Abstract
With the rising number of patients with human immunodeficiency virus (HIV)/AIDS in developing countries, the control of mycobacteria is of growing importance. Previous studies have shown that rodents and insectivores are carriers of mycobacteria. However, it is not clear how widespread mycobacteria are in these animals and what their role is in spreading them. Therefore, the prevalence of mycobacteria in rodents and insectivores was studied in and around Morogoro, Tanzania. Live rodents were trapped, with three types of live traps, in three habitats. Pieces of organs were pooled per habitat, species, and organ type (stratified pooling); these sample pools were examined for the presence of mycobacteria by PCR, microscopy, and culture methods. The mycobacterial isolates were identified using phenotypic techniques and sequencing. In total, 708 small mammals were collected, 31 of which were shrews. By pool prevalence estimation, 2.65% of the animals were carriers of mycobacteria, with a higher prevalence in the urban areas and in Cricetomys gambianus and the insectivore Crocidura hirta. Nontuberculous mycobacteria (Mycobacterium chimaera, M. intracellulare, M. arupense, M. parascrofulaceum, and Mycobacterium spp.) were isolated from C. gambianus, Mastomys natalensis, and C. hirta. This study is the first to report findings of mycobacteria in African rodents and insectivores and the first in mycobacterial ecology to estimate the prevalence of mycobacteria after stratified pool screening. The fact that small mammals in urban areas carry more mycobacteria than those in the fields and that potentially pathogenic mycobacteria were isolated identifies a risk for other animals and humans, especially HIV/AIDS patients, that have a weakened immune system.
Wild and commensal rodents and insectivores can be a reservoir for pathogens that can cause diseases in humans. Examples are leptospirosis, plague, toxoplasmosis, and Lassa fever (14). The fact that diseases can be transmitted between rodents and humans emphasizes the risk associated with the close contact between humans and commensal or peridomestic rodents and insectivores. Information about the prevalence of various infections in rodents and insectivores is needed to estimate the risk for humans. From this information we can deduce whether intervention is necessary, how this intervention has to take place, and when or where it can be applied (14). A group of less-investigated potential pathogens in wild rodents and insectivores are the mycobacteria.
The genus Mycobacterium comprises more than 120 named species recognized currently, several of which are pathogenic; most of them cause opportunistic infections (34). The pathogenic species are responsible for some important diseases in humans and animals, in the developed world, as well as in developing countries, namely, tuberculosis, leprosy, and Buruli ulcer. With the rising numbers of human immunodeficiency virus (HIV)/AIDS patients, there has been a rising incidence of tuberculosis and other mycobacterial diseases in many parts of the developed and developing world, because HIV/AIDS patients are more sensitive to mycobacterial diseases due to a weakened immune system (7). Therefore, the control of mycobacteria, including the control of their reservoir and transmission, is very important, especially in developing countries.
Wild and domestic animals are known to carry mycobacteria; several pathogenic mycobacterial species have already been found in animals (2, 8, 9, 17, 38). Mycobacterioses can as such occur as zoonoses in humans.
Rodents and insectivores can be a source of mycobacteria. The occurrence of mycobacteria in rodents was first reported by Wells and Oxon (38), with a prevalence of 9 to 31% of Mycobacterium microti in rodents. Mycobacteria were isolated by Lapage from the lungs, spleen, liver, kidney, and lymph glands of 8 out of 500 shrews with tuberculous pathomorphological changes (1.6%) caught in 1946 in Great Britain (19). Several mycobacteria were also found in insectivores and rodents in the Czech Republic with a prevalence of 9.8% (12), and M. microti was found in a wild rodent population in England with a prevalence of 8% (4). M. bovis was found in 5% of hedgehogs (21) and in 0.6 to 2.8% of rodents and insectivores in the southwest region of England (8). If and how rodents and insectivores can transmit mycobacteria to humans is not yet known. However, cases of M. microti infection in humans have already been observed (35).
In the present study the prevalence of mycobacteria in rodents and insectivores caught in and around houses and on fields in urban and peri-urban Morogoro, Tanzania, was determined using direct smear examination (DSE) after acid-fast staining techniques, PCR, and culture methods. Because of time and material limiting factors, we used a stratified pool screening approach, after which the pool prevalence was estimated.
MATERIALS AND METHODS
Trapping of rodents.
The trapping of animals took place in July and August 2003. We trapped rodents and insectivores in urban and peri-urban Morogoro, a medium large city in Tanzania, 200 km west of Dar es Salaam.
Rodents were caught in 16 different locations divided into three habitat types, namely, in or near human residences (“urban areas”), swampy fields, and dry fields, using three types of live traps: Sherman LFA Live Traps (7.5 by 9.0 by 23.0 cm; HB Sherman Traps, Inc., Tallahassee, FL), box traps (self-made live trap consisting of a wooden box with a wired window and an iron door controlled by the mechanism of a snap trap), and big wire cage traps. Peanut butter with maize bran was used as bait in the Sherman taps and box traps; the big wire cage traps were baited with one-third of a fresh maize cob.
Sample collection.
The animals were processed according to a standard protocol. In brief, the trapping site and the preliminary identification to the genus level were recorded for each animal before the animal was anesthetized with ether; blood samples were then taken. Dissections were performed after the animals were killed with chloroform, the samples were labeled, and the external characteristics and measurements were recorded. Samples were taken for the detection of several pathogens. All organs were checked for the presence of tuberculous pathomorphological changes. For mycobacterial detection, a piece of liver (±0.5 cm3), half of the spleen (the size depended on the species), a piece of lung (±0.1 cm3), and the mesenteric lymph nodes (again, the size depended on the species) were removed and kept at −20°C for microbiological analysis at the Institute of Tropical Medicine, Antwerp, Belgium. The carcasses were kept in formalin and sent to the University of Antwerp for further identification to the species level.
Pooling of samples and detection and identification of mycobacteria.
The organs were homogenized in 2 ml of phosphate-buffered saline in a sterile mortar, and the same organs were pooled per one to six individuals of the same species caught at the same location (stratified pooling). Individual animals were arranged per species and per trapping site. Groups of one to six animals of the same species and the same trapping sites were randomly formed (depending on how many animals of the same species were trapped at the same trapping site). This resulted in 155 groups of individuals. For each group, the four different organ homogenates (liver, spleen, lungs, and mesenteric lymph nodes) collected from each animal were pooled separately, resulting in 620 pools to be tested (from 2,832 individual samples). In the results it is important to note that when we speak of the results in terms of “groups,” we mean the combined results of the four organs, whereas when we talk about “pools” we are referring to the results of the pooled organs (which can be a pool of livers, spleens, lungs, or mesenteric lymph nodes). This means that when two pools that originated from the same group of animals are positive (but from different organs), this group of animals can only count as one positive group.
In order to reduce the overgrowth of microorganisms growing faster than mycobacteria which could be present in the collected samples, 1 ml of each pool was decontaminated for 20 min with 3 ml of 1 N HCl and neutralized with 3 ml of 1 N NaOH. After centrifugation, the pellet was resuspended in 2 ml of sterile distilled water and used for DSE, culture, and PCR (30).
Inoculation was done on four different in-house-prepared culture media: Löwenstein-Jensen medium, Stonebrink medium, Ogawa-Mycobactine medium and Löwenstein-paratuberculosis medium (25). Cultivation took place at 37°C (30) for at least 6 months and for up to 10 months.
DNA was extracted by using the method of Mangiapan et al. (22) and amplified in a nested PCR specific for all mycobacteria except for M. nonchromogenicum, as previously described by Portaels et al. (27), with the modified primers P1 (5′-TGCTTAACACATGCAAGTCG-3′) and P2 (5′-TGAGATTTCACGAACAACGC-3′) in the first run and the primers P7 (5′-CATGCAAGTCGAACGGAAAGG-3′) and P16 (5′-AAGCCGTGAGATTTCACGAACA-3′) in the second run targeting the 16S rRNA gene.
DSE was done using two acid-fast staining methods: the Ziehl-Neelsen method and auramine staining (30).
The mycobacteria isolated on culture were checked for acid fastness by Ziehl-Neelsen staining and identified to the species level by biochemical methods (36) and by sequencing the 16S rRNA gene (36) and, for some isolates, the 16S-23S rRNA gene internal transcribed spacer 1 (31). Mycobacteria detected by PCR were tested with a PCR using specific primers for M. tuberculosis complex (MTBC; P1 and P2 in the first run, P3 [5′-AACCCGGACCTTCGTCGATG-3′] and P9 [5′-CATGTCTTGTGGTGGAAAGCGC-3′] in the second run) (28), since we were especially interested in members of the MTBC.
Statistical methods.
The estimation of prevalence by pool screening with pools of equal size has a long history (1, 5, 16). More recently, the theory has been extended to unequal pool sizes that allow estimation while reducing the amount of specimen handling required (16). The standard probability model for both estimation and testing treats each pool tested as an independent trial with a success probability equal to [1 − (1 − p)n], where p is the unknown prevalence of infection or disease in an individual from the vector or reservoir species and n is the size of the pool collected. If we let Xi denote the result of testing the ith pool (Xi = 1 if the pool tests positive and 0 if not) and let ni equal the size of the ith pool, then the estimate of the prevalence, p, is found by the standard statistical approach of maximum-likelihood estimation. (The details of these calculations are described in references 1 and 16.) Tests of specific hypotheses are easily constructed by using a likelihood ratio approach (20), which is how the tests reported here were performed. The null hypothesis is rejected at a significance level of P < 0.05.
RESULTS
Trapping results.
In 5,368 trapping nights, 708 small mammals were caught, 677 of which were rodents and 31 of which were shrews (insectivores). The multimammate mouse Mastomys natalensis (Smith 1834), which was the most common species, was mainly found in the fields, while in and around the houses mostly the black rat Rattus rattus (Linnaeus 1758), Mus spp. (the house mouse M. musculus Linnaeus, 1758 and Mus [Nannomys] spp.), and the Gambian rat Cricetomys gambianus Waterhouse 1840 were caught. Only low numbers of the brown rat Rattus norvegicus (Berkenhout 1769) were caught in three urban trapping sites. A gerbil species, Tatera vicina (Matschie 1911), was found only on the dry fields. Other rarely caught species (a ticket rat species, Grammomys surdaster Mathey 1971; the single-striped grass mouse Lemniscomys rosalia [Thomas 1904]; and the Tanzanian shaggy marsh rat Dasymys sua [Verheyen et al. 2003]) were only caught in the fields. The lesser red musk shrew Crocidura hirta (Peters 1852) was found mainly in the fields but also in and around houses. None of the animals showed any external or internal signs of tuberculous pathomorphological changes.
DSE.
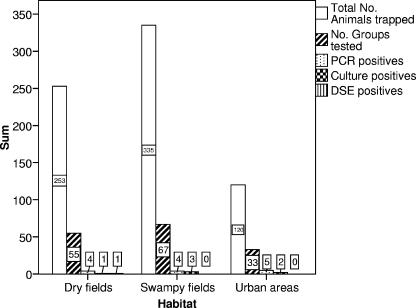
Only one pool was found to be positive for acid-fast bacilli. One globus with acid-fast bacilli was seen in a smear from a liver of a C. gambianus caught in a tree on a dry field (Fig. 1 and 2).
FIG. 1.
Number of trapped animals, groups tested, and groups that were positive for PCR with primers specific for mycobacteria in general, culture, and DSE in the three different habitats.
FIG. 2.
Number of trapped animals, groups tested, and groups that were positive for PCR with primers specific for mycobacteria in general, culture, and DSE for C. gambianus, C. hirta, and other rodents. The fact that the rodent C. gambianus carries significantly more mycobacteria than other rodents and that the insectivore C. hirta carries significantly more than rodents is indicated with an asterisk.
PCR.
PCR with Mycobacterium genus-specific primers (P1 and P2 in the first run, and P7 and P16 in the second run) resulted in 15 positives out of 620 pools of organs (2.42%). The habitat and species of the organs that were positive are indicated in Table 1. None of these were positive for PCR with primers specific for MTBC. Only one of the PCR positive pools was also positive in culture. In terms of groups, 13 of 155 groups of animals tested positive (8.39%) (Fig. 1 and 2): in two groups two pools of different organs tested positive. The estimated prevalence of positivity for mycobacteria by PCR was 1.89%.
TABLE 1.
Pools that were found to be PCR and/or culture positive
| Habitat | Speciesa | Organb | No. of animals/pool | PCR positive | Culture positive (ITM no.) | Identification of culture isolates | Identification method usedc
|
||
|---|---|---|---|---|---|---|---|---|---|
| PhI | 16S rRNA | ITS-1 | |||||||
| Wet field | C. hirta | MLN | 5 | Yes | No | NAd | |||
| Wet field | G. surdaster | Spleen | 5 | Yes | No | NA | |||
| Wet field | M. natalensis | Lung | 5 | No | 05001105 | M. intracellulare | x | x | x |
| Wet field | M. natalensis | MLN | 5 | No | 05001106 | Mycobacterium sp. strain IWGMT 90093 | x | x | |
| Wet field | M. natalensis | Lung | 6 | No | 04000693 | M. arupense | x | x | |
| Wet field | M. natalensis | Lung | 5 | Yes | No | NA | |||
| Wet field | M. natalensis | Lung | 1 | Yes | No | NA | |||
| Dry field | C. hirta | Lung | 1 | No | 04001436 | Mycobacterium sp. strain JLS | x | x | |
| Dry field | M. natalensis | Lung | 5 | Yes | No | NA | |||
| Dry field | M. natalensis | Spleen | 6 | Yes | No | NA | |||
| Dry field | M. natalensis | Liver | 5 | Yes | No | NA | |||
| Dry field | T. vicina | MLN | 6 | Yes | No | NA | |||
| Urban | C. gambianus | Spleen | 4 | No | 04002873 | M. parascrofulaceum | x | x | |
| Urban | C. gambianus* | Liver | 4 | Yes | 04001190 | M. arupense | x | x | |
| Urban | C. gambianus* | MLN | 4 | No | 04002874 | M. chimaera | x | x | x |
| Urban | C. gambianus* | Lung | 4 | Yes | No | NA | |||
| Urban | C. gambianus | Lung | 4 | Yes | No | NA | |||
| Urban | C. hirta† | Lung | 1 | Yes | No | NA | |||
| Urban | C. hirta† | MLN | 1 | Yes | No | NA | |||
| Urban | M. musculus | Spleen | 4 | Yes | No | NA | |||
| Urban | M. musculus | Spleen | 3 | Yes | No | NA | |||
*, the pool of livers, the pool of mesenteric lymph nodes, and the pool of lungs originated from the same group of animals; †, the lungs and mesenteric lymph nodes originated from the same animal.
MLN, mesenteric lymph nodes.
PhI, phenotypic identification; 16S rRNA, 16S rRNA gene sequencing; ITS-1, 16S-23S rRNA gene internal transcribed spacer 1 sequencing.
NA, not applicable.
Culture.
Of 620 pools of organs, 7 resulted in a positive culture of acid-fast bacilli (1.13%). The habitat and species of the organs that were positive are indicated in Table 1. In all cases except one, only a few colonies were observed. The pool of livers from C. gambianus caught in an urban area grew about 15 colonies and was the only pool that was positive for both culture and PCR. In terms of groups, 6 of 155 groups of animals tested positive (3.87%) (Fig. 1 and 2): in one group, two pools of different organs tested positive. The estimated prevalence by culture was 0.86%. The identification of the mycobacteria is shown in Table 1.
Pool prevalence estimation and hypothesis testing.
With pool prevalence estimation, we could estimate the prevalence based on the results of pooled samples. We estimated prevalences of 1.89% by PCR, 0.86% by culture, and 2.65% by combining the PCR and culture results.
We comment below only on the results obtained in the analysis of the combined PCR and culture results. We made the same comparisons for the PCR and culture results separately, and the same trends were observed in these analyses (data not shown).
For the combined PCR and culture results, no significant difference was seen between the three different habitat types, namely, urban (residential) areas, dry fields, and swampy fields (P = 0.229). When we compared urban trapping sites versus all fields (both dry and swampy fields), we observed a 2.5-fold higher prevalence in the urban trapping sites, although this difference was not significant (P = 0.09). This may be due to the relatively small sample size from the urban trapping sites compared to that of the fields (Fig. 1).
When we compared the results for the rodent species (without taking into consideration the insectivores), C. gambianus had a 7.9-fold higher prevalence of mycobacteria than the other rodents combined (P = 0.004). When we compared the results for the insectivore C. hirta versus all rodents, the insectivore C. hirta has a 4.8-fold higher prevalence than the rodents (P = 0.016). The numbers of trapped animals, groups tested, and groups that were positive for PCR, culture, and DSE for C. gambianus, C. hirta and the other rodents are shown in Fig. 2.
DISCUSSION
Detection methods for mycobacteria.
Only one pool was positive for microscopy (DSE). This was not confirmed by culture or by PCR, although the estimated detection limit of microscopy (104 bacilli/ml) is considerably less than that of culture (102 bacilli/ml) and of the PCR setting used in the present study (nested 16S rRNA-PCR after Mangiapan DNA extraction: between 2 × 102 and 2 × 103 bacilli/ml) (24). The positive DSE result is probably due to acid-fast bacteria other than mycobacteria (e.g., Corynebacterium, Gordona, Nocardia, Rhodococcus, and Tsukamurella spp.) or to artifacts (36); therefore, we did not include this result in our analysis.
Only one pool tested positive for both PCR and culture. Culture is more sensitive than 16S rRNA-PCR, which explains the fact that six pools resulted in a positive culture but a negative PCR (24). Fourteen pools resulted in a positive PCR but a negative culture. This can be due to the following reasons. First, not all mycobacteria grow at 37°C (19a). In the present study we did not incubate at other temperatures because our goal was to detect MTBC or other pathogenic mycobacteria, most of which grow at 37°C (with the notable exception of M. ulcerans, but in Tanzania no cases of Buruli ulcer have been observed thus far). Second, the DNA of both dead and live mycobacteria is detected by PCR. Mycobacteria killed by the immune system of the animals or during the transport, storage, and decontamination of samples do not grow on culture but can still be detected by PCR (13). Storage of samples at −20°C has an effect on the viability of the mycobacteria, with a variable loss in viability of up to 100% for different mycobacterial strains (18). Acid decontamination has an effect on the viability of mycobacteria as well. The decontamination method used in the present study was tested on different strains of M. ulcerans. This method reduced the viability of some strains with 77 to 98%, while other strains had a delay of about 1 week compared to the control (23). Third, a number of pathogenic and nonpathogenic mycobacteria do not grow on the culture media used in the present study or do not grow in vitro at all. Examples include M. lepraemurium, M. genavense, and M. leprae, causing disease in rodents, birds, and humans, among others (26).
The aim of the present study was to determine the total prevalence of all mycobacteria (both pathogenic and nonpathogenic) in rodents and insectivores. Since culture and PCR are complementary methods, the combined results of both methods were used in estimating the prevalence and in subsequent hypothesis testing.
Mycobacterial species isolated.
In the present study, several different culture media were used to increase the chance of isolating different mycobacterial species: Löwenstein-Jensen medium for mycobacteria in general; Stonebrink medium, which favors the growth of M. bovis; and Ogawa-Mycobactine medium or Löwenstein-paratuberculosis medium for members of the M. avium complex (MAC).
Although we did not find members of the MTBC in rodents and insectivores, we did isolate some potentially pathogenic mycobacterial species. These mycobacteria are not highly invasive organisms (17), but they can infect people with lowered immune status due to HIV/AIDS, cancer, or toxoplasmosis (7).
In our study two members of MAC were isolated: M. intracellulare from a pool of lungs from M. natalensis trapped in a swampy field and M. chimaera from a pool of mesenteric lymph nodes from C. gambianus trapped in an urban area. M. intracellulare is known to cause lung disease in HIV-negative patients with or without predisposing factors (7). The lungs of at least one animal belonging to the positive group yielded a low number of colonies on all three culture media. The lungs did not show any sign of infection. Whether this animal was infected or colonized by M. intracellulare is difficult to say. M. intracellulare tends to form biofilms in watery environments (11). Since these animals were caught in a swampy field, the probable portal of entry is through inhalation of contaminated aerosols. The second MAC isolate, M. chimaera, has only recently been described by Tortoli et al. (33) as a new member of MAC. The mesenteric lymph nodes of at least one animal from this group of animals were colonized by M. chimaera, since only a few colonies were obtained from the pool of organs. Since this species has only been described recently, the natural reservoir of M. chimaera is not known. It is probably in the environment, such as for M. avium. The results of a recent study based on M. avium sensitin skin tests indicated that soil is a reservoir for MAC associated with human infection (29).
Three other potentially pathogenic strains were isolated: two M. arupense strains and one M. parascrofulaceum strain. These nontuberculous mycobacteria are of less clinical importance, but they have caused disease in several human cases (34).
Habitat.
The rapid urbanization in Africa leads to an economic decline with a lot of people living in overcrowded slums and shantytowns. A lack of sanitary infrastructure, e.g., bad sewerage and bad waste disposal systems, leads to poor hygienic conditions, which attract insects and rodents (14). When these rodents are carriers of pathogenic or opportunistic mycobacteria, in urban areas the close contact between rodents and humans facilitates the transmission of these mycobacteria.
A study of the environmental flora in the Democratic Republic of Congo gave comparable results, with a higher prevalence of some mycobacterial species in urban areas than in nonurban areas (26).
Rodents and insectivores as reservoir or transmitters of mycobacteria.
In general, rodents and insectivores can come into contact with mycobacteria through the environment by feeding, contact with soil, and contact with the feces of wild and domestic animals or humans. These mycobacteria can pass through the stomach of these animals without being digested since they are resistant to acid. Pathogenic and opportunistic mycobacteria can pass through the stomach and can survive in tissues and organs. In this way they can be spread over long distances with the migration of these animals (12).
Isolation rates of NTM in the environment vary. Portaels et al. (26) isolated 956 mycobacterial strains from 1,664 samples of water, mud, soil, grasses, and fish in Zaire, with seasonality depending on the strain types. MAC was found in 15% of the water samples taken in Zaire and Kenya (37), and NTM were detected by PCR or culture in 64% of the soil samples and 79% of the water samples obtained in Northern Malawi, with a higher frequency in the dry season than in the wet season (6). These frequencies are much higher than the frequency of mycobacteria in the small mammals observed in the present study. Therefore, we do not present enough evidence here to consider rodents a reservoir for mycobacteria. More targeted studies are currently ongoing, including studies to assess the presence of mycobacteria in rodent feces.
Nevertheless, rodents are known to transmit diseases through different transmission pathways. Mycobacteria were found in all four organs that were analyzed and could be excreted by the rodents and insectivores, e.g., in their feces. Considering what is currently known about transmission of diseases by rodents and of diseases in general (32), several ways of transmission of mycobacteria are possible: through direct contact with rodent excreta, through ingestion of food or water contaminated with rodent excreta or through ingestion of the animal itself, through inhaling aerosolized rodent excreta, through rodent bites, or through ectoparasites.
Pool prevalence estimation.
Since the end of the 1990s, the method of pooling samples for prevalence estimation of diseases in epidemiological or reservoir studies in the environment, (wild) animals, and even humans has been used by an increasing number of researchers (16). For this reason, research has been done on testing pooled samples to estimate the prevalence. In mycobacterial epidemiology, this method has mainly been applied to test pools of fecal and milk samples of cattle for paratuberculosis caused by M. avium subsp. paratuberculosis (15). In mycobacterial ecology, to our knowledge, only one published study used the pool screening approach (10). Although that study used the pool screening method, it has not used the statistical tools that are available to estimate the prevalence using data obtained from pooled sample testing. The present study is the first to estimate the pool prevalence of mycobacterial infections in wild animals using the statistical tools that have been developed for this purpose.
It is both costly and time-consuming to screen environmental samples for mycobacterial presence. In reservoir studies the aim is not to identify the individual animals that are infected or diseased but to have an assessment of the presence or absence of the infectious agent in a certain environment or host and, if present, its prevalence. In this way, individual test data of all samples are not required, and pooling samples can be a way to lower the costs and the time effort of such studies. If pooling is done in a stratified way (e.g., per habitat, per species, and/or per organ), it is still possible with the available statistical tools to show a difference between different environments (e.g., in the present study, urban versus fields), different animal groups (in the present study, insectivores versus rodents), or different rodent species (in the present study, C. gambianus versus other rodents) to determine the possible animal species or environments that can be reservoirs.
Supplementary Material
Acknowledgments
This research was supported by a Ph.D. grant from the Flemish Interuniversity Council, the Directorate-General for Development Cooperation (Brussels, Belgium), the Damien Foundation, and the European Union (Ratzooman Project, INCO-Dev ICA-CT-2002-10056).
We thank Krista Fissette, Pim De Rijk, and Natalie Van Houtte for excellent technical assistance; the staff of SUA Pest Management Centre for their support during the field work; and Erik Verheyen for help in identifying the rodents.
Footnotes
Published ahead of print on 7 December 2007.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Barker, J. T. 2000. Statistical estimators of infection potential based on PCR pool screening with unequal pool sizes. Ph.D. thesis. University of Alabama, Birmingham.
- 2.Bercovier, H., and V. Vincent. 2001. Mycobacterial infections in domestic and wild animals due to Mycobacterium marinum, M. fortuitum, M. chelonae, M. porcinum, M. farcinogenes, M. smegmatis, M. scrofulaceum, M. xenopi, M. kansasii, M. simiae, and M. genavense. Rev. Sci. Tech. Off. Int. Epizoot. 20:265-290. [DOI] [PubMed] [Google Scholar]
- 3.Reference deleted.
- 4.Cavanagh, R., M. Begon, M. Bennett, T. Ergon, I. M. Graham, P. E. W. De Haas, C. A. Hart, M. Koedam, K. Kremer, X. Lambin, P. Roholl, and D. van Soolingen. 2002. Mycobacterium microti infection (vole tuberculosis) in wild rodent populations. J. Clin. Microbiol. 40:3281-3285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chiang, C. L., and W. C. Reeves. 1962. Statistical estimation of virus infection rates in mosquito vector populations. Am. J. Hyg. 75:377-391. [DOI] [PubMed] [Google Scholar]
- 6.Chilima, B. Z., I. M. Clark, S. Floyd, P. E. M. Fine, and P. R. Hirch. 2006. Distribution of environmental mycobacteria in Karonga District, northern Malawi. Appl. Environ. Microbiol. 72:2343-2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Collins, F. M. 1989. Mycobacterial disease, immunosuppression, and acquired immunodeficiency syndrome. Clin. Microbiol. Rev. 2:360-377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Delahay, R. J., G. C. Smith, A. M. Barlow, N. Walker, A. Harris, R. S. Clifton-Hadley, and C. L. Cheeseman. 2007. Bovine tuberculosis infection in wild mammals in the South-West region of England: a survey of prevalence and a semiquantitative assessment of the relative risks to cattle. Vet. J. 173:287-301. [DOI] [PubMed] [Google Scholar]
- 9.de Lisle, G. W., C. G. Mickintosh, and R. G. Bengis. 2001. Mycobacterium bovis in free-living and captive wildlife, including farmed deer. Rev. Sci. Tech. Off. Int. Epizoot. 20:86-111. [DOI] [PubMed] [Google Scholar]
- 10.de Lisle, G. W., G. F. Yates, P. Caley, and R. J. Corboy. 2004. Surveillance of wildlife for Mycobacterium bovis infection using culture of pooled tissue samples from ferrets (Mustela furo). N. Z. Vet. J. 53:14-18. [DOI] [PubMed] [Google Scholar]
- 11.Falkinham, J. O., III, C. D. Norton, and M. W. LeChevallier. 2001. Factors influencing numbers of Mycobacterium avium, Mycobacterium intracellulare, and other mycobacteria in drinking water distribution systems. Appl. Environ. Microbiol. 67:1225-1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fischer, O., L. Matlova, J. Bartl, L. Dvorska, I. Melicharek, and I. Pavlik. 2000. Findings of mycobacteria in insectivores and small rodents. Folia Microbiol. 45:147-152. [DOI] [PubMed] [Google Scholar]
- 13.Fukushima, R., S. Kawakami, H. Iinuma, and K. Okinaga. 2003. Molecular diagnosis of infectious disease, abstr. Nippon Ganka Gakkai Zasshi 104:518-522. [PubMed] [Google Scholar]
- 14.Gratz, N. G. 1997. The burden of rodent-borne diseases in Africa south of the Sahara. Belg. J. Zool. 127(Suppl. 1):71-84. [Google Scholar]
- 15.Kallis, C. H. J., J. W. Hesselink, H. W. Barkema, and M. T. Collins. 2000. Culture of strategically pooled bovine fecal samples as a method to screen herds for paratuberculosis. J. Vet. Diagn. Investig. 12:547-551. [DOI] [PubMed] [Google Scholar]
- 16.Katholi, C. R., L. Toe', A. Merriweather, and T. R. Unnasch. 1995. Determining the prevalence of Onchocerca volvulus infection in vector populations by PCR screening of pools of black flies. J. Infect. Dis. 172:1414-1417. [DOI] [PubMed] [Google Scholar]
- 17.Kiehn, T. E., and M. H. White. 1998. The changing nature of nontuberculous mycobacteriology, p. 207-219. In W. M. Scheld, D. Armstrong, and J. M. Hughes (ed.), Emerging infections 1. ASM Press, Washington, DC.
- 18.Kim, T. H., and G. P. Kubica. 1973. Preservation of mycobacteria: 100% viability of suspensions stored at −70°C. Appl. Microbiol. 25:956-960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lapage, G. 1947. Tuberculosis of voles and shrews. Nature 160:168. [DOI] [PubMed] [Google Scholar]
- 19a.Leão, S. C., A. Martin, G. I. Meija, J. C. Palomino, J. Robledo, M. A. da Silva Telles, and F. Portaels. 2004. Practical handbook for the phenotypic and genotypic identification of mycobacteria. Vanden Broele, Brugges, Belgium.
- 20.Lehmann, E. L., and J. T. Romano. 2006. Testing statistical hypotheses, 3rd ed. Springer Verlag, New York, NY.
- 21.Lugton, I. W., A. C. Johnstone, and R. S. Morris. 1995. Mycobacterium bovis infection in New Zealand hedgehogs (Erinaceus europaeus). N. Z. Vet. J. 43:342-345. [DOI] [PubMed] [Google Scholar]
- 22.Mangiapan, G., M. Vokurka, L. Schouls, J. Cadranel, D. Lecossier, J. Van Embden, and A. J. Hance. 1996. Sequence capture-PCR improves detection of mycobacterial DNA in clinical specimens. J. Clin. Microbiol. 34:1209-1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Palomino, J. C., and F. Portaels. 1998. Effects of decontamination methods and culture conditions on viability of Mycobacterium ulcerans in the BACTEC system. J. Clin. Microbiol. 36:402-408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pfaller, M. A. 1994. Application of new technology to the detection, identification and antimicrobial susceptibility testing of mycobacteria. Am. J. Clin. Pathol. 101:329-337. [DOI] [PubMed] [Google Scholar]
- 25.Portaels, F., A. De Muynck, and M. P. Sylla. 1988. Selective isolation of mycobacteria from soil: a statistical analysis approach. J. Gen. Microbiol. 134:849-855. [DOI] [PubMed] [Google Scholar]
- 26.Portaels, F. 1995. Epidemiology of mycobacterial diseases. Clin. Dermatol. 13:207-222. [DOI] [PubMed] [Google Scholar]
- 27.Portaels, F., L. Realini, L. Bauwens, B. Hirschel, W. M. Meyers, and W. de Meurichy. 1996. Mycobacteriosis caused by Mycobacterium genavense in birds kept in a zoo: 11-year survey. J. Clin. Microbiol. 34:319-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Proaño-Perez, F., L. Rigouts, J. Brandt, P. Dorny, J. Ron, M.-A. Chavez, R. Rodriguez, K. Fissette, A. Van Aerde, F. Portaels, and W. Benitez-Ortiz. 2006. Preliminary observations on Mycobacterium spp. in dairy cattle in Ecuador. Am. J. Trop. Med. Hyg. 75:318-323. [PubMed] [Google Scholar]
- 29.Reed, C., C. R. Von Reyn, S. Chamblee, T. V. Ellerbrock, J. W. Johnson, B. J. March, L. S. Johnson, R. J. Trenschel, and C. R. Horsburgh, Jr. 2006. Environmental risk factors for infection with Mycobacterium avium complex. Am. J. Epidemiol. 164:32-40. [DOI] [PubMed] [Google Scholar]
- 30.Rieder, H. L., T. M. Chonde, H. Myking, R. Urbanczik, A. Laszlo, S. J. Kim, A. Van Deun, and A. Trébucq. 1998. The Public Health Service National Tuberculosis Reference Laboratory and the National Laboratory Network. International Union against Tuberculosis and Lung Disease, Paris, France.
- 31.Roth, A., M. Fischer, M. E. Hamid, S. Michalke, W. Ludwig, and H. Mauch. 1998. Differentiation of phylogenetically related slowly growing mycobacteria based on 16S-23S rRNA gene internal transcribed spacer sequences. J. Clin. Microbiol. 36:139-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shakespeare, M. 2001. Zoonoses. Pharmaceutical Press, London, United Kingdom.
- 33.Tortoli, E., L. Rindi, M. J. Garcia, P. Chiaradonna, R. Dei, C. Garzelli, R. M. Kroppenstedt, N. Lari, R. Mattei, A. Mariottini, G. Mazzarelli, M. I. Murcia, A. Nanetti, P. Piccoli, and C. Scarparo. 2004. Proposal to elevate the genetic variant MAC-A, included in the Mycobacterium avium complex, to species rank as Mycobacterium chimaera sp. nov. Int. J. Syst. Evol. Microbiol. 54:1277-1285. [DOI] [PubMed] [Google Scholar]
- 34.Tortoli, E. 2006. The new mycobacteria: an update. FEMS Immunol. Med. Microbiol. 48:159-178. [DOI] [PubMed] [Google Scholar]
- 35.Van Soolingen, D., A. G. M. Van der Zanden, P. E. W. De Haas, G. T. Noordhoek, A. Kiers, N. A. Foudraine, F. Portaels, A. H. J. Kolk, K. Kremer, and J. D. A. Van Embden. 1998. Diagnosis of Mycobacterium microti infections among humans by using novel genetic markers. J. Clin. Microbiol. 36:1840-1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vincent Lévy-Frébault, V., and F. Portaels. 1992. Proposed minimal standards for the genus Mycobacterium and for description of new slowly growing Mycobacterium species. Int. J. Syst. Bacteriol. 42:315-323. [DOI] [PubMed] [Google Scholar]
- 37.Von Reyn, C. F., R. D. Waddell, T. Eaton, R. D. Arbeit, J. N. Maslow, T. W. Barber, R. J. Brindle, C. F. Gilks, J. Lumio, J. Lähdevirta, A. Ranki, D. Dawson, and J. O. Falkinham III. 1993. Isolation of Mycobacterium avium complex from water in the United States, Finland, Zaire, and Kenya. J. Clin. Microbiol. 31:3227-3230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wells, A. Q., and D. M. Oxon. 1937. Tuberculosis in wild voles. Lancet i:1221. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.