Abstract
A single-chain Fv (scFv) antibody was developed and applied for efficient and specific detection of Bacillus anthracis spores. The antibody was isolated from a phage display library prepared from spleens of mice immunized with a water-soluble extract of the outer membrane of the B. anthracis spore (exosporium). The library (7 × 106 PFU) was biopanned against live, native B. anthracis ATCC Δ14185 spores suspended in solution, resulting in the isolation of a unique soluble scFv antibody. The antibody was affinity purified and its affinity constant (3 × 108 ± 1 × 108 M−1) determined via flow cytometry (FCM). Preliminary characterization of scFv specificity indicated that the scFv antibody does not cross-react with representatives of some phylogenetically related Bacillus spores. The potential use of scFv antibodies in detection platforms was demonstrated by the successful application of the soluble purified scFv antibody in enzyme-linked immunosorbent assays, immunofluorescence assays, and FCM.
Bacillus anthracis spores, the primary infectious agents causing anthrax, are probably the most likely candidates for a biological terrorist assault. Therefore, rapid detection of spores is critical for a timely response and successful treatment of the disease. Various immunoassays based on the high specificity of antibodies have been developed for the rapid detection of several pathogens. Over the past decade, recombinant technology enabled the production of engineered antibody fragments such as Fabs or single-chain Fv (scFv) antibodies. An scFv antibody is a small engineered antibody in which the variable heavy chain and light chain of the antibody molecule are connected by a short, flexible polypeptide linker. Phage display technology enables the presentation of scFv antibody on the phage surface and has been used successfully for the isolation of specific scFv antibodies from animal repertoire libraries via several enrichment cycles.
Using scFv antibodies for antigen detection has several advantages. scFv antibodies can be produced in large quantities in bacterial expression systems, with high reproducibility at low cost, and can be manipulated genetically for improved specificity and affinity (5, 15, 17). The recombinant antibody can also be fused to marker molecules for detection purposes (25). Recombinant antibodies have been developed for treatment of anthrax infection (26) and disease detection in clinical applications (25). However, the use of recombinant antibodies for detection of B. anthracis spores has not been described. In this study, we describe the construction of an scFv antibody and its successful application for B. anthracis spore detection.
MATERIALS AND METHODS
Bacteria.
B. anthracis Δ14185 is a nontoxinogenic, nonencapsulated (Tox− Cap−) derivative of ATCC 14185 (4) (Bacillus Genetic Stock Center); Bacillus subtilis DSM 675 and Bacillus cereus 569 are from the Israel Institute for Biological Research collection. Escherichia coli TG1 was part of the RPAS expression module (GE Healthcare UK Limited, Little Chalfont, Buckinghamshire, United Kingdom).
Bacillus cultures and sporulation.
Spores of all strains were produced in SSM sporulation medium, as previously described (6).
Immunization.
Female BALB/c mice were immunized subcutaneously with 1 × 107 CFU of irradiated (15 min at maximum intensity in a microwave oven) B. anthracis spores or with 10 μg of a soluble exosporium fraction (4). Immunizations were carried out every 2 weeks in incomplete Freund's adjuvant until no change in antibody titer was observed (four or five injections). Enzyme-linked immunosorbent assay (ELISA) was carried out against live spores. Mice were sacrificed (4 days after the last booster), and spleens were removed directly to liquid nitrogen.
ELISA for immunized mouse antibody titer determination.
ELISA plates were coated with 1 × 108 CFU/ml urografin-purified (20) B. anthracis Δ14185 spores in carbonate-bicarbonate buffer (C-3041; Sigma-Aldrich, St. Louis, MO) and incubated overnight at 4°C. Plates were then washed three times with PBS-T (phosphate-buffered saline [PBS] containing 0.05% [vol/vol] Tween 20) and blocked for 1 h with PBS-2% bovine serum albumin (BSA)-0.05% Tween 20 at 37°C. After additional washes, immunized mouse serum, diluted in blocking buffer, was applied for another hour, to be detected by alkaline phosphatase-anti-mouse immunoglobulin G antibodies (A-4312; Sigma). Antibody titers were calculated as reciprocal geometric mean titers. Values of at least twice the background signal were considered positive.
Construction of scFv library.
Total RNA was extracted from homogenized spleen tissues (homogenization was carried out under liquid nitrogen), using TRI reagent (TR118; MRC Molecular Research Center) according to the manufacturer's instructions. cDNA templates were prepared using Moloney murine leukemia virus reverse transcriptase (Promega, Madison, WI), applying a two-step reverse transcription-PCR protocol recommended by the manufacturer. Primers were used for the amplification and assembly of heavy- and light-chain DNAs as described previously (3). scFv DNA was purified, digested with SfiI and NotI, and ligated (T4 DNA ligase; NEB, Beverly, MA) into NotI/SfiI-linearized phagemid pCANTAB-5E (Pharmacia) containing an E tag sequence in frame. The recombinant phagemid was introduced into competent E. coli TG1 cells by electroporation, and library size was determined by plating. Diversity was analyzed by fingerprinting (BstNI digestion).
Biopanning against live spores in suspension.
The scFv phage library was biopanned for binders against live spores in suspension as follows. The library was amplified and rescued as described previously (3). Phage particles (1 × 1012) were challenged with 1 × 109 CFU of live avirulent B. anthracis Δ14185 spores suspended in 1 ml PBS containing 2% skim milk (Difco-Becton Dickinson, Sparks, MD). After 1.5 h at room temperature with gentle agitation, the spores were spun down (5 min, 4°C, 12,000 × g) and washed 10 times (PBS containing 0.05% [vol/vol] Tween 20). The spore-phage complex was then allowed to infect E. coli TG1 host cells directly, with no elution step, in order to amplify the selected spore-binding phage. The enriched library was then plated on ampicillin (100 μg/ml)- and glucose (0.4%)-containing plates, rescued, and used for further panning cycles (no B. anthracis contamination of the resulting enriched libraries was observed). Further cycles were carried out essentially as described above, with decreasing spore titers (down to 1 × 108 CFU) and increasing wash cycles (up to 10× PBS plus 0.05% Tween 20 and 5× PBS). For cycle n, the phage enrichment factor was calculated as follows: (Outn/Inn)/(Out1/In1). Spore-binding activity following library enrichment was monitored by phage ELISA as described in the subsequent section.
Screening for specific binders by phage ELISA.
Rescue of individual phage colonies and testing for spore-binding activity was carried out as described previously (3). The same ELISA was also used for monitoring enrichment of specific binders following panning cycles. ELISA plates were coated with B. anthracis Δ14185 spores. Blocking was carried out with PBSM (PBS plus 2% skim milk) for 2 hours at 37°C. BSA was used as the control antigen. Anti-M-13 horseradish peroxidase (HRP)-conjugated antibodies (Pharmacia) were used as reporting antibodies (1:5,000) in the assay. Clones were considered positive if they demonstrated at least twice the signal developed with the control antigen.
Production and screening of soluble scFv antibody fragments by ELISA.
Soluble scFv antibodies were produced in E. coli TG1 cells as described previously (3). Screening for spore-binding soluble scFv antibodies was carried out by ELISA, after cold osmotic shock of each clone (RPAS purification module; Amersham Pharmacia Biotech, Buckinghamshire, England). The ELISA was carried out essentially as described in the previous section, with the following modification: anti-E-tag HRP-conjugated antibodies (Pharmacia) were used as reporting antibodies (1:2,000) in the assay. Clones were considered positive if they demonstrated at least twice the signal developed with the control antigen.
Purification of soluble scFv antibody fragments.
Large-scale production of positive scFv antibody clones as E. coli periplasmic extracts was carried out as outlined for the RPAS purification module (Amersham Pharmacia Biotech, Buckinghamshire, England). Isopropyl-β-d-thiogalactopyranoside (IPTG; final concentration, 0.75 mM) was given at an optical density at 600 nm of 1.5, and the culture was harvested after 20 h at 30°C. The resulting pellet was resuspended in 0.5 M phosphate buffer, pH 8, and sonicated on ice (eight pulses of 30 seconds each, at maximum capacity) in a Vibra Cell sonicator (Sonics and Materials Inc., Newtown, CT). Purification of scFv antibodies from the periplasmic extract of E. coli was carried out using a commercial anti-E-tag affinity column (Amersham Pharmacia Biotech, Buckinghamshire, England) according to the manufacturer's instructions.
Western blotting for antigenic determinant (epitope) evaluation.
A soluble exosporium fraction was resolved in 10% sodium dodecyl sulfate (SDS)-polyacrylamide gels. Gels were either stained with Coomassie blue or blotted onto a nitrocellulose filter (polyvinylidene difluoride; Roche Diagnostics GmbH, Mannheim, Germany). The filter was probed with the soluble scFv antibody, which in turn was detected with an anti-E-tag HRP-conjugated antibody (Pharmacia) (1:3,000). An E. coli periplasmic extract was used as a negative control sample after induction of an irrelevant scFv antibody. Buffers and incubation periods were as outlined in a commercial Western blotting kit (BM chemiluminescence Western blotting kit; Roche Diagnostics GmbH, Mannheim, Germany).
Determination of affinity constant and specificity profile.
The affinity of the purified scFv antibody for B. anthracis spores was determined using flow cytometry (FCM) on a FACSCalibur machine (Becton Dickinson Immuno Cytometry Systems, Mississauga, Ontario, Canada), as described previously (18, 27). Each test tube contained B. anthracis spores (1 × 106 CFU/ml) and anti-E-tag (Pharmacia) and fluorescein isothiocyanate (FITC)-conjugated anti-mouse (F2266; Sigma-Aldrich, St. Louis, MO) (2 μg/ml) antibodies. The purified scFv antibody, diluted in assay buffer (PBS supplemented with 1% BSA, 0.05% Tween 20, and 0.02% azide), was added to each test tube to a final concentration of 0 to 135 nM. Spore-related fluorescence was determined after 30 min of incubation. Analysis was performed using CellQuest Pro (Becton Dickinson) and FlowJo software. The FCM setup for spore analysis was described elsewhere (27). For specificity determination, different spores (B. anthracis, B. cereus 569, and B. subtilis DSM 675) were applied in the assay (5 × 106 CFU/ml), with 1 nM scFv antibody.
“Sandwich” ELISA.
ELISA plates were coated with 10 μg/ml of rabbit anti-exosporium antibodies diluted in carbonate-bicarbonate buffer (C-3041; Sigma-Aldrich, St. Louis, MO). After an overnight incubation (4°C), ELISA plates were washed (3× PBS supplemented with 0.05% Tween 20) and blocked for 1 h with PBS-2% BSA-0.05% Tween 20 at 37°C. B. anthracis spores were then loaded into each plate at different concentrations and incubated for 1 h at room temperature, after which the plate was rewashed. The plate was then mounted with purified scFv antibodies, and these were detected with anti-E-tag-HRP (1:2,000). The assay dilution buffer was PBS supplemented with 6% Similac Top2 (Abbott Ireland, Cavan, Ireland) and 0.05% Tween 20. B. subtilis DSM 675 was used as a negative control in the assay. The presence of B. subtilis DSM 675 spores on the plate was verified by polyclonal rabbit anti-B. subtilis antibodies. A signal was considered positive if it demonstrated twice the blank (no spores) signal.
IFA.
An indirect immunofluorescence assay (IFA) was carried out with B. anthracis or B. subtilis DSM 675 spores (1 × 109 CFU/ml) air dried on multispot slides. Slides were incubated for 30 min (37°C, humid incubator) with scFv antibody-containing periplasmic extracts diluted 1:1 in assay buffer (PBS supplemented with 2% BSA and 0.05% Tween 20). The slides were rinsed with water, dried, and reincubated with anti-E-tag (Pharmacia) (1:100) for an additional 30 min. After an additional step of rinsing and drying, the slides were mounted with anti-mouse FITC-conjugated antibody (F2266; Sigma-Aldrich, St. Louis, MO) (1:500) for another 30 min. The slides were then examined under fluorescent illumination with a Nikon phase-contrast microscope (Nikon Eclipse E400). An E. coli periplasmic extract of an unrelated scFv antibody was used to eliminate possible nonspecific interactions of E. coli proteins with adhered spores.
RESULTS
Library construction.
The first step in the preparation of a high-quality immunized phage display library involves the optimization of an immunization strategy for high antibody titers. To this end, two immunization strategies were evaluated. The first utilized the intact antigen inactivated B. anthracis spores. The second consisted of a partial spore extract, i.e., a water-soluble fraction of the outer membrane of B. anthracis spores (exosporium). The two reagents were used for immunization of two BALB/c mouse groups. Antibody titers, determined by ELISA with live spores, were 10,000 and 100,000, respectively, indicating a higher humoral response induced by the soluble fraction. Hence, RNAs, used as a template for library construction, were extracted from the spleens of mice immunized with a soluble exosporium fraction. An scFv antibody phage display library was constructed from cDNA templates essentially as described previously (3). After primary and secondary amplifications of the heavy- and light-chain genes by PCR, the linker-containing fragments were assembled by overlap extension PCR, resulting in scFv DNA fragments with an expected length of ∼750 bp. The fragments were NotI/SfiI digested and ligated into NotI/SfiI-linearized phagemid pCANTAB-5E (Pharmacia). The antibodies were fused to the pIII phage surface protein and expressed with an E tag sequence in frame. The recombinant phagemids were introduced into competent E. coli TG1 cells, with a transformed rate of 7 × 106. Library quality was assessed by PCR for full-length inserts and fingerprinting (BstNI digestion of full-length inserts) of 30 randomly selected antibodies. The library was found to contain >90% intact (∼750-bp) unique (different digestion patterns) antibodies.
Biopanning.
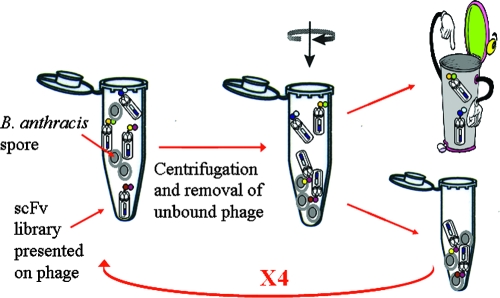
The biopanning process, outlined in Fig. 1, included the introduction of the entire phage library into live B. anthracis spores in solution. Thus, antibodies were challenged by the native antigen. After four panning cycles, with decreasing spore concentrations and increasing wash cycles, an enrichment factor (calculated as described in Materials and Methods) of 2,000 was obtained. The antibody population consisted of 90% intact (as determined by full-length insert PCR) and 60% diverse (fingerprinting of the antibody fragments) antibodies. Phage, after each enrichment cycle, were tested for spore-binding ability by phage ELISA as follows. Phage rescue was carried out with adhered spores and detected with anti-M13 HRP-conjugated antibodies. The final enriched library exhibited an enhanced (14 times the signal of the original library) specific response to B. anthracis spores.
FIG. 1.
Schematic representation of solution-based panning. A phage display scFv antibody library (7 × 106 PFU) was panned against live B. anthracis Δ14185 spores in suspension. After centrifugation, unbound phage were washed away, while sedimented spore-bound phage were used for the following panning cycles.
Selection of anti-spore binding antibodies.
Individual clones of the enriched library were rescued and examined for spore-binding activity by phage ELISA, and 80% of them were found to be positive. All binders were induced (IPTG) for soluble expression of the scFv antibody, and a periplasmic extract was prepared by cold osmotic shock. ELISA for spore-binding activity was performed against adhered spores. Soluble scFv antibodies were detected with an anti-E-tag HRP-conjugated antibody. Fingerprinting (BstNI digestion) of the resulting antibodies revealed four unique patterns. The expression of intact antibodies in the periplasmic fraction of E. coli was verified by Western blotting. A clear and strong signal for the putative scFv protein (≈30 kDa) was detected, using anti-E-tag antibodies, in the induced TG1 periplasmic samples carrying the selected scFv antibody clones. No band was observed for the uninduced E. coli periplasmic extract (results not shown). The four selected clones exhibited similar responses with B. anthracis spores. Consequently, only one of the antibodies was further characterized. Expression of the selected scFv antibody was carried out in E. coli TG1 cells. The scFv antibody was purified from 1 liter of cell culture on a commercial anti-E-tag column, with a yield of about 0.2 to 0.3 mg/liter. The purity of the resulting scFv protein was verified with a Coomassie blue-stained SDS-polyacrylamide gel (results not shown).
Characterization of scFv antibody.
The soluble scFv antibody was incubated with the exosporium fraction in order to elucidate an antigen recognition pattern. To this end, the soluble exosporium was resolved in 10% SDS-polyacrylamide gels that were either stained with Coomassie blue or blotted onto a nitrocellulose filter. The filter was then probed with the soluble scFv antibody, which was detected in turn with an anti-E-tag HRP-conjugated antibody. A clear, high-molecular-mass (>250 kDa) band was detected (Fig. 2), indicating a strong response of the scFv antibody to a high-molecular-weight fraction of the B. anthracis exosporium. This high-molecular-weight band is probably the Bcl1 protein, known as the most immunogenic antigen of the exosporium fraction (14, 20, 22). The affinity constant of the purified scFv antibody was determined via FCM as demonstrated previously (18, 27). In short, mean spore fluorescence at steady state was measured as a function of the antibody concentration. The results revealed a positive correlation between spore fluorescence and scFv antibody concentration, until a point at which the fluorescence intensity per particle no longer changed. This point is defined as the saturation point and reflects the maximum number of labeled antibody sites per spore, assuming quantitative adsorption of the ligand to the particle. The results could therefore be represented as a Langmuir adsorption curve, from which the affinity constant of the scFv antibody for spores was extracted. An affinity constant of 3 × 108 ± 1 × 108 M−1 was attained, in accordance with affinity constants obtained by other groups for immune phage display libraries (1, 19). Affinity constants in this range are characteristic of secondary immune response antibodies. Further characterization of the purified scFv antibody included a preliminary specificity profile determination. To this end, two other Bacillus species, the phylogenetically closely related species B. cereus and the somewhat distant relative B. subtilis DSM 675, were subjected to FCM analysis (10). As indicated in Fig. 3, the purified scFv antibody reacted specifically with B. anthracis spores, with no cross-reactivity with any of the other tested spores (at a concentration of 5 × 106 CFU/ml spores).
FIG. 2.
SDS-polyacrylamide gel electrophoresis and Western blot analysis of the soluble exosporium fraction. Coomassie blue staining (A) and Western blot analysis with the purified scFv antibody (B) of the soluble exosporium fraction. HRP-conjugated anti-E-tag antibody was used as a secondary antibody for colorimetric visualization.
FIG. 3.
FCM analysis of scFv antibody specificity profile. Spores of B. anthracis, B. subtilis, or B. cereus (5 × 106 CFU/ml) were challenged with the purified scFv antibody (1 nM). The scFv antibody was detected by anti-E-tag followed by an anti-mouse-FITC antibody. Plot histograms were gated to the spore population, as determined by light scatter parameters (side and forward scatter), and were analyzed by the FlowJo program.
Application of the purified scFv antibody to B. anthracis detection.
The scFv antibody was applied in a “sandwich” ELISA for detection of B. anthracis spores. To this end, different concentrations of B. anthracis spores were captured by anti-exosporium antibodies and detected with the purified scFv antibody followed by an HRP-conjugated anti-E-tag antibody (Fig. 4A). The soluble scFv antibody afforded a strong ELISA signal with live spores, with a detection limit of 2 × 106 CFU/ml (background plus 3 standard deviations), no less than the one obtained when polyclonal antibodies, previously prepared by our group (27), were implemented in the same ELISA format (11). No signal was observed with B. subtilis DSM 675 spores, which were used as control spores in the experiment. The binding of the soluble scFv antibody to B. anthracis spores was also demonstrated with immunofluorescence microscopy. B. anthracis spores were strongly detected (Fig. 4B), whereas no signal was observed on B. subtilis DSM 675 spores. No detectable binding to either of the spores was observed with a control unrelated scFv antibody produced from an unbiased clone of the original library (results not shown). Visualization of spores by IFA validated our previous results, indicating that the scFv antibody interacts with an immunodominant extracellular protein on the spore surface, which is probably the known protein Bcl1.
FIG. 4.
scFv antibody application in specific detection of B. anthracis spores. Analysis of B. anthracis and B. subtilis spores was done by “sandwich” ELISA (A) and IFA (B).
DISCUSSION
Anthrax is an acute infectious disease caused by the spore-forming organism B. anthracis. The potential use of B. anthracis spores as a biological weapon and the success of biological attacks in the past highlight the need for rapid detection of spores. In this work, we report the isolation of a specific, highly reactive scFv antibody suitable for specific spore detection from a phage display library. The purified soluble antibody was successfully implemented in several detection platforms.
A phage display library was constructed from spleens of mice immunized with a soluble exosporium fraction. This soluble fraction, comprised of proteins from the outermost layer of the spore, was found to induce a greater humoral response in mice than that induced by irradiated spores. This may indicate an advantage for the native, soluble, more accessible antigen versus its particulate form (irradiated spores) for immunization. In fact, the use of selected fractions of viral or bacterial lysates, consisting of membrane-bound or membrane-associated proteins as immunogens, was applied by other groups for efficient detection of intact live agents in clinical and environmental specimens (12). The same exosporium fraction was applied in the past for the successful preparation of polyclonal anti-B. anthracis spore antibodies in rabbits (27), resulting in highly reactive antibodies. The constructed library contained 7 × 106 independent scFv recombinants, a population characteristic of immunized phage display libraries (9, 24). Following library construction, rescued phage were challenged with live B. anthracis spores in solution and used to infect E. coli logarithmic-phase cells with no elution step. This methodology enabled the isolation of four unique scFv antibodies which were highly reactive against B. anthracis spores. Solution-based screening was applied in the past by several groups for the successful isolation of scFv antibodies (2, 23). Previous observations indicate that solution-based screening has some advantages over solid-based screening, particularly for the selection of monomeric over dimeric scFv antibodies (15) and for better recognition of the native antigen (16). One of the unique isolated scFv antibodies was expressed, purified, and characterized. The purified scFv antibody demonstrated an affinity constant of 3 × 108 ± 1 × 108 M−1 for B. anthracis spores (determined by FCM). Affinity constants in this range are characteristic of antibodies produced in the secondary immune response and were determined (by surface plasmon resonance) for other scFv antibodies isolated from immune phage display libraries (1, 19). The scFv antibody produced in this work was implemented successfully in several detection platforms, including IFA, ELISA, and FCM. Preliminary characterization has shown that the antibody has no cross-reactivity with other phylogenetically related Bacillus spores (Fig. 3), demonstrating improved specificity compared to that of polyclonal antibodies generated in our group by means of the same exosporium fraction (27). As demonstrated in our previous work, implementation of the polyclonal antibodies for specific detection of B. anthracis was achieved solely by using a double-staining method (fluorescence resonance energy transfer), whereas the scFv antibody enabled direct specific detection. Additional comparison of the specificity profile via ELISA (11) revealed a clear signal of the polyclonal antibodies on Bacillus megaterium and Bacillus thuringiensis subsp. thuringiensis (at ∼107 CFU/ml), whereas the scFv antibody did not detect any of the bacilli, even at ∼109 CFU/ml. As far as one can compare, the observed specificity was similar to that in other cases where monoclonal antibodies were implemented for B. anthracis spore detection (13, 21). However, in contrast to monoclonal antibodies, the cloned scFv antibody enables the possible use of direct genetic manipulations for the improvement of desired antibody characteristics (5, 15, 17). Moreover, genetic tagging options enable the construction of a bispecific scFv antibody suitable for direct application in detection platforms (25). The utilization of recombinant antibodies for the detection of biological warfare agents has been reported scarcely. Recombinant Fab antibodies isolated from a phage display library were incorporated in several tests for the detection of botulinum toxin, with a detection limit of 10 to 20 ng/ml (7, 8). Phage display was also utilized for the preparation of scFv antibodies against the biological warfare agent Brucella melitensis (9). The isolated phage antibodies were used for specific antigen detection, in contrast to an existing diagnostic monoclonal antibody. Another work described the development of scFv and Fab antibodies against B. subtilis as a model for B. anthracis (28).
In conclusion, to our knowledge, this work is the first in which an scFv antibody was developed and implemented for specific, efficient, and sensitive detection of B. anthracis spores.
Footnotes
Published ahead of print on 26 October 2007.
REFERENCES
- 1.Amersdorfer, P., C. Wong, S. Chen, T. Smith, S. Deshpande, R. Sheridan, R. Finnern, and J. D. Marks. 1997. Molecular characterization of murine humoral immune response to botulinum neurotoxin type A binding domain as assessed by using phage antibody libraries. Infect. Immun. 65:3743-3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Benhar, I., I. Eshkenazi, T. Neufeld, J. Opatowsky, S. Shaky, and J. Rishpon. 2001. Recombinant single chain antibodies in bioelectrochemical sensors. Talanta 55:899-907. [DOI] [PubMed] [Google Scholar]
- 3.Benhar, I., and Y. Reiter. 2002. Phage display of single-chain antibody constructs, p. 10.19B.1-10.19B.31. In R. Coico (ed.), Current protocols in immunology. John Wiley & Sons, Inc., New York, NY. [DOI] [PubMed]
- 4.Cohen, S., I. Mendelson, Z. Altboum, D. Kobiler, E. Elhanany, T. Bino, M. Leitner, I. Inbar, H. Rosenberg, Y. Gozes, R. Barak, M. Fisher, C. Kronman, B. Velan, and A. Shafferman. 2000. Attenuated nontoxinogenic and nonencapsulated recombinant Bacillus anthracis spore vaccines protect against anthrax. Infect. Immun. 68:4549-4558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coia, G., P. J. Hudson, and R. A. Irving. 2001. Protein affinity maturation in vivo using E. coli mutator cells. J. Immunol. Methods 251:187-193. [DOI] [PubMed] [Google Scholar]
- 6.Elhanany, E., R. Barak, M. Fisher, D. Kobiler, and Z. Altboum. 2001. Detection of specific Bacillus anthracis spore biomarkers by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Rapid Commun. Mass Spectrom. 15:2110-2116. [DOI] [PubMed] [Google Scholar]
- 7.Emanuel, P., T. O'Brien, J. Burans, B. R. DasGupta, J. J. Valdes, and M. Eldefrawi. 1996. Directing antigen specificity towards botulinum neurotoxin with combinatorial phage display libraries. J. Immunol. Methods 193:189-197. [DOI] [PubMed] [Google Scholar]
- 8.Emanuel, P. A., J. Dang, J. S. Gebhardt, J. Aldrich, E. A. Garber, H. Kulaga, P. Stopa, J. J. Valdes, and A. Dion-Schultz. 2000. Recombinant antibodies: a new reagent for biological agent detection. Biosens. Bioelectron. 14:751-759. [DOI] [PubMed] [Google Scholar]
- 9.Hayhurst, A., S. Happe, R. Mabry, Z. Koch, B. L. Iverson, and G. Georgiou. 2003. Isolation and expression of recombinant antibody fragments to the biological warfare pathogen Brucella melitensis. J. Immunol. Methods 276:185-196. [DOI] [PubMed] [Google Scholar]
- 10.Keim, P., A. Kalif, L. Schupp, K. Hill, S. E. Travis, K. Richmond, D. M. Adair, M. Hugh-Jones, C. R. Kuske, and P. M. Jackson. 1997. Molecular evolution and diversity in Bacillus anthracis detection by amplified fragment length polymorphism markers. J. Bacteriol. 179:818-824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mechaly, A., E. Zahavi, and M. Fisher. 2006. Abstr. Detection Technologies 2006, San Diego, CA, p. 364.
- 12.Peruski, A. H., and L. F. Peruski, Jr. 2003. Immunological methods for detection and identification of infectious disease and biological warfare agents. Clin. Diagn. Lab. Immunol. 10:506-513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Phillips, A. P., A. M. Campbell, and R. Quinn. 1988. Monoclonal antibodies against spore antigens of Bacillus anthracis. FEMS Microbiol. Immunol. 47:169-178. [DOI] [PubMed] [Google Scholar]
- 14.Redmond, C., L. W. J. Baillie, S. Hibbs, A. J. G. Moir, and A. Moir. 2004. Identification of proteins in the exosporium of Bacillus anthracis. Microbiology 150:355-363. [DOI] [PubMed] [Google Scholar]
- 15.Schier, R., J. Bye, G. Apell, A. McCall, G. P. Adams, M. Malmqvist, L. M. Weiner, and J. D. Marks. 1996. Isolation of high-affinity monomeric human anti-c-erbB-2 single chain Fv using affinity-driven selection. J. Mol. Biol. 255:28-43. [DOI] [PubMed] [Google Scholar]
- 16.Schier, R., J. D. Marks, E. J. Wolf, G. Apell, C. Wong, J. E. McCartney, M. A. Bookman, J. S. Huston, L. L. Houston, L. M. Weiner, and G. P. Adams. 1995. In vitro and in vivo characterization of a human anti-c-erbB-2 single-chain Fv isolated from a filamentous phage antibody library. Immunotechnology 1:73-81. [DOI] [PubMed] [Google Scholar]
- 17.Schier, R., A. McCall, G. P. Adams, K. W. Marshall, H. Merritt, M. Yim, R. S. Crawford, L. M. Weiner, C. Marks, and J. D. Marks. 1996. Isolation of picomolar affinity anti-c-erbB-2 single-chain Fv by molecular evolution of the complementarity determining regions in the center of the antibody binding site. J. Mol. Biol. 263:551-567. [DOI] [PubMed] [Google Scholar]
- 18.Siiman, O., and A. Burshteyn. 2000. Cell surface receptor-antibody association constants and enumeration of receptor sites for monoclonal antibodies. Cytometry 40:316-326. [PubMed] [Google Scholar]
- 19.Stacy, J. E., L. Kausmally, B. Simonsen, S. H. Nordgard, L. Alsoe, T. E. Michaelsen, and O. H. Brekke. 2003. Direct isolation of recombinant human antibodies against group B Neisseria meningitidis scFv expression libraries. J. Immunol. Methods 283:247-259. [DOI] [PubMed] [Google Scholar]
- 20.Steichen, C., P. Chen, J. F. Kearney, and C. L. Turnbough, Jr. 2003. Identification of the immunodominant protein and other proteins of the Bacillus anthracis exosporium. J. Bacteriol. 185:1903-1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Swiecki, M. K., M. W. Lisanby, F. Shu, C. L. Turnbough, Jr., and J. F. Kearney. 2006. Monoclonal antibodies for Bacillus anthracis spore detection and functional analyses of spore germination and outgrowth. J. Immunol. 176:6076-6084. [DOI] [PubMed] [Google Scholar]
- 22.Sylvestre, P., E. Couture-Tosi, and M. Mock. 2002. A collagen like surface glycoprotein is a structural component of the Bacillus anthracis exosporium. Mol. Microbiol. 45:169-178. [DOI] [PubMed] [Google Scholar]
- 23.Turnbough, C. L., Jr. 2003. Discovery of phage display peptide ligands for species-specific detection of Bacillus spores. J. Microbiol. Methods 53:263-271. [DOI] [PubMed] [Google Scholar]
- 24.Vukovic, P., K. Chen, X. Q. Liu, M. Foley, A. Boyd, D. Kaslow, and M. F. Good. 2002. Single-chain antibodies produced by phage display against the C-terminal 19 kDa region of merozoite surface protein-1 of Plasmodium yoelii reduce parasite growth following challenge. Vaccine 20:2826-2835. [DOI] [PubMed] [Google Scholar]
- 25.Wang, S.-H., J.-B. Zhang, Z.-P. Zhang, Y.-F. Zhou, R.-F. Yang, J. Chen, Y.-C. Guo, F. You, and X.-E. Zhang. 2006. Construction of single chain variable fragment (scFv) and biscFv-alkaline phosphatase fusion protein for detection of Bacillus anthracis. Anal. Chem. 78:997-1004. [DOI] [PubMed] [Google Scholar]
- 26.Wild, M. A., H. Xin, T. Maruyama, M. J. Nolan, P. M. Calveley, J. D. Malone, M. R. Wallace, and K. S. Bowdish. 2003. Human antibodies from immunized donors are protective against anthrax toxin in vivo. Nat. Biotechnol. 21:1305-1306. [DOI] [PubMed] [Google Scholar]
- 27.Zahavy, E., M. Fisher, A. Bromberg, and U. Olshevsky. 2003. Detection of frequency resonance energy transfer pair on double-labeled microsphere and Bacillus anthracis spores by flow cytometry. Appl. Environ. Microbiol. 69:2330-2339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhou, B., P. Wirsching, and K. D. Janda. 2002. Human antibodies against spores of the genus Bacillus: a model study for detection of and protection against anthrax and the bioterrorist threat. Proc. Natl. Acad. Sci. USA 99:5241-5246. [DOI] [PMC free article] [PubMed] [Google Scholar]