Abstract
The mouse Bapx1 gene is homologous to the Drosophila homeobox containing bagpipe (bap) gene. A shared characteristic of the genes in these two organisms is expression in gut mesoderm. In Drosophila, bap functions to specify the formation of the musculature of the midgut. To determine the function of the mammalian cognate, we targeted a mutation into the Bapx1 locus. Bapx1, similar to Drosophila, does have a conspicuous role in gut mesoderm; however, this appears to be restricted to development of the spleen. In addition, Bapx1 has a major role in the development of the axial skeleton. Loss of Bapx1 affects the distribution of sclerotomal cells, markedly reducing the number that appear ventromedially around the notochord. Subsequently, the structures in the midaxial region, the intervertebral discs, and centra of the vertebral bodies, fail to form. Abnormalities are also found in those bones of the basal skull (basioccipital and basisphenoid bones) associated with the notochord. We postulate that Bapx1 confers the capacity of cells to interact with the notochord, effecting inductive interactions essential for development of the vertebral column and chondrocranium.
The mouse and human Bapx1 genes (1, 2) are homeobox-containing genes that belong to the NK-2 family first described in Drosophila (3). This Drosophila gene family is comprised of three members: the tinman (tin), ventral nervous system defective (vnd), and bagpipe (bap) genes (reviewed in ref. 4). Bapx1 is most similar to the Drosophila bap gene and, along with two other closely related vertebrate genes, Xenopus Xbap (5) and the urodele Nkx-3.2 (S. Nicolas and Y. Le Parco, GenBank accession no. 488714), appears to form a distinct subgroup within the NK-2 family (1, 4).
Bapx1 expression in the mouse (1) is detected initially at E8.5 in the splanchnic mesoderm adjacent to the prospective gut endoderm and in the sclerotomal portion of the somites. At E10.5, Bapx1 is detected in limb mesenchyme and first branchial arch that becomes restricted to the precursor of Meckel’s cartilage. In Drosophila, bap is required for the specification of the visceral mesoderm during midgut musculature formation (6). Genetic lesions within this gene show a reduction or deletion in the visceral musculature. The role of bap in gut musculature and the expression of Bapx1 in splanchnic mesoderm surrounding the gut led to the suggestion that the Drosophila and mouse genes may have similar roles during gut development (1). Here, we inactivate the Bapx1 gene in mouse and show that this gene does have a role in splanchnic mesoderm; however, this role appears to be restricted to the early development of the spleen. In addition, this gene plays a major role in the development of the axial skeleton and the base of the chondrocranium.
MATERIALS AND METHODS
Targeting Strategy.
To obtain a genomic clone, a λget library (7) was screened. A clone containing an insert of approximately 7 kb, stretching 5′ from the KpnI site at nucleotide 1,272 to the vector cloning site, was subcloned (in pBluescript II S/K), digested with RsrII at nucleotide 710, and the XhoI-BamHI fragment of pMC1neo-polyA (Stratagene) was inserted. The final clone was designated pBapneo45.
Approximately 100 μg of the pBapneo45 insert was electroporated into 2 × 107 E14 embryonic stem (ES) cells (a kind gift of Austin Smith). Individual neomycin-resistant clones (309) were picked and grown, of which 173 were analyzed (9). Genomic DNA was extracted (10), digested with SacI, run on a 1% agarose gel, and blotted onto Zeta-Probe GT membranes (Bio-Rad) for Southern analysis. The membranes were hybridized with a probe external to the DNA within the targeting construct; i.e., representing 3′ Bapx1 nucleotides 1,275 to 1,472 (Fig. 1A). The wild-type allele gives a band of 2.5 kb, whereas the correctly targeted allele is represented by a band 1.1 kb larger. Five clones were identified as correctly targeted and were reanalyzed by using the external 3′ probe and an internal neomycin probe. Two of these clones were prepared for karyotyping and then injected into blastocysts (8).
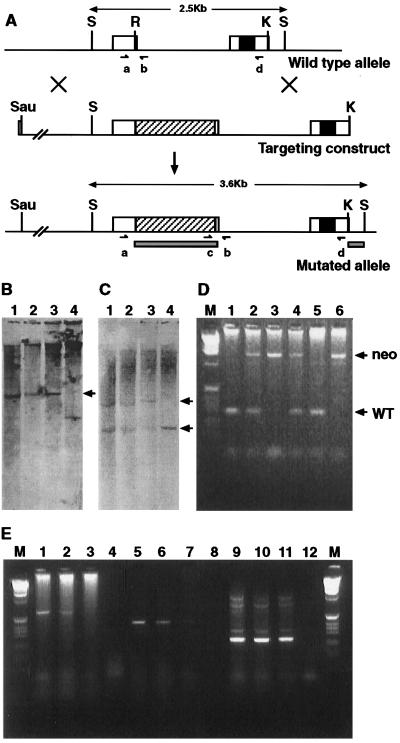
Figure 1.
Diagrammatic representation of the genomic organization and analysis of the targeted mutation. (A) Top line shows the Bapx1 genomic organization, the open boxes representing the two exons, the filled-in box the homeobox. The middle line is the targeting construct, an approximate 7-kb fragment. The diagonally hatched bar shows the location of pMC1neo-polyA at the RsrII site. The bottom line represents the genomic organization after correct targeting. The solid bars show the position of the two probes used for Southern blot analysis, and the oligonucleotides used for genotyping are represented by a, b, c, and d. Abbreviations for restrictions sites are: S, SacI; R, RsrII; and K, KpnI. (B and C) Southern analysis of four ES cell clones after digestion with SacI. B was probed with the pMC1neo-polyA probe, and lanes 1, 2, and 3 show the expected 3.6-kb band (arrow) for the correctly targeted allele, and 4 shows the incorrectly targeted control. C was probed with the 3′ probe, external to the targeting construct (the right hand solid box in A). All four lanes show the wild-type 2.5-kb band (bottom arrow), whereas only lanes 1, 2, and 3 show the additional 3.6-kb band generated from the mutated allele (top arrow). D shows the results of PCR genotyping on the yolk sacs from a number of F2 progeny from a heterozygous intercross. The primers used are marked as a and b in Fig. 1A. These give rise to a 280-bp fragment from the wild-type allele (arrow marked WT) and 1.4 kb from the mutated allele (arrow marked neo). Thus the genotypes of the embryos are: lane1 +/+; lane 2 +/−; lane 3 −/−; lane 4 +/−; lane 5 +/+, lane 6 −/−. Analysis of expression from the mutant allele is shown in E. Embryonic RNA samples used were wild type in lanes 1, 5, and 9; Bapx1+/− in lanes 2, 6, and 10, and Bapx1−/− in lanes 3, 7, and 11. The no-RNA controls are in lanes 4, 8, and 12. Lanes 1–4 are reverse transcription–PCR products from 5′ of the neo insertion (primer a) to 3′ of the homeobox (primer d) and show no full-length transcripts in the Bapx1−/− track (lane 3). Lanes 5–8 use primers 3′ of the neo insertion to 3′ of the homeobox (primer d) and show reduced levels of transcription in the Bapx1−/− track (lane7). Lanes 9–12 show the product from the hypoxanthine phosphoribosyltransferase control primers.
The resulting male chimeras were crossed to C57BL/6 females, and the progeny was analyzed by PCR. Primers used are from the 5′ exon of Bapx1 (CCGAACCAGAACAGCCGTGG) (a in Fig. 1A) and the 5′ end of the intron (CAGCCCCCTTCCTGGAGAAC) (b in Fig. 1A). These PCR reactions, done as described in the manufacturer’s recommendations (Perkin–Elmer) (except the reactions were supplemented with 10% DMSO), give rise to a wild-type product of 280 bp and a targeted product of 1.4 kb (Fig. 1D). The presence of the neomycin-containing allele was initially confirmed by using a primer from within the 5′ end of neomycin (CATCGCCTTCTATCGCCTTC) (c in Fig. 1A) in conjunction with the Bapx1 intron primer. Neomycin-positive F1 progeny were then intercrossed and the F2 generation examined at weaning, birth, and as embryos.
To look at expression from the mutant allele, RNA was extracted from E11.5 embryos with RNAzol (Biogenesis, Bournemouth, U.K.) for reverse transcription–PCR. First Strand cDNA Synthesis Kit (Amersham Pharmacia Biotech) was used for cDNA synthesis and PCR according to the manufacturer’s instructions. Primer combinations used were either a and d in Fig. 1 (35 cycles) or a primer within exon 1 of BapX1 but 3′ of the RsrII site (AGCGAGACGTCAGCCAGCG) with primer d. As a positive control, primers within the HPRT locus (CACAGGACTAGAACACCTGC and GCTGGTGAAAAGGACCTCT) (30 cycles) were used. In all cases, primer combinations cross an intron, and the band obtained represents the correctly spliced message.
Analysis of Embryos.
Yolk-sac DNA (9) was used to genotype each embryo by PCR, as described above.
In preparation for skeletal staining, E17.5 and E18.5 fetuses were fixed in 100% ethanol, skinned, and eviscerated, then stained with alizarin red and alcian blue overnight at 37°C. These were cleared for 10 hr in 1% KOH then for several days in 50:50 1% KOH/glycerol before photography.
Embryos for histology were fixed in 4% paraformaldehyde, sectioned at 20 μm on a cryostat (E18.5), and stained with haemotoxylin and eosin.
Whole-Mount in Situ Hybridization.
Whole-mount in situ hybridizations were performed as in Hammond et al. (10). The probes used for the analyses were Hox11 (nucleotides 83–614 [11]), myogenin (P. Soriano), and Pax1 (P. Gruss).
RESULTS
Genetic Inactivation of Bapx1.
A 7-kb genomic fragment extending from the 5′ flanking DNA and containing the 5′ exon, the single intron, and part of the 3′ exon, was used to create the construct for homologous recombination in ES cells (Fig. 1A). The strategy pursued was to insert the neomycin dominant selectable gene, pMC1Neo-polyA, into the unique RsrII site of the coding region of the 5′ exon (Fig. 1A). Of 173 ES cell clones analyzed, five proved to have the construct correctly targeted into the homologous site (Fig. 1 B–D). Two independent ES lines, both of which showed the normal chromosomal karyotype, were selected to create chimeric mice. Mice derived from the two ES cell lines passed the mutant allele to offspring, and mouse lines were then derived.
Appropriate genomic integration of the construct is predicted to have a 2-fold effect on normal production of the Bapx1 gene product. First, insertion of the pMCNeo-polyA fragment into the first exon (Fig. 1A) places two stop codons into the reading frame that would result in a truncated translation product. Second, the pMC1Neo coupled to the SV40 Poly(A) addition signal would be predicted to disrupt transcription. Analysis of transcription from the mutated allele showed undetectable levels of the full length read-through transcript (Fig. 1E). Downstream of the pMC1Neo insert, very low production of spliced mRNA was found. Thus the insertion interferes with normal transcription; however, we cannot exclude the occurrence of an anomalous splicing event that may yield a translation product with altered activity.
Analysis of 253 intercross progeny showed no Bapx1−/− homozygous mutants at weaning. However, there was no reduction in the number of Bapx1 heterozygotes showing a ratio of 84 wild type to 169 heterozygotes. At stages just before birth, at E17.5 and E18.5, there was no apparent reduction in the number of homozygous fetuses. Analysis of 111 fetuses showed approximately the expected ratio of genotypes (30 wild type:49 Bapx1+/−:32 Bapx1−/−); thus the Bapx1 mutation results in neonatal lethality.
External examination of live mice revealed detectable differences between the wild-type and heterozygous littermates. Forty-six percent of the heterozygotes (54 of 116) displayed kinked tails, a trait lacking in the wild-type littermates.
Bapx1 Mutant Mice Lack Ventral Vertebral Elements.
At E18.5, Bapx1−/− homozygous fetuses appeared slightly shorter and broader than wild-type mice. Because Bapx1 is expressed in sclerotome from an early stage (1), it seemed likely that the axial skeleton was defective. Skeletal analysis showed that the shorter embryos were characterized by ribs that were tightly spaced, appearing more splayed (Fig. 2 A–D), but which otherwise showed no abnormalities such as fusions or disruptions. The sterna of Bapx1−/− fetuses were unaffected, and the size and number of sternabra were the same as in wild type (Fig. 2 C and D). The vertebrae also appeared more closely spaced along the whole of the vertebral column. This was particularly pronounced for the vertebrae in the cervical region (Fig. 2 E and F).
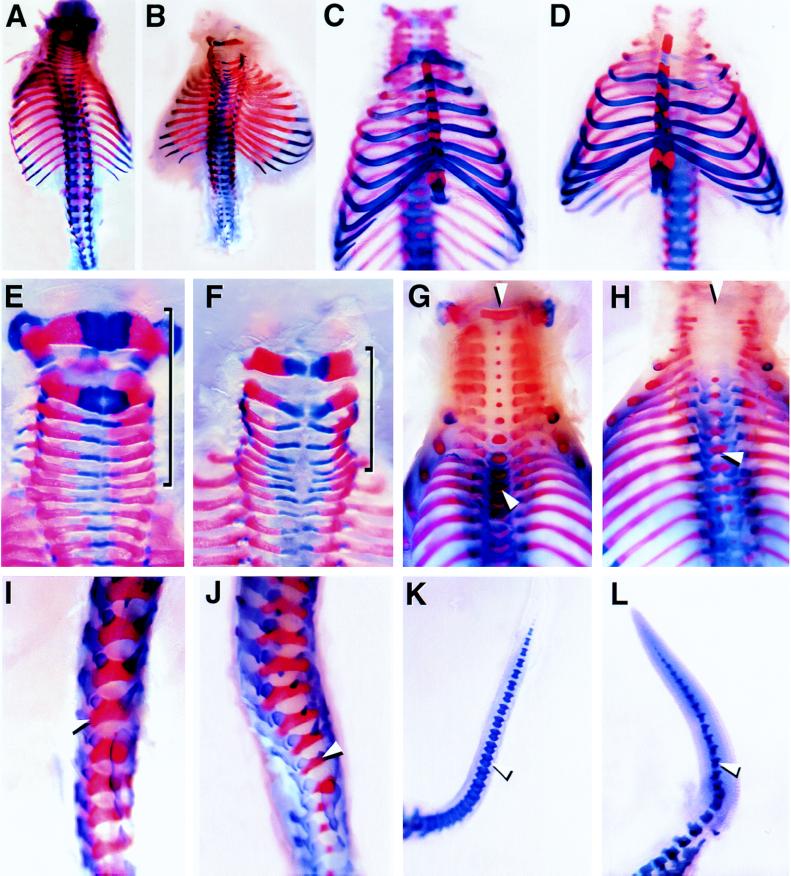
Figure 2.
Skeletal preparations of E18.5 fetuses. Wild-type (A, C, E, G, I, and K) and Bapx1−/− mutant (B, D, F, H, J, and L) fetuses were prepared such that the axial skeleton could be examined. In a dorsal view, the mutant fetus B has a slightly shortened skeleton, more highly compact vertebrae, and lateral extension of the ribs as compared with wild type (A). C and D are viewed ventrally, which shows that the ribs and sternum are unaffected in the mutant (D). Wild-type (E and G) and mutant (F and H) cervical vertebrae are shown dorsally (E and F) and ventrally (G and H). In E and F, the cervical vertebrae are highlighted by the bracket showing the compression of the mutant vertebrae (F). In G and H, the midline is indicated by the upper white arrowhead showing the medial ossification centers (red button of stain). The midline clefting seen in the mutant thoracic vertebrae is shown by the lower white arrowhead. A lateral view of the lumbar (I and J) region shows that the elements that form the neural arches are more narrow in the mutant (J). The arrowhead points to the last lumbar vertebra for comparison. The fetal tails are shown in K and L. The mutant tail (L) is shorter and thicker, and the vertebrae are missing the anterior processes (arrowhead).
Wild-type animals revealed in each vertebra an area of ossification in the ventromedial region (red midline stain in Fig. 2G). In Bapx1−/− embryos, this ossification center was not apparent in most of the vertebrae (Fig. 2H). Some midthoracic vertebrae showed medial ossification centers; however, these occurred at the point of midline fusion of the neural arches. (Fig. 2H). These thoracic vertebrae showed a medial clefting (lower arrow in Fig. 2H) immediately ventral to the region where the centra normally form. The tail vertebrae were also defective in the mutant and appeared small with the anterior processes missing (Fig. 2 K and L).
Transverse sections of E18.5 fetuses showed that the ventromedial elements of each vertebra, i.e., the vertebral bodies and intervertebral discs, were absent in Bapx1−/− fetuses. In the cervical region (Fig. 3 A and B), all elements at the midline were missing, and the neural arches did not fuse at the midline. In the thoracic region (Fig. 3 C and D), in contrast, the vertebral bodies showed a narrow ventral bridge linking the lateral vertebral elements of the neural arches (arrow in Fig. 3D). This dorsal bridge resulted in the clefting seen in Fig. 2H. Thus in these vertebrae only the centra are lacking. In wild-type animals, the intervertebral discs are distinguished by the presence of the notochord-derived nucleus pulposus (Fig. 3 A and C). In Bapx1−/− fetuses, these ventromedial elements were not found, and only a remnant of the notochord persisted throughout (arrowhead in Fig. 3 B and D).
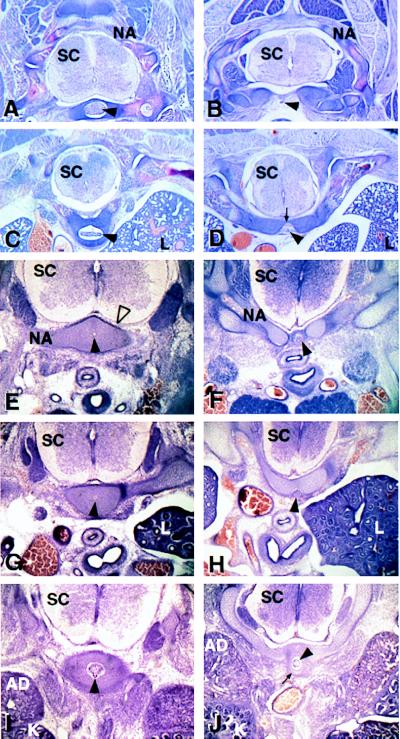
Figure 3.
Histological analysis of wild-type and mutant embryos. Transverse sections are shown for E18.5 (A–D) and E14.5 (E–J) wild-type (A, C, E, G, and I) and Bapx1−/− (B, D, F, H, and J) embryos. A, B, E, and F are sections at the level of the cervical vertebrae, C, D, G, and H, the thoracic vertebrae, and I and J, the lumbar vertebrae. At E18.5, the notochord has given rise to the nucleus pulposum (arrowhead in A and C) surrounded by the vertebral body. In the mutant (B and D), these elements are missing, and the arrowheads indicate the vestigial notochord. Note the ventral bridges of the neural arches (arrow in D). At E14.5, cells in the wild type have organized at the ventral midline around the notochord (black arrowhead) at all axial levels (E, G, and I). In the mutant, few cells organize around the notochord in the cervical and thoracic vertebrae (F and H). In the lumbar region (J), more cells are apparent (arrow), but are not well organized. SC, spinal cord; NA, neural arch; L, lung; Ad, Adrenal; and K, Kidney.
Because Bapx1 is expressed in the early sclerotome, an earlier developmental stage was examined. At E14.5 (Fig. 3 E–J), the cells that composed the lateral elements of the neural arches had condensed in both wild-type and mutant embryos; however, the organization around the notochord differed. At this stage, the wild-type embryos showed an organized condensation of sclerotomal cells around the notochord (Fig. 3 E, G, and I), which in the cervical and upper thoracic region of mutants was lacking (Fig. 3 F and H). In the lower vertebrae, more cells accumulated at the midline near to the notochord; however, this tissue was still markedly reduced in volume and appeared disorganized (Fig. 3J).
Abnormal Notochord and Basal Cranial Bones in Bapx1 Mutants.
The basioccipital and the basisphenoid bones are located at the base of the skull and are derived from mesenchyme associated with the notochord. The basioccipital bone forms from the occipital somites, whereas the development of the posterior basisphenoid does not depend on somites but is derived from the cephalic parachordal mesenchyme (12). At E14.5 the number of cells at the midline of the basioccipital bone was markedly reduced (Fig. 4C and arrowhead in D). By E18.5, the basioccipital bone was misshapen and the posterior basisphenoid bone was dramatically smaller (Fig. 4 A and B).
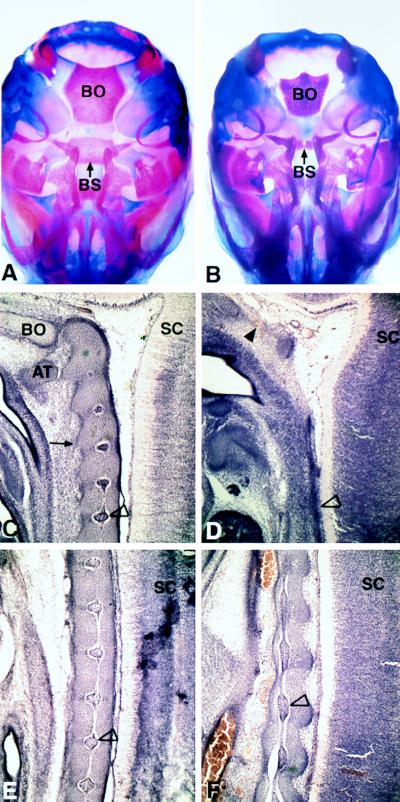
Figure 4.
Analysis of the chondrocranium (A and B at E18.5) and notochord (C–F at E14.5) Ventral views of the skull of wild type (A) and mutant (B) show a decrease in size of the basioccipital bone (BO) and posterior basisphenoid bone (BS). C–F show sagittal sections of wild-type (C and E) and mutant (D and F) embryos near the midline of the axial skeleton through the notochord region. C and D are sections from the cervical region. The characteristic notochordal swellings are shown in C (open arrowhead indicates characteristic swelling). The thin mutant notochord is indicated by the open arrowhead in D. The condensing cells of the anterior located basioccipital bone (in C, designated BO) are missing in the mutant (region indicted by filled arrowhead in D). More posteriorly, the periodic notochordal swellings in wild type (open arrowhead in E) are abnormal in the mutant (F). AT, anterior arch of the atlas; SC, spinal cord; BO, basioccipital; BS, posterior basisphenoid.
Sagittal sections of the wild-type embryo showed the normal notochordal pattern of swellings embedded in the densely packed precursors of the intervertebral discs and constrictions residing in the vertebral bodies (Fig. 4 C and E). In the posterior region of mutant embryos, the notochord did not manifest in this repetitive pattern, and the swollen nodes were either broadened (Fig. 4F) or, in some embryos, did not appear. This abnormal pattern correlated with a reduction in number of ventral cells and a disruption of the normal boundaries between the vertebral bodies and intervertebral discs that surround the notochord (Fig. 4F). In the cervical region, the notochord showed no periodic swellings and had no organized arrangement of cells surrounding it (Fig. 4D).
Bapx1 Mutation Disrupts Spleen Formation.
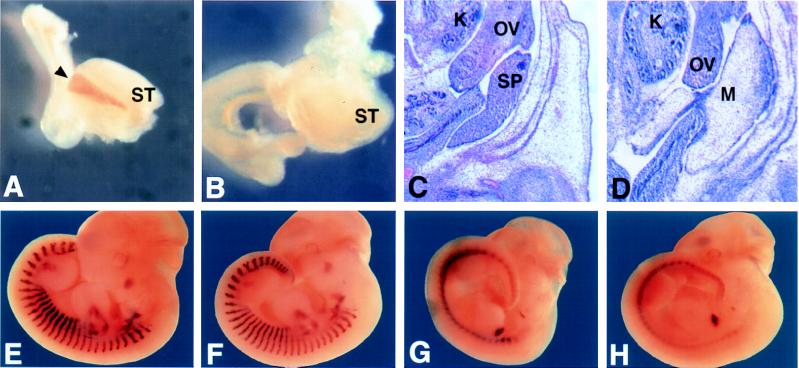
The spleen was apparent in the E14.5 wild-type embryo as densely packed cells situated in splanchnic mesoderm derived tissue that lies near the stomach (Fig. 5C). In the Bapx1−/− embryos, the corresponding region of the mesenchyme showed no evidence of splenic cells (Fig. 5D). In addition, expression of Hox11, an early spleen-specific gene expression marker (11, 13), was not detected in the Bapx1−/− mutant at E12 (Fig. 5 A and B), demonstrating that spleen development is disrupted at an early stage.
Figure 5.
Analysis of the spleen and expression of Hox11, myogenin, and Pax1. Expression of Hox11 at E12 was examined in stomach and surrounding tissue from dissected embryos. The spleen of wild-type (A) embryos is observed as a brown stripe next to the stomach (black arrowhead), which is missing in Bapx1−/− (B). Histological examination in transverse sections shows densely packed cells of the spleen in wild type (in C, designated SP), which is not observed in a comparable region of mesentery in the mutant (D). Wild type (E and F) and mutant (G and H) show no differences in the expression of myogenin (lateral stripes in E and F) or Pax1 (lateral periodic spots in G and H). ST, stomach; K, kidney; OV, ovary; SP, spleen; M, mesentary.
The Pax2 Gene Is Not Regulated by Bapx1.
Pax1, which has a very similar mutant phenotype (14, 15), is expressed in the sclerotomal cells at the same early stages as Bapx1 (1). At E11.5, we assayed for Pax1 expression in the Bapx1 mutant to examine their relationship. There is no detectable difference in Pax1 expression between wild-type (Fig. 5G) and mutant (Fig. 5H) embryos. In addition, the defective sclerotome appears to have little or no effect on the neighboring myotome as shown by the expression of myogenin (Fig. 5 E and F). We also examined the expression of Gli2 in the sclerotome and Shh in the notochord but did not detect any differences (data not shown).
DISCUSSION
Spleen Deficiency.
Spleen deficiencies are rare in mouse and thus far only two other asplenic mutations have been described: dominant hemimelia (Dh) (16) and the Hox11 targeted mutation (11, 13). The absence of a morphologically distinguishable spleen and the lack of Hox11 expression show that Bapx1 is a third gene fundamental to early spleen development.
A possible evolutionary link between Bapx1 and the Drosophila homologue bap has been suggested (1). The Drosophila bap gene affects specification of the visceral mesoderm resulting in reduction or loss of visceral musculature (6). Bapx1 is expressed broadly in the splanchnic mesoderm that in addition to spleen development contributes to the gut musculature (see ref. 17 for review). However, Bapx1 did not have an overt effect on formation of gut musculature, suggesting there is no equivalent function for the mouse and Drosophila genes. Bapx1 and bap, therefore, affect cognate tissue types, i.e., gut mesoderm, but with differing consequences. In addition to a restricted role in splanchnic mesoderm, Bapx1 is also expressed in the limb and Meckel’s cartilage (1), where there is no apparent phenotype. Thus the possibility exists that other bagpipe-related genes are present that compensate for the loss of Bapx1.
Vertebral and Cranial Abnormalities.
Abnormalities within the axial skeleton of the Bapx1 mutants were restricted to the ventromedial elements of the vertebrae that develop surrounding the notochord. The defects decrease in severity in a rostral-to-caudal direction. At E14.5, the cervical vertebrae contain very few midline sclerotomal cells. In the mid to lower thoracic/lumbar region, more sclerotomal cells appear at the ventral midline; however, these cells are markedly decreased in number and are disorganized. By E18.5, the ventromedial elements of each vertebra are completely absent. In the cervical vertebrae, the whole of the vertebral midline is missing. More posteriorly in many of the thoracic and lumbar vertebrae, the ventral neural arches fuse at the midline, suggesting that the deficiency is exclusive to the centra in these vertebrae.
In the basal plate of the chondrocranium, two bones are affected by the Bapx1 mutation. The first, the basioccipital bone, is deficient in midline cells at E14.5, which by E18.5 leads to reduction in the size of the bone. Sclerotome from the occipital somites is responsible for the basioccipital bone; however, formation of this cranial element is distinctly different from the vertebrae (12). The second cranial bone affected by Bapx1−/− is the posterior basisphenoid bone. This bone is not derived from somitic mesoderm, but from the cephalic parachordal mesoderm.
A feature shared by the ventromedial vertebral components and these two cranial bones is that, initially, development occurs in association with the notochord. The notochord is the primary embryonic component implicated in inductive signaling to somites in the derivation of the sclerotome (19, 20). By implication, the notochord signals to the parachordal mesoderm in formation of the basisphenoid bone. We argue that Bapx1 functions in specifying an identity to the medial cells, whether of sclerotomal or cephalic mesodermal origin, such that these cells are responsive to the notochord.
A signaling molecule produced by the notochord is Shh (19, 20). Two genes, Pax1 (19, 21) and Gli2 (22), both expressed in sclerotome, may function in Shh inductive signaling from the notochord. Mutations in either of these two genes show phenotypes similar to the Bapx1−/− in the axial skeleton (14, 15, 22). Additionally, Gli2 loss of function disrupts development of the basioccipital and basisphenoid bones in a manner similar to Bapx1−/− (22). Evidence suggests that Pax1 acts as a mediator of the Shh inductive signal in sclerotomal cells (18, 19, 23). Gli2, like the Drosophila homolog ci, appears to participate in the hedgehog-signaling cascade (22). The similarity in phenotype between the Bapx1 deficiency and the Gli2 and Pax1 mutations suggests that Bapx1 acts similarly to mediate the Shh notochordal signal. It is apparent that Bapx1 does not regulate Pax1 and Gli2 expression, suggesting that Bapx1 is acting either downstream or in a parallel pathway to both these genes.
It is clear that the structure of the notochord is also abnormal in Bapx−/− animals. At E14.5, the periodic notochordal pattern is disrupted, and by E18.5 the enlarged nucleus pulposus of each intervertebral disc is missing. We examined Shh expression in the notochord at E11.5 (a stage relevant to Shh signaling) and did not observe differences in expression. Because Bapx1 is not detectably expressed in the notochord, we suggest that the disruption of the notochord is caused by lack of surrounding Bapx1 expressing sclerotomal cells. We suggest that these sclerotomal-derived cells are necessary for the modeling of the notochordal pattern, perhaps by signaling back to the notochord.
Acknowledgments
The authors thank Austin Smith for the kind gift of the E14 ES cells and Philippe Soriano, Peter Gruss, and Jacob Hecksher-Sørenson for the in situ probes. We also thank the Medical Research Council–Human Genetics Unit Photography and Illustration Department for their expertise, Vince Ranaldi and Andy Wilson for their expert technical assistance, and Duncan Davidson for helpful discussion.
ABBREVIATION
- ES
embryonic stem
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
References
- 1.Tribioli C, Frasch M, Lufkin T. Mech Dev. 1997;65:145–162. doi: 10.1016/s0925-4773(97)00067-1. [DOI] [PubMed] [Google Scholar]
- 2.Tribioli C, Lufkin T. Gene. 1997;203:225–233. doi: 10.1016/s0378-1119(97)00520-9. [DOI] [PubMed] [Google Scholar]
- 3.Kim Y, Nirenberg M. Proc Natl Acad Sci USA. 1989;86:7716–7720. doi: 10.1073/pnas.86.20.7716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Harvey R P. Dev Biol. 1996;178:203–216. doi: 10.1006/dbio.1996.0212. [DOI] [PubMed] [Google Scholar]
- 5.Newman C S, Grow M W, Cleaver O, Chia F, Krieg P. Dev Biol. 1997;181:223–233. doi: 10.1006/dbio.1996.8416. [DOI] [PubMed] [Google Scholar]
- 6.Azpiazu N, Frasch M. Genes Dev. 1993;7:1325–1340. doi: 10.1101/gad.7.7b.1325. [DOI] [PubMed] [Google Scholar]
- 7.Nehls M, Pfeifer D, Boehm T. Oncogene. 1994;9:2169–2175. [PubMed] [Google Scholar]
- 8.Dorin J R, Dickinson P, Alton E W, Smith S N, Geddes D M, Stevenson B J, Kimber W L, Fleming S, Clarke A R, Hooper M L, et al. Nature (London) 1992;359:211–215. doi: 10.1038/359211a0. [DOI] [PubMed] [Google Scholar]
- 9.Laird P W, Zijderveld A, Linders K, Rudnicki M A, Jaenisch R, Berns A. Nucleic Acids Res. 1991;19:4293. doi: 10.1093/nar/19.15.4293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hammond K L, Hanson I M, Brown A G, Lettice L A, Hill R E. Mech Dev. 1998;74:121–132. doi: 10.1016/s0925-4773(98)00071-9. [DOI] [PubMed] [Google Scholar]
- 11.Roberts C W M, Shutter J R, Korsmeyer S J. Nature (London) 1994;368:747–749. doi: 10.1038/368747a0. [DOI] [PubMed] [Google Scholar]
- 12.Couly G F, Coltey P M, Le Douarin N M. Development (Cambridge, UK) 1993;117:409–429. doi: 10.1242/dev.117.2.409. [DOI] [PubMed] [Google Scholar]
- 13.Dear T N, Colledge W H, Carlton M B, Lavenir I, Larson T, Smith A J, Warren A J, Evans M J, Sofroniew M V, Rabbitts T H. Development (Cambridge, UK) 1995;121:2909–2915. doi: 10.1242/dev.121.9.2909. [DOI] [PubMed] [Google Scholar]
- 14.Wallin J, Wilting J, Koseki H, Fritsch R, Christ B, Balling R. Development (Cambridge, UK) 1994;120:1109–1121. doi: 10.1242/dev.120.5.1109. [DOI] [PubMed] [Google Scholar]
- 15.Wilm B, Dahl E, Peters H, Balling R, Imai K. Proc Natl Acad Sci USA. 1998;95:8692–8697. doi: 10.1073/pnas.95.15.8692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Green M C. Dev Biol. 1967;15:62–89. doi: 10.1016/0012-1606(67)90006-1. [DOI] [PubMed] [Google Scholar]
- 17.Yasugi S. Dev Growth Differ. 1993;35:1–9. doi: 10.1111/j.1440-169X.1993.00001.x. [DOI] [PubMed] [Google Scholar]
- 18.Koseki H, Wallin J, Wilting J, Mizutani Y, Kispert A, Ebensperger C, Herrmann B G, Christ B, Balling R. Development (Cambridge, UK) 1993;119:649–660. doi: 10.1242/dev.119.3.649. [DOI] [PubMed] [Google Scholar]
- 19.Fan C M, Tessier-Lavigne M. Cell. 1994;79:1175–1186. doi: 10.1016/0092-8674(94)90009-4. [DOI] [PubMed] [Google Scholar]
- 20.Johnson R L, Laufer E, Riddle R D, Tabin C. Cell. 1994;79:1165–1173. doi: 10.1016/0092-8674(94)90008-6. [DOI] [PubMed] [Google Scholar]
- 21.Fan C M, Porter J A, Chiang C, Chang D T, Beachy P A, Tessier-Lavigne M. Cell. 1995;81:457–465. doi: 10.1016/0092-8674(95)90398-4. [DOI] [PubMed] [Google Scholar]
- 22.Mo R, Freer A M, Zinyk D L, Crackower M A, Michaud J, Heng H H-Q, Chik K W, Shi X-M, Tsui L-C, Cheng S H, et al. Development (Cambridge, UK) 1997;124:113–123. doi: 10.1242/dev.124.1.113. [DOI] [PubMed] [Google Scholar]
- 23.Balling R, Neubuser A, Christ B. Cell Dev Biol. 1996;7:129–136. doi: 10.1006/dbio.1996.0227. [DOI] [PubMed] [Google Scholar]