-8674(00)80501-2. [DOI] [PubMed] [Google Scholar]
- 9.Li P, Nijhawan D, Budihardjo I, Srinivasula S M, Ahmad M, Alnemri E S, Wang X. Cell. 1997;91:479–489. doi: 10.1016/s0092-8674(00)80434-1. [DOI] [PubMed] [Google Scholar]
- 10.Liu X, Kim C N, Yang J, Jemmerson R, Wang X. Cell. 1996;86:147–157. doi: 10.1016/s0092-8674(00)80085-9. [DOI] [PubMed] [Google Scholar]
- 11.Zou H, Li Y, Liu X, Wang X. J Biol Chem. 1999;274:11549–11556. doi: 10.1074/jbc.274.17.11549. [DOI] [PubMed] [Google Scholar]
- 12.Vaux D L, Cory S, Adams J M. Nature (London) 1988;335:440–442. doi: 10.1038/335440a0. [DOI] [PubMed] [Google Scholar]
- 13.Adams J M, Cory S. Science. 1998;281:1322–1326. doi: 10.1126/science.281.5381.1322. [DOI] [PubMed] [Google Scholar]
- 14.Cecconi F, Alvarez-Bolado G, Meyer B I, Roth K A, Gruss P. Cell. 1998;94:727–737. doi: 10.1016/s0092-8674(00)81732-8. [DOI] [PubMed] [Google Scholar]
- 15.Yoshida H, Kong Y-Y, Yoshida R, Elia A J, Hakem A, Hakem R, Penninger J M, Mak T W. Cell. 1998;94:739–750. doi: 10.1016/s0092-8674(00)81733-x. [DOI] [PubMed] [Google Scholar]
- 16.Kuida K, Haydar T F, Kuan C-Y, Gu Y, Taya C, Karasuyama H, Su M S-S, Rakic P, Flavell R A. Cell. 1998;94:325–337. doi: 10.1016/s0092-8674(00)81476-2. [DOI] [PubMed] [Google Scholar]
- 17.Hakem R, Hakem A, Duncan G S, Henderson J T, Woo M, Soengas M S, Elia A, de la Pompa J L, Kagi D, Khoo W, et al. Cell. 1998;94:339–352. doi: 10.1016/s0092-8674(00)81477-4. [DOI] [PubMed] [Google Scholar]
- 18.Zamzami N, Brenner C, Marzo I, Susin S A, Kroemer G. Oncogene. 1998;16:2265–2282. doi: 10.1038/sj.onc.1201989. [DOI] [PubMed] [Google Scholar]
- 19.Green D R, Reed J C. Science. 1998;281:1309–1311. doi: 10.1126/science.281.5381.1309. [DOI] [PubMed] [Google Scholar]
- 20.Kluck R M, Bossy-Wetzel E, Green D R, Newmeyer D D. Science. 1997;275:1132–1136. doi: 10.1126/science.275.5303.1132. [DOI] [PubMed] [Google Scholar]
- 21.Hengartner M O. Nature (London) 1998;391:441–442. doi: 10.1038/35036. [DOI] [PubMed] [Google Scholar]
- 22.Spector M S, Desnoyers S, Hoeppner D J, Hengartner M O. Nature (London) 1997;385:653–656. doi: 10.1038/385653a0. [DOI] [PubMed] [Google Scholar]
- 23.Wu D, Wallen H D, Nuñez G. Science. 1997;275:1126–1129. doi: 10.1126/science.275.5303.1126. [DOI] [PubMed] [Google Scholar]
- 24.Chinnaiyan A M, O’Rourke K, Lane B R, Dixit V M. Science. 1997;275:1122–1126. doi: 10.1126/science.275.5303.1122. [DOI] [PubMed] [Google Scholar]
- 25.Seshagiri S, Miller L K. Curr Biol. 1997;7:455–460. doi: 10.1016/s0960-9822(06)00216-8. [DOI] [PubMed] [Google Scholar]
- 26.del Peso L, González V M, Núñez G. J Biol Chem. 1998;273:33495–33500. doi: 10.1074/jbc.273.50.33495. [DOI] [PubMed] [Google Scholar]
- 27.Pan G H, O’Rourke K, Dixit V M. J Biol Chem. 1998;273:5841–5845. doi: 10.1074/jbc.273.10.5841. [DOI] [PubMed] [Google Scholar]
- 28.Hu Y, Benedict M A, Wu D, Inohara N, Núñez G. Proc Natl Acad Sci USA. 1998;95:4386–4391. doi: 10.1073/pnas.95.8.4386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Inohara N, Gourley T S, Carrio R, Muñiz M, Merino J, Garcia I, Koseki T, Hu Y, Chen S, Núñez G. J Biol Chem. 1998;273:32479–32486. doi: 10.1074/jbc.273.49.32479. [DOI] [PubMed] [Google Scholar]
- 30.Song Q, Kuang Y, Dixit V M, Vincenz C. EMBO J. 1999;18:167–178. doi: 10.1093/emboj/18.1.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang D C S, Cory S, Strasser A. Oncogene. 1997;14:405–414. doi: 10.1038/sj.onc.1200848. [DOI] [PubMed] [Google Scholar]
- 32.Huang D C S, O’Reilly L A, Strasser A, Cory S. EMBO J. 1997;16:4628–4638. doi: 10.1093/emboj/16.15.4628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huang D C S, Adams J M, Cory S. EMBO J. 1998;17:1029–1039. doi: 10.1093/emboj/17.4.1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Horton R M, Ho S N, Pullen J K, Hunt H D, Cai Z, Pease L R. Methods Enzymol. 1993;217:270–279. doi: 10.1016/0076-6879(93)17067-f. [DOI] [PubMed] [Google Scholar]
- 35.Strasser A, Harris A W, Huang D C S, Krammer P H, Cory S. EMBO J. 1995;14:6136–6147. doi: 10.1002/j.1460-2075.1995.tb00304.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pezzella F, Tse A G D, Cordell J L, Pulford K A F, Gatter K C, Mason D Y. Am J Pathol. 1990;137:225–232. [PMC free article] [PubMed] [Google Scholar]
- 37.Veis D J, Sentman C L, Bach E A, Korsmeyer S J. J Immunol. 1993;151:2546–2554. [PubMed] [Google Scholar]
- 38.Chinnaiyan A M, Chaudhary D, O’Rourke K, Koonin E V, Dixit V M. Nature (London) 1997;388:728–729. doi: 10.1038/41913. [DOI] [PubMed] [Google Scholar]
- 39.Srinivasula S M, Ahmad M, Fernandes-Alnemri T, Alnemri E S. Mol Cell. 1998;1:949–957. doi: 10.1016/s1097-2765(00)80095-7. [DOI] [PubMed] [Google Scholar]
- 40.Hu Y, Ding L, Spencer D M, Núñez G. J Biol Chem. 1998;273:33489–33494. doi: 10.1074/jbc.273.50.33489. [DOI] [PubMed] [Google Scholar]
- 41.Hsu Y T, Youle R J. J Biol Chem. 1997;272:13829–13834. doi: 10.1074/jbc.272.21.13829. [DOI] [PubMed] [Google Scholar]
- 42.Wolter K G, Hsu Y T, Smith C L, Nechushtan A, Xi X G, Youle R J. J Cell Biol. 1997;139:1281–1292. doi: 10.1083/jcb.139.5.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fang G, Chang B S, Kim C N, Perkins C, Thompson C B, Bhalla K N. Cancer Res. 1998;58:3202–3208. [PubMed] [Google Scholar]
- 44.Imai Y, Kimura T, Murakami A, Yajima N, Sakamaki K, Yonehara S. Nature (London) 1999;398:777–785. doi: 10.1038/19709. [DOI] [PubMed] [Google Scholar]
- 45.Bertin J, Nir W-J, Fischer C M, Tayber O V, Errada P R, Grant J R, Keilty J J, Gosselin M L, Robison K E, Wong G H W, et al. J Biol Chem. 1999;274:12955–12958. doi: 10.1074/jbc.274.19.12955. [DOI] [PubMed] [Google Scholar]
- 46.Yang J, Liu X, Bhalla K, Kim C N, Ibrado A M, Cai J, Peng T-I, Jones D P, Wang X. Science. 1997;275:1129–1132. doi: 10.1126/science.275.5303.1129. [DOI] [PubMed] [Google Scholar]
- 47.Kharbanda S, Pandey P, Schofield L, Israels S, Roncinske R, Yoshida K, Bharti A, Yuan Z-M, Saxena S, Weichselbaum R, et al. Proc Natl Acad Sci USA. 1997;94:6939–6942. doi: 10.1073/pnas.94.13.6939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bossy-Wetzel E, Newmeyer D D, Green D R. EMBO J. 1998;17:37–49. doi: 10.1093/emboj/17.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Brustugun O T, Fladmark K E, Døskeland S O, Orrenius S, Zhivotovsky B. Cell Death Differ. 1998;5:660–668. doi: 10.1038/sj.cdd.4400399. [DOI] [PubMed] [Google Scholar]
- 50.Li F, Srinivasan A, Wang Y, Armstrong R C, Tomaselli K J, Fritz L C. J Biol Chem. 1997;272:30299–30305. doi: 10.1074/jbc.272.48.30299. [DOI] [PubMed] [Google Scholar]
- 51.Martinou I, Desagher S, Eskes R, Antonsson B, André E, Fakan S, Martinou J-C. J Cell Biol. 1999;144:883–889. doi: 10.1083/jcb.144.5.883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Holinger E P, Chittenden T, Lutz R J. J Biol Chem. 1999;274:13298–13304. doi: 10.1074/jbc.274.19.13298. [DOI] [PubMed] [Google Scholar]
- 53.Cosulich S C, Savory P J, Clarke P R. Curr Biol. 1999;9:147–150. doi: 10.1016/s0960-9822(99)80068-2. [DOI] [PubMed] [Google Scholar]