Abstract
Culturing a population of Saccharomyces cerevisiae for many generations under conditions to which it is not optimally adapted selects for fitter genetic variants. This simple experimental design provides a tractable model of adaptive evolution under natural selection. Beginning with a clonal, founding population, independently evolved strains were obtained from three independent cultures after continuous aerobic growth in glucose-limited chemostats for more than 250 generations. DNA microarrays were used to compare genome-wide patterns of gene expression in the evolved strains and the parental strain. Several hundred genes were found to have significantly altered expression in the evolved strains. Many of these genes showed similar alterations in their expression in all three evolved strains. Genes with altered expression in the three evolved strains included genes involved in glycolysis, the tricarboxylic acid cycle, oxidative phosphorylation, and metabolite transport. These results are consistent with physiological observations and indicate that increased fitness is acquired by altering regulation of central metabolism such that less glucose is fermented and more glucose is completely oxidized.
Keywords: adaptive evolution, DNA microarrays, glucose metabolism, chemostat
Evolutionary theory has long held that the process of adaptation is driven by the competition for limiting resources. Among heterotrophic microorganisms in nature, the availability of reduced carbon places stringent limitations on the ability of these organisms to multiply. As a result, the machinery of central metabolism is tuned to exploit reduced carbon resources in natural environments, where they vary greatly in both form and abundance.
Saccharomyces cerevisiae is by far the most important domesticated microorganism; it has been exploited for baking and alcohol production by virtually every human society. Its utility in these respects results from the ability to ferment sugar even in the presence of oxygen. Thus, S. cerevisiae exhibits considerable flexibility in the coordinated regulation of the enzymes that mediate fermentative and oxidative metabolism. Maximal growth rate is achieved on high concentrations of glucose, under which conditions nearly all of it is fermented, even in the presence of oxygen (1–3). In other words, S. cerevisiae displays only a minimal “Pasteur effect,” i.e., aerobic inhibition of glycolysis.
Two kinds of regulation are known that may account for much of this flexibility. First, respiratory capacity is limited by regulation in the presence of fermentable carbon sources. Yeast readily increase their respiratory capacity when provided only with nonfermentable carbon sources, in part by increasing transcription of genes encoding, among others, the mitochondrial enzymes required for respiration (4). Second, S. cerevisiae has evolved an elaborate regulatory network for sensing glucose, its preferred carbon source. In the presence of glucose, expression of genes for catabolism of other carbon sources, even those that can be fermented, is repressed (5). Both mechanisms probably contribute to the pattern of carbon utilization characteristic of S. cerevisiae, a pattern that couples maximal growth rate to incomplete oxidation of glucose.
Much of what we understand about the physiology of these processes is derived from the study of yeast cultured under continuous nutrient limitation, a condition achieved by growth in a chemostat. Fresh medium with a limiting concentration of glucose is pumped through the growth chamber at a constant rate, and a constant efflux of cells and medium from the growth chamber occurs at the same rate. Yeast grown in a glucose-limited chemostat quickly reach a steady state. The identity of the limiting nutrient is easily confirmed: modest changes in limiting nutrient concentration result in a simple, predictable change in the steady-state density of the organisms (6, 7).
Chemostat experiments that extend for fewer than 20 generations allow for the fine-scale investigation of physiological responses to defined conditions (8). Experiments of longer duration become the study of evolution in action. When a yeast population is propagated asexually under continuous long-term nutrient limitation, it characteristically undergoes a series of adaptive shifts whereby clones of higher fitness successively replace one another over time. Such shifts have been observed to occur on the order of once every 50 generations (9, 10). They can readily be recognized by the relatively sudden disappearance of neutral mutations that have been accumulating in the population, as each new, fitter mutant outgrows the population having the progenitor genotype (11, 12).
MATERIALS AND METHODS
Strains, Media, Culture Conditions, and Sampling.
The parental strain used in these experiments, CP1AB, is an isogenic diploid derived from the haploid Saccharomyces cerevisiae strain XC500A by Paquin and Adams (9); its genotype is a/α, gal2/gal2, mel/mel, mal/mal. Three long-term chemostats were inoculated with this parental diploid yeast strain, culminating in three independently evolved populations referred to as E1, E2, and E3. The parental strain (CP1AB, referred to herein as P) and E1 (28–15-L4) have been described (9, 13). All three populations were subjected to the same continuous aerobic culture at 30°C in 200-ml vessels on glucose-limited (0.08%) minimal medium (13) at a dilution rate of 0.2 h−1. E1 was cultivated for ≈500 generations, E2 and E3 for ≈250 generations.
For the purpose of yield determinations, steady-state metabolite concentrations and expression analyses, single-colony isolates of the P, E1, E2, and E3 populations were grown to steady state in short-term (10–15 generation) chemostat cultures under conditions identical to those used for the long-term evolutionary selections. Yield cell number and yield biomass dry weight were assayed as described (13). Equilibrium substrate concentration, s, in the chemostat, was determined by spectrophotometric assay of 0.2-μm-filtered chemostat cultures by using the coupled hexokinase/glucose-6-phosphate dehydrogenase assay (Boehringer Mannheim kit no. 716251). Ethanol concentrations in filtered media were measured spectrophotometrically by using the coupled alcohol dehydrogenase/acetaldehyde dehydrogenase assay (Boehringer Mannheim kit no.176290). For expression analyses, culture volumes of ≈150 ml were fast-filtered by using a 0.45-μm nylon membrane and immediately frozen in liquid N2. Cells were kept at −70°C until processed for RNA.
Gene Expression Analyses.
To characterize changes in the pattern of gene expression in the three evolved strains, mRNA levels for virtually every yeast gene were compared by using DNA microarrays. As described (4, 14, 15), these arrays were made by robotically spotting PCR-amplified copies of each yeast ORF onto glass microscope slides. RNA extracted from samples taken from short-term chemostat cultures was used to generate fluorescently labeled probe; the mRNA samples from the evolved strains were labeled with Cy5 (red) and those from the parental strain with Cy3 (green). Mixtures of Cy5- and Cy3-labeled probes were used to hybridize to three arrays (i.e., E1/parent; E2/parent, and E3/parent), and the red and green fluorescence intensities determined. The complete data set can be downloaded as tab-delimited tables of supplemental data from the PNAS web site (www.pnas.org) or can be explored at http://genome-www.stanford.edu/evolution/.
As a control, we compared transcript levels between RNA samples prepared from two growths of a single-colony isolate of the E1 population that were independently cultured for 10 generations in a chemostat. One sample was labeled with Cy5 and the other with Cy3. When these were mixed and hybridized to the array, only 0.19% (12/6,124) of the genes showed as much as 2-fold differential expression, meaning that the much more numerous changes in expression levels described below are due to genetic differences and not just experimental variation among isolates, chemostats, or sample preparation.
Microscopy.
Cells were cultured aerobically in the chemostat with glucose limitation, harvested, and fixed in 4% formaldehyde PBS (pH 7.2) for 60 min. Cells were washed three times in PBS and resuspended in mounting medium (glycerol, 0.1 mg⋅ml−1 p-phenylenediamine, 10% PBS, pH 8.0) containing 50 ng⋅ml−1 4′,6′-diamino-2-phenylindole (DAPI). Imaging and analysis were performed by using a photometrix PXL cooled CCD camera attached to the Delta Vision deconvolution system (Applied Precision, Issaquah, WA) microscope (Applied Imaging Systems). Twenty 0.2-μm sections were deconvoluted for the DAPI images.
RESULTS AND DISCUSSION
The growth characteristics of the three independently evolved strains were very similar (Table 1). Each of the three produced, at steady state, ≈4-fold more cells (3-fold greater biomass) per gram of glucose; each left ≈3-fold less glucose unused. Most significantly, perhaps, all three evolved strains produced at least an order of magnitude less ethanol than the parental strain. Functionally, then, all three strains had evolved to improve the efficiency with which they use glucose, the limiting resource. The low ethanol production suggests that the evolved strains have curtailed the metabolism of pyruvate into ethanol in favor of respiration. To confirm that the differences seen were not just because of differences in cell density, the same parameters were measured with cell density of the parental strain matched to that of the evolved strains by increasing the concentration of glucose in the feed medium (Table 1). These experiments resulted in a proportional increase in steady-state ethanol concentration.
Table 1.
Metabolites and cell yield
| Strain | Media glucose, concentration | Steady-state A600* | Residual glucose, g/liter | EtOH g/liter | Yield,† cell no. | Yield,‡ biomass | No. of independent cultures |
|---|---|---|---|---|---|---|---|
| P | 0.8§ | 0.61 ± 0.09¶ | 0.17 ± 0.07 | 0.27 ± 0.12 | 0.94 | 0.19 | 5 |
| 2.4§ | 1.60 ± 0.09 | 0.18 ± 0.24 | 0.79 ± 0.21 | 1.02 | 0.14 | 5 | |
| E1 | 0.8 | 1.67 ± 0.12 | 0.04 ± 0.01 | 0.01 ± 0.01 | 3.43 | 0.59 | 4 |
| E2 | 0.8 | 1.70 ± 0.18 | 0.06 ± 0.01 | 0.03 ± 0.02 | 4.34 | 0.67 | 4 |
| E3 | 0.8 | 1.59 ± 0.18 | 0.05 ± 0.03 | 0.02 ± 0.02 | 4.49 | 0.45 | 4 |
Steady state was at a dilution of 0.2 h−1.
Units for cell no. yield is 1010 cells per gram of glucose.
Units for biomass yield is grams of dry cell weight per gram of glucose.
Glucose concentration in the feed medium reservoir was varied.
Values are expressed as mean ± SD.
When the three evolved strains were each compared in turn to the parent strain, it became clear that the patterns of gene expression in the evolved strains were remarkably similar to one another. Nearly 10% (608/6,124) of the genes on the array showed at least a 2-fold change in transcript abundance between at least one of the evolved strains and their common ancestor. When the results from the three evolved strains were averaged, 3.0% (184/6,124) of yeast genes still showed at least a 2-fold difference between the average transcript level in the evolved strains and the level in the parental strain. Of these, virtually all showed a consistent alteration of at least 2-fold in at least two comparisons between evolved and parent. Half of these genes (1.4%, or 88/6,124) have well characterized functional descriptions in the Saccharomyces Genome Database (http://genome-www.stanford.edu/Saccharomyces). We discuss only this subset of 88 genes below, but the data for each comparison and the entire body of data for all genes can be viewed on our web site or downloaded as tables from the PNAS web site.
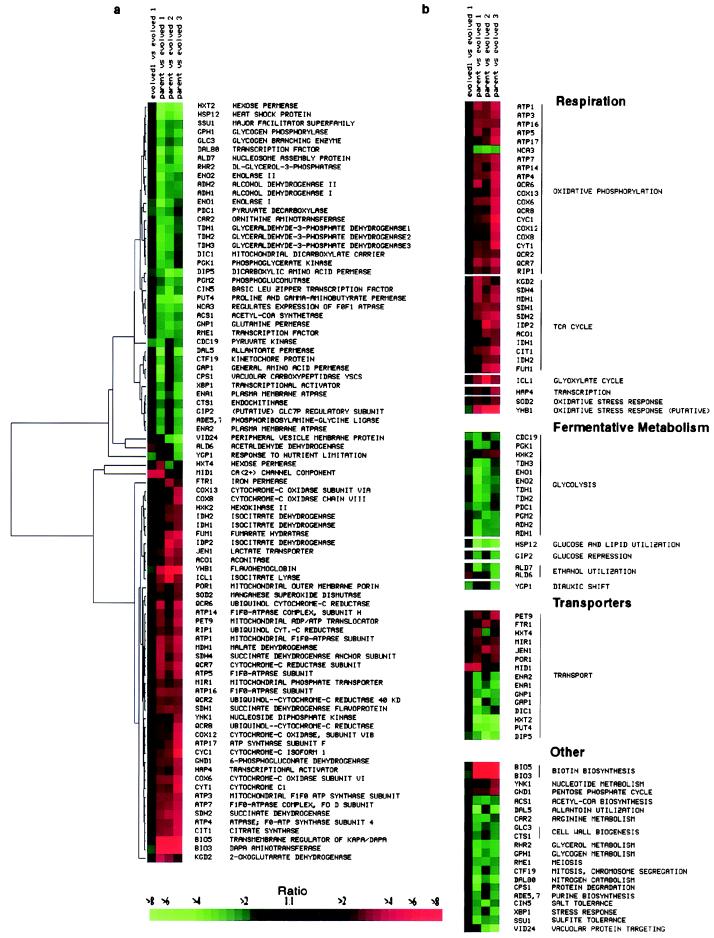
To help identify similarities and differences in the expression profiles among the evolved and the parental strains, the gene expression patterns were subjected to cluster analyses, as described by Eisen et al. (16). Fig. 1 shows two representations of the results for the 88 named genes whose transcript levels differed from those in the parental strain by at least 2-fold in at least two of the evolved strains. In Fig. 1a, the genes are ordered by means of a hierarchical clustering algorithm, by which genes with the most similar expression patterns are arranged adjacent to each other (Fig. 1a). In Fig. 1b, genes with similar functional descriptors (as assigned by the Saccharomyces Genome Database) are grouped together (Fig. 1b). Most of the genes of known function that were consistently and substantially altered in the three evolved strains have functions in glycolysis, ATP synthesis, oxidative phosphorylation (and associated mitochondrial structural proteins), the tricarboxylic acid cycle, or transport, not only of sugars but also other metabolites. There was a striking functional coherence among the recurrent alterations in gene expression in this model of evolution. It should nevertheless be noted that there were also clear differences among the evolved strains—expression of many genes varied substantially from the parental strain in only one of the three evolved strains (data not shown but available on our web site, http://genome-www.stanford.edu/evolution/).
Figure 1.
Graphical representations of the differences in transcript abundance between the evolved strains and the parental strain for the 88 functionally characterized genes whose transcripts differed by at least 2-fold on two or more microarrays. The ratio of transcript levels in the evolved strain compared with the parental strain, measured for each gene, is represented by the color of the corresponding box in this graphical display (see scale). Red represents a higher level of expression in the evolved strain, and green represents a higher level of expression in the parental strain. The color saturation represents the magnitude of the expression ratio, as indicated by the scale at the bottom of the figure. Black indicates no detectable difference in expression levels; gray denotes a missing observation. (a) Hierarchical clustering of gene expression. Relationships in expression patterns among the genes are represented by the tree at the left of the display, with branch lengths indicative of the magnitude of the differences between gene expression patterns (16). (b) Expression patterns organized according to functionally defined categories.
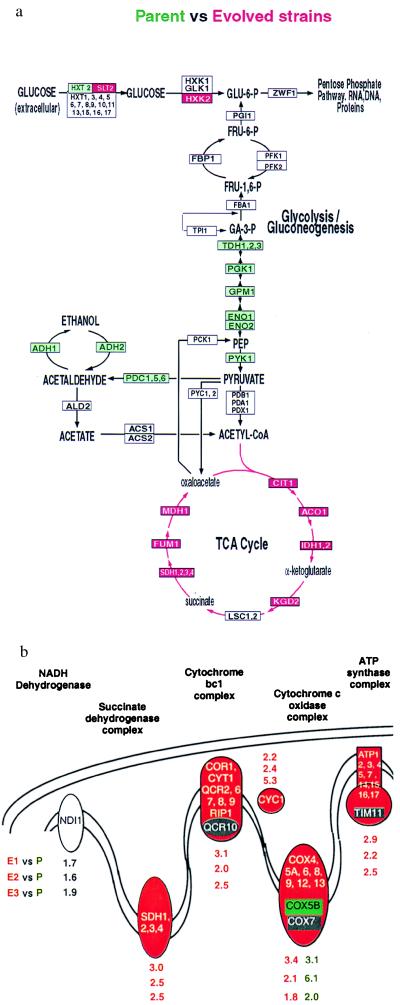
Expression of the genes encoding glycolytic enzymes was generally decreased relative to the parental strain. Particularly noteworthy were the consistent decreases observed in the terminal steps leading to the production of ethanol. Conversely, expression of genes specifying the enzymes of the tricarboxylic acid cycle, ATP generation, and oxidative phosphorylation was increased (Fig. 2). The recurrent alterations in genes encoding transporters differed from gene to gene: some increased, and others decreased expression. The overall pattern (Fig. 2) is a blueprint for efficient utilization of glucose as an energy source under low substrate concentrations; the characteristic changes in the transcriptional program in the evolved cells appear to be directed at maximizing ATP production per mole of glucose.
Figure 2.
(a) Alterations in gene expression consistent in two or more experiments are projected onto metabolic maps of central carbon metabolism. Increases (red) and decreases (green) of 2-fold or greater in transcripts encoding the enzymes that mediate the steps in these pathways are represented by the color of the corresponding box. (b) Average expression ratios for sets of genes encoding the mitochondrial protein complexes involved in oxidative phosphorylation. The expression ratios for COX5B, the differentially expressed isozyme of COX5A, are indicated separately from the other components of cytochrome c oxidase complex.
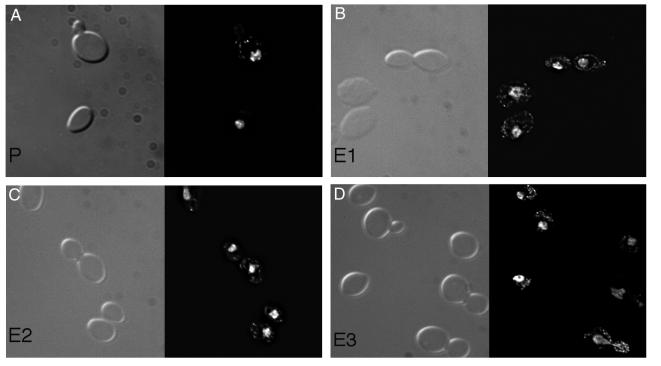
A large fraction of the functionally characterized genes whose expression was increased in the evolved strains encode mitochondrial proteins involved in respiration. Although the nuclear genes encoding mitochondrial proteins are represented on our arrays, those encoded in the mitochondrial genome are not. Given that many of the proteins encoded by the mitochondrial genome function in oxidative phosphorylation, one might have expected to see these transcripts overproduced in the evolved strains relative to the parent. Increase in mtDNA copy number has been previously reported on transfer of yeast to nonfermentable carbon sources (17), presumably in response to increased demand for mitochondrial function. We examined the possibility that the evolved strains, growing in the chemostat under carbon limitation, might have more mtDNA than the parental strain under the same conditions by staining with DAPI (Fig. 3). The results showed qualitatively that the evolved strains contain more mtDNA staining than the parental strains.
Figure 3.
Cells were cultured aerobically in the chemostat at 30°C with glucose limitation (0.08%), harvested, and fixed in formaldehyde. Cells were stained with DAPI and photographed with a Delta Vision microscope (Applied Imaging Systems) to compare the abundance of mitochondrial DNA in the parental and evolved strains. (A) Parental strain. (B, C, and D) Isolates from the three evolved populations E1, E2, and E3, respectively.
The overall picture that emerged is that selection over hundreds of generations under glucose limitation in the chemostat resulted in similar improvements in the efficiency of glucose utilization in three independent evolutionary experiments. In each case, the magnitude and the nature of the improvement were similar, as were the major classes of genes whose level of expression (measured as transcript abundance) was significantly altered. It is worth noting that none of these changes appeared to impair growth in glucose-sufficient conditions in batch culture: the parental strain and E1 showed no differences in yield and maximal growth rate (13). One way of stating the nature of the change in physiology is that the evolved strains displayed an enhanced, classical Pasteur effect, such that fermentation of glucose was effectively inhibited, in the presence of oxygen, in favor of glucose oxidation.
This physiological result was completely in accord with our understanding of energy metabolism in yeast. The yield of ATP per mole of glucose has long been known to be much higher when glucose is oxidized than when it is fermented. Our experiments invite comparison with those of DeRisi et al. (4), who used DNA microarrays to follow gene expression during the diauxic shift observed when S. cerevisiae growing on glucose in batch culture exhaust the glucose (largely through fermentation to ethanol) and begin to oxidize the ethanol. Many of the same genes (notably those involved in glycolysis, the tricarboxylic acid cycle, ATP generation, oxidative phosphorylation, and the associated mitochondrial structural proteins) showed parallel changes in expression levels in the two experimental situations. In one experiment, the changes resulted from an evolutionary adaptation to growth in limiting glucose, and in the other, a physiological adaptation to a gradual reduction in the glucose available in the environment. A remarkable feature of both experiments was the degree to which the systematic nature of metabolic adaptation, whether evolutionary or physiological, was accurately represented in the pattern of alteration in gene expression at the mRNA level (data not shown but available on our web site). This was not a result we could confidently have predicted, and we believe that it may point to an important principle. The same regulatory systems may well be critical in producing the results in both experiments: for instance, expression of the well characterized transcriptional activator HAP4 (18) was found to be increased as glucose disappeared in the diauxic shift and in our evolved strains. One should exercise caution, however, in comparing these experiments in too much detail. The growth conditions were very different: our experiments used a defined medium, and cells were growing in a steady state, whereas DeRisi et al. (4) studied batch cultures in a rich medium under changing conditions (e.g., in pH, nitrogen sources, and ethanol, as well as glucose). Some of the observed differences between the metabolic adaptation to the diauxic shift and the evolutionary adaptation to glucose limitation are clearly reflective of these differences in the growth conditions: for instance, in batch culture, DeRisi et al. (4) noted substantial increases in expression of ALD2 and ACS1, both essential for ethanol utilization. In our data set, neither gene is increased in the evolved strains (ALD2 is actually decreased almost by half in two of the strains).
The mechanistic basis for the changes in the evolved strains is still obscure. The number of mutations that contribute to the changes is likely to be quite limited. Previous experiments by Paquin and Adams (9) using the same parental strain cultured under similar conditions showed approximately six different adaptive sweeps in ≈305 generations, during which the population was repeatedly overtaken by a new, “more fit” strain. The identity of the mutated genes is not directly addressed by our experiments, which measured the result (reproducible changes in expression of a subset of ≈100 genes) and not the cause. The consistent, coherent alterations in transcription of large sets of functionally related genes strongly suggests that among this small set of mutations that accounted for the increased fitness of the evolved strains, some directly affected the regulation of genes that had altered expression. A parsimonious model might involve mutations in a few key regulators of these genes. At the same time, it is very likely that many of the changes we observed were indirect consequences of changes in physiology wrought by one or more of the causative mutations. Furthermore, because the altered physiology of the evolved strains resulted in changes in their environments in the chemostats, some of the changes we observed may have been secondary to these environmental differences.
In this paper, we have focused on the behavior of genes whose function has been well characterized. It is important to emphasize that about an equal number of uncharacterized genes had reproducibly changed expression patterns in the evolved strains. Many of these may well turn out to have roles in metabolism, and their behavior in these experiments (or others like them) could well provide important clues to their function. An example of this is the ORF YPR020W. Its expression was increased in all three evolved strains, and its expression pattern in general was similar to that of other genes for oxidative phosphorylation. It was first described in the databases as encoding a protein with weak similarity to a subunit of the bovine F1Fo-ATP synthase and very recently was shown to encode subunit γ of the yeast mitochondrial F1Fo-ATP synthase and was therefore renamed ATP19 (19).
The strong similarity among the evolved strains in global gene expression patterns suggests that the mechanism by which they are achieved may also be similar. However, such an inference must await explicit genetic analysis of the causative mutations. All of the strains used in these experiments were (and remained) diploid, making genetic analysis of the changes less than straightforward.
Finally, the similarity in the evolved strains could imply that further large-scale “improvements” in physiological performance in glucose-limited chemostats may not be possible through selection of modest duration in the chemostat. In addition to the similarities shown in Table 1 and Fig. 1, this idea is supported by the fact that the E1 clone had experienced an additional 200 generations in continuous culture over the other two evolved strains (13). An alternative model is that there may be very few routes by which to reach this altered phenotype by means of a small number of mutational steps. If this is so, mutations of certain key genes, perhaps encoding the regulators of families of genes involved in energy metabolism, may be common to all such short pathways. Genetic studies of evolved strains will be required to shed more light on this question.
Supplementary Material
Acknowledgments
We thank G. Sherlock for preparing the web site; K. Schwartz, J. Binkley, J. Mulholland, S. Palmieri, J. DeRisi, L. Brennan-Jobs, and J. Evans for their technical assistance; and J. Adams for providing the strains P (CP1AB) and E1. This paper is dedicated to Barbara Powell. This work was supported by funding to R.F.R. from the University of Idaho Research Council and an Exceptional Opportunity Grant from the M. J. Murdock Charitable Trust and by grants to P.O.B. from the National Human Genome Research Institute (NHGRI) (HG00450 and HG 00983) and by the Howard Hughes Medical Institute (HHMI) and to D.B. from the National Institutes of Health (GM 46406 and CA77097). T.L.F. was supported in part by an Institutional Training Grant in Genome Sciences (T32 HG00044) from the NHGRI. P.O.B. is an associate investigator of HHMI.
ABBREVIATION
- DAPI
4′,6′-diamino-2-phenylindole
References
- 1.De Deken R H. J Gen Microbiol. 1966;44:149–156. doi: 10.1099/00221287-44-2-149. [DOI] [PubMed] [Google Scholar]
- 2.Lagunas R. Mol Cell Biochem. 1979;2:139–146S. doi: 10.1007/BF00215362. [DOI] [PubMed] [Google Scholar]
- 3.Lagunas R. Yeast. 1986;2:221–228. doi: 10.1002/yea.320020403. [DOI] [PubMed] [Google Scholar]
- 4.DeRisi J L, Iyer V R, Brown P O. Science. 1997;278:680–686. doi: 10.1126/science.278.5338.680. [DOI] [PubMed] [Google Scholar]
- 5.Ronne M. Trends Genet. 1995;11:12–17. doi: 10.1016/s0168-9525(00)88980-5. [DOI] [PubMed] [Google Scholar]
- 6.Monod J. Annu Rev Microbiol. 1949;3:371. [Google Scholar]
- 7.Novick A. Annu Rev Microbiol. 1955;9:97–110. doi: 10.1146/annurev.mi.09.100155.000525. [DOI] [PubMed] [Google Scholar]
- 8.Kubitschek H E. Introduction to Research with Continuous Cultures. Englewood Cliffs, NJ: Prentice-Hall; 1970. [Google Scholar]
- 9.Paquin C, Adams J. Nature (London) 1983;302:495–500. doi: 10.1038/302495a0. [DOI] [PubMed] [Google Scholar]
- 10.Horiuchi T, Tomizawa J, Novick A. Biochem Biophys Acta. 1962;55:152–163. doi: 10.1016/0006-3002(62)90941-1. [DOI] [PubMed] [Google Scholar]
- 11.Novick A, Szilard L. Proc Natl Acad Sci USA. 1950;36:708–719. doi: 10.1073/pnas.36.12.708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fox M S. Genetics. 1998;148:1415–1418. doi: 10.1093/genetics/148.4.1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brown C J, Todd K M, Rosenzweig R F. Mol Biol Evol. 1998;15:931–942. doi: 10.1093/oxfordjournals.molbev.a026009. [DOI] [PubMed] [Google Scholar]
- 14.Chu S, DeRisi J, Eisen M, Mulholland J, Botstein D, Brown P O, Herskowitz I. Science. 1998;282:699–705. doi: 10.1126/science.282.5389.699. [DOI] [PubMed] [Google Scholar]
- 15.Spellman P T, Sherlock G, Zhang M Q, Iyer V R, Anders K, Eisen M B, Brown P O, Botstein D, Futcher B. Mol Biol Cell. 1998;9:3273–3297. doi: 10.1091/mbc.9.12.3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eisen M B, Spellman P T, Brown P O, Botstein D. Proc Natl Acad Sci USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ulery T L, Jang S H, Jaehning J A. Mol Cell Biol. 1994;14:1160–1170. doi: 10.1128/mcb.14.2.1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Forsburg S L, Guarente L. Genes Dev. 1989;3:1166–1178. doi: 10.1101/gad.3.8.1166. [DOI] [PubMed] [Google Scholar]
- 19.Boyle G, Roucou X, Nagley P, Devenish R J, Prescott M. Eur J Biochem. 1999;262:315–323. doi: 10.1046/j.1432-1327.1999.00345.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.