Abstract
In the present study we characterized 47 food-borne isolates of Bacillus cereus using multilocus sequence typing (MLST). Newly determined sequences were combined with sequences available in public data banks in order to produce the largest data set possible. Phylogenetic analysis was performed on a total of 296 strains for which MLST sequence information is available, and three main lineages—I, II, and III—within the B. cereus complex were identified. With few exceptions, all food-borne isolates were in group I. The occurrence of horizontal gene transfer (HGT) among various strains was analyzed by several statistical methods, providing evidence of widespread lateral gene transfer within B. cereus. We also investigated the occurrence of toxin-encoding genes, focusing on their evolutionary history within B. cereus. Several patterns were identified, indicating a pivotal role of HGT in the evolution of toxin-encoding genes. Our results indicate that HGT is an important element in shaping the population structure of the B. cereus complex. The results presented here also provide strong evidence of reticulate evolution within the B. cereus complex.
The Bacillus cereus complex comprises five closely related species of gram-positive, spore-forming bacteria. They have traditionally been classified as different species based on their distinct virulence characteristics. B. cereus, the species giving the name to the group, is widely distributed in the environment and has been associated with food poisoning and soft tissue infections (22). Bacillus thuringiensis, another species in the complex, is an insect pathogen that has occasionally been indicated as responsible for human infections similar to those caused by B. cereus strains (9, 32). Bacillus anthracis, a pathogen of humans and other mammals, results in another pathogenic phenotype (45). Bacillus mycoides is a fourth taxon that can be differentiated from B. cereus by its distinctive rhizoid colony morphology and is not considered to be a human pathogen. A fifth species, named Bacillus weihenstephanensis, was recently created to accommodate some psychrotolerant strains (40). The validity of B. cereus, B. thuringiensis, and B. anthracis being classified as distinct species has been questioned by some authors that consider them a single species (29). B. thuringiensis is the most diverse species within the B. cereus group, and its strains have been classified into 84 serovars (serovarieties) (39, 54). The taxonomic value of these serovars is currently not settled. Although there is a very clear division of the virulence characteristics and the known virulence genes, the genetic relationships of the species themselves is not immediately apparent from genomic data.
Comprehensive studies using a large range of biochemical and molecular biology techniques have been carried out to test the validity of the species status of the various isolates and to elucidate the relationships among them (7, 8, 28, 29, 31, 36, 37, 52, 58, 60). All of these studies, together with complete genome comparisons (25), have revealed extensive genomic similarity with only sets of genes being identified as species specific, indicating that the genetic content of the genomic backbone alone does not adequately define the different virulence and ecological characteristics of the species. The population structure within the B. cereus group has been addressed in several studies, but their results were inconclusive. Some studies provided evidence for a horizontally spread gene transfer (28, 37), while other analyses suggested more typical clonal behavior (29-31, 52, 58, 60). Such marked differences could be due to the different approaches used, as well as taxon sampling.
Several B. cereus strains have been identified as the cause of two kinds of gastrointestinal disease associated with very different types of toxins, namely, emetic syndrome and diarrheal poisoning (57). Emetic syndrome is caused by a heat-stable peptide toxin named cereulide (13), which is preformed in food. Recently, the peptide-synthetase genes responsible for the nonribosomal production of cereulide (ces genes) were identified and characterized. A molecular assay for the detection of emetic toxin producers was also developed (14, 16, 17). The other gastrointestinal disease caused by B. cereus is diarrheal poisoning. This is due to heat-labile enterotoxins produced during vegetative growth of B. cereus in the small intestine. At present, three different toxins have been associated with food poisoning outbreaks: the protein complex hemolysin BL (HBL [6]), the nonhemolytic enterotoxin (NHE [23]), and the single protein cytotoxin CytK (41). Two additional single cytotoxin proteins produced by B. cereus, bceT and entFM, have been described (1, 4, 27). Immunological assays are commercially available for the detection of NHE and HBL, as well as monoclonal antibodies targeting these enterotoxin complexes (10, 11), whereas no such tools are yet available for CytK or cereulide. This could be because studies on the B. cereus toxins have been focused on the nhe and hbl genes (20, 24, 26, 49, 53). Despite the recognition of B. cereus as a food-borne pathogen over 50 years ago and the identification of several enterotoxin genes, its virulence mechanisms have still not been fully elucidated. The in vivo roles of the putative virulence factors produced by B. cereus have not been characterized, except for HBL, which is associated with diarrheal food poisoning (6). Based on their biological activities, these factors are likely to be involved in B. cereus infection and illnesses. NHE, a homolog of HBL, likely possesses biological activities similar to those of HBL and could be a factor in the diarrheal syndrome; however, this hypothesis has not been tested (41).
Multilocus sequence typing (MLST) studies have previously been used to examine the phylogeny of the B. cereus complex (5, 30, 31), identifying three distinct lineages. These lineages largely correspond to the distribution of the species. The clinical isolates used in these studies (5, 30), however, comprised largely clinical isolates from soft tissue infections. Therefore, the relationship of food-borne isolates to the inferred phylogeny is not known. In the present study, we analyzed the MLST profile of 47 strains isolated from different types of food. We analyzed the MLST profiles of our samples in combination with others available in the public database in order to elucidate the phylogenetic relationships among different species included in the B. cereus complex. The presence or absence of toxin genes was also studied to better understand the evolutionary history of this group of proteins, which has received only limited attention thus far. The population structure of the newly characterized isolates, together with strains available in the literature, was surveyed. Finally, we investigated the occurrence of horizontal gene transfer (HGT) in the B. cereus complex and its effect on the evolution of this group of bacteria.
MATERIALS AND METHODS
Bacterial isolates and DNA extraction.
The foodstuff was analyzed for B. cereus on mannitol-yeast extract-polymyxin-agar (Oxoid, Basingstoke, United Kingdom) according to ISO 7932 (3) using replicate plates. All isolates were confirmed as Bacillus spp. by Gram staining, catalase reaction, and conventional biochemical tests (API 50CHB; bioMérieux, Marcy l'Etoile, France). A total of 47 pure cultures of B. cereus were isolated (Table 1). The complete set of B. cereus in the present study represents all isolates analyzed by our food-microbiological laboratory in 2005. The geographical origins were as follows: isolates 1 to 26 were from southern Italy, isolates 27 to 32 and 41 to 45 were from northwestern Italy, and the remaining isolates were from northeastern Italy. Bacterial cultures were grown overnight in nutrient broth (5 ml) aimed at DNA isolation. Chromosomal DNA was prepared from a 1.0-ml culture aliquot by using a Spin Bacteria DNA isolation kit (Invitek, Berlin, Germany) according to the manufacturer's instructions.
TABLE 1.
New isolates of B. cereus characterized in the present study
| Isolate | Sourcea | MLSTb
|
Presence (+) or absence (-) of toxin gene
|
||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ST | glpF | gmk | ilvD | pta | pur | pycA | tpi | hblA | hblC | hblD | nheA | nheB | nheC | entFM | bceT | cytK1 | cytK2 | em | ITS | ||
| BC14 | EGP | 12 | 15 | 7 | 7 | 2 | 7 | 10 | 13 | + | + | + | + | + | + | + | - | - | - | - | + |
| BC26 | EGP | 15 | 9 | 8 | 16 | 13 | 2 | 16 | 9 | + | + | + | + | + | + | - | + | + | - | - | + |
| BC4 | EGP | 24 | 12 | 8 | 9 | 14 | 11 | 12 | 10 | + | + | + | + | + | + | + | - | + | - | - | + |
| BC8 | EGP | 24 | 12 | 8 | 9 | 14 | 11 | 12 | 10 | + | + | + | + | + | + | + | - | + | + | - | + |
| BC27 | REP | 24 | 12 | 8 | 9 | 14 | 11 | 12 | 10 | + | + | + | + | + | + | + | - | + | + | - | + |
| BC37 | IDP | 24 | 12 | 8 | 9 | 14 | 11 | 12 | 10 | + | + | + | + | + | - | + | - | - | + | - | + |
| BC7 | EGP | 26 | 3 | 2 | 31 | 5 | 16 | 3 | 4 | - | - | - | + | + | + | + | - | - | - | + | + |
| BC9 | EGP | 142 | 13 | 8 | 8 | 11 | 9 | 12 | 7 | + | + | + | + | + | + | + | + | + | + | - | + |
| BC17 | EGP | 142 | 13 | 8 | 8 | 11 | 9 | 12 | 7 | + | + | + | + | + | + | + | + | + | - | - | + |
| BC29 | REP | 142 | 13 | 8 | 8 | 11 | 9 | 12 | 7 | + | + | + | + | + | + | + | + | + | - | - | + |
| BC32 | REP | 142 | 13 | 8 | 8 | 11 | 9 | 12 | 7 | + | + | + | + | + | + | + | + | + | - | - | + |
| BC30 | REP | 160 | 73 | 43 | 70 | 60 | 64 | 63 | 32 | - | - | - | + | + | + | + | + | - | - | - | + |
| BC28 | REP | 205 | 19 | 2 | 21 | 5 | 19 | 3 | 2 | - | - | - | + | + | + | + | - | - | - | - | + |
| BC1 | EGP | 369 | 48 | 30 | 33 | 37 | 44 | 103, EF421124 | 5 | - | - | - | + | + | + | + | - | - | - | - | + |
| BC24 | EGP | 369 | 48 | 30 | 33 | 37 | 44 | 103, EF421124 | 5 | - | - | - | + | + | + | + | - | - | - | - | + |
| BC2 | EGP | 370 | 14 | 8 | 43 | 11 | 115, EF421121 | 104, EF421125 | 85, EF421127 | + | + | + | + | + | + | + | + | - | + | - | + |
| BC6 | EGP | 370 | 14 | 8 | 43 | 11 | 115, EF421121 | 104, EF421125 | 85, EF421127 | + | + | + | + | + | + | + | + | - | + | - | + |
| BC15 | EGP | 370 | 14 | 8 | 43 | 11 | 115, EF421121 | 104, EF421125 | 85, EF421127 | + | + | + | + | + | + | + | + | - | + | - | + |
| BC16 | EGP | 370 | 14 | 8 | 43 | 11 | 115, EF421121 | 104, EF421125 | 85, EF421127 | + | + | + | + | + | + | + | + | + | - | - | + |
| BC19 | EGP | 370 | 14 | 8 | 43 | 11 | 115, EF421121 | 104, EF421125 | 85, EF421127 | + | + | + | + | + | - | + | + | - | - | - | + |
| BC23 | EGP | 370 | 14 | 8 | 43 | 11 | 115, EF421121 | 104, EF421125 | 85 | + | + | + | + | + | + | + | + | + | + | - | + |
| BC3 | EGP | 371 | 21 | 2 | 19 | 5 | 32 | 18 | 2 | + | + | - | + | + | + | + | - | - | - | - | + |
| BC11 | EGP | 371 | 21 | 2 | 19 | 5 | 32 | 18 | 2 | - | + | - | + | + | + | + | - | + | + | - | + |
| BC25 | EGP | 371 | 21 | 2 | 19 | 5 | 32 | 18 | 2 | - | + | - | + | + | + | + | - | - | + | - | + |
| BC21 | EGP | 371 | 21 | 2 | 19 | 5 | 32 | 18 | 2 | - | - | - | + | + | + | + | - | - | + | - | + |
| BC5 | EGP | 372 | 14 | 8 | 9 | 28 | 4 | 12 | 86, EF421128 | + | + | + | + | + | + | + | + | + | + | - | + |
| BC10 | EGP | 373 | 22 | 9 | 18 | 18 | 46 | 19 | 7 | + | + | + | + | + | + | + | - | - | + | - | + |
| BC13 | EGP | 374 | 29 | 2 | 24 | 5 | 32 | 3 | 2 | + | + | + | + | + | + | + | - | - | - | - | + |
| BC20 | EGP | 374 | 29 | 2 | 24 | 5 | 32 | 3 | 2 | + | + | + | + | + | + | + | - | - | - | - | + |
| BC22 | EGP | 374 | 29 | 2 | 24 | 5 | 32 | 3 | 2 | + | + | + | + | + | + | + | - | + | - | - | + |
| BC18 | EGP | 375 | 110 | 34 | 111 | 102 | 112 | 96 | 26 | + | + | + | + | + | + | + | - | - | - | - | + |
| BC33 | IDP | 376 | 112, EF421112 | 8 | 14 | 12 | 2 | 50 | 7 | + | + | + | + | + | + | + | + | - | + | - | + |
| BC36 | IDP | 376 | 8 | 8 | 14 | 12 | 2 | 50 | 7 | + | + | - | + | + | + | + | + | - | + | - | + |
| BC38 | IDP | 376 | 8 | 8 | 14 | 12 | 2 | 50 | 7 | + | + | + | + | + | + | + | + | - | + | - | + |
| BC39 | RWF | 376 | 8 | 8 | 14 | 12 | 2 | 50 | 7 | + | + | + | + | + | + | + | + | + | + | - | + |
| BC40 | RWF | 376 | 8 | 8 | 14 | 12 | 2 | 50 | 7 | + | + | + | + | + | + | + | + | - | + | - | + |
| BC46 | FDP | 376 | 8 | 8 | 14 | 12 | 2 | 50 | 7 | + | + | + | + | + | + | + | + | - | + | - | + |
| BC47 | FDP | 376 | 8 | 8 | 14 | 12 | 2 | 50 | 7 | + | + | + | + | + | + | + | + | + | - | - | + |
| BC48 | FDP | 376 | 8 | 8 | 14 | 12 | 2 | 50 | 7 | + | + | + | + | + | + | + | + | - | + | - | + |
| BC49 | FDP | 376 | 8 | 8 | 14 | 12 | 2 | 50 | 7 | + | + | + | + | + | + | + | + | + | + | - | + |
| BC34 | IDP | 377 | 8 | 70, EF421114 | 114, EF421115 | 110, EF421118 | 116, EF421122 | 105, EF421126 | 32 | - | - | - | + | + | + | + | - | - | - | - | + |
| BC35 | IDP | 377 | 112, EF421112 | 70, EF421114 | 114, EF421115 | 110, EF421118 | 116, EF421122 | 105, EF421126 | 32 | - | - | - | + | + | + | + | - | - | - | - | + |
| BC41 | FDP | 378 | 113, EF421113 | 31 | 115, EF421116 | 43 | 117, EF421123 | 53 | 37 | + | + | + | + | + | + | + | - | - | - | - | + |
| BC45 | FDP | 379 | 33 | 17 | 13 | 112, EF421120 | 29 | 17 | 7 | - | - | - | + | + | + | + | - | - | - | - | + |
| BC42 | FDP | 387 | 81 | 53 | 117, EF421117 | 111, EF421119 | 113 | 93 | 80 | + | + | + | + | + | - | + | - | - | - | - | + |
| BC43 | FDP | 387 | 81 | 53 | 117, EF421117 | 111, EF421119 | 113 | 93 | 80 | + | + | + | + | + | + | + | - | - | - | - | + |
| BC44 | FDP | 387 | 81 | 53 | 117, EF421117 | 111, EF421119 | 113 | 93 | 80 | + | + | + | + | + | + | + | - | - | - | - | + |
EGP, egg product; REP, ready-to-eat product; IDP, industrial dairy product; RWF, raw fish; FDP, farm dairy product.
Boldface numbers indicate new alleles. GenBank accession numbers (EF numbers) are provided for new allele sequences.
MLST gene amplification, sequencing, and characterization.
The MLST approach (31) was applied to the food-borne isolates using primers and conditions reported in the primers section of the B. cereus MLST database (http://pubmlst.org/bcereus/). Portions of glpF, gmk, ilvD, pta, pur, pycA, and tpi housekeeping genes were amplified by using the DNA extracted from the pure culture of each of the 47 B. cereus strains as a template (Table 1). PCR amplifications were performed on a Primus thermal cycler (Euroclone, Celbio, Milan, Italy) by using the following cycle: an initial denaturation 2 min at 94°C and 20 s at 94°C, followed by 30 s at 55°C and 30 s at 72°C in a final volume of 20 μl. The amplification mix contained 1 U of GoTaq polymerase (Promega), 1× GoTaq buffer, 1.5 μM MgCl2, 140 μM concentrations of each deoxynucleoside triphosphate, and 0.125 μM concentrations of the forward and the reverse primers. These amplification conditions allowed for the direct sequencing of amplification products without any additional purification. DNA sequencing was performed on both strands by using the ABI Prism dye terminator cycle sequencing core kit (PE Applied Biosystems). Electrophoresis was performed on an ABI Prism model 377 (version 2.1.1) automated sequencer. The sequence of each allele was compared to those available in the MLST B. cereus database by using the BLAST program (2).
New allele sequences were submitted to GenBank and to the B. cereus MLST database. Accession numbers, allele codes, and ST numbers are provided in Table 1.
Molecular phylogenetic analysis.
Publicly available MLST sequences (downloaded from http://pubmlst.org/bcereus/version on 11 October 2006) were combined with new data obtained in the present study in order to produce the multiple alignment used for the phylogenetic analysis. Different sequence types (STs) were numbered according to the MLST scheme developed for B. cereus (33). Multiple alignments containing the concatenated sequences (referred to hereafter as the MLST data set) were straightforward and performed according to the MLST scheme (33). All analyzed MLST sequences were the same length. The gene order in the MLST data set was as follows: glpF, gmk, ilvD, pta, pur, pycA, and tpi (33). This order differs from the common gene order of glpF, ilvD, pur, gmk, pycA, tpi, and pta observed in all complete genomes (available in GenBank) sequenced thus far for species of the B. cereus complex.
A phylogenetic tree was inferred by using the neighbor-joining (NJ) method (55). The NJ analysis was performed by using the MEGA 3.1 program (38) applying the Tamura and Nei evolutionary model (46). Nonparametric bootstrap resampling (BT) (19) was performed to test the robustness of the tree topology (1,000 replicates).
Toxin genes analysis.
Portions of the hblA, hblC, hblD, nheA, nheB, nheC, entFM, bceT, and cytK genes and the emetic-specific sequence (em) were amplified using the set of primers listed in Yang et al. (61) with the 16S to 23S rRNA internal transcribed sequence (ITS) used as an internal control. The amplification was performed in quadruplex (mix A) or pentaplex (mix B and mix C) reactions with the primer pairs reported in Table 2. Amplifications were performed on a Primus thermal cycler (Euroclone, Celbio, Milan, Italy) under the following conditions: an initial denaturation for 2 min at 94°C and 30 s at 94°C, followed by 30 s at 56°C and 45 s at 72°C, in a final volume of 20 μl. The amplification mix contained 1 U of GoTaq polymerase (Promega, Madison, WI), 1× GoTaq buffer, 1.5 mM MgCl2, 200 μM concentrations of each deoxynucleoside triphosphate, and variable numbers and quantities of primers (Table 2). The result of each amplification mix was visualized on a 1.8% agarose gel stained with ethidium bromide. All samples were analyzed with two independent PCRs.
TABLE 2.
PCR portions of toxin genes
| PCR | Length (bp) | Mix | Final concn (μM) of each primer |
|---|---|---|---|
| HblA | 237 | C | 0.4 |
| HblC | 386 | A | 0.5 |
| HblD | 430 | C | 0.4 |
| NheA | 475 | B | 0.25 |
| NheB | 328 | C | 0.25 |
| NheC | 557 | A | 0.25 |
| EntFM | 290 | B | 0.25 |
| BceT | 701 | C | 0.7 |
| CytK1 | 800 | A | 0.25 |
| CytK2 | 800 | B | 0.25 |
| em | 635 | B | 0.25 |
| ITS | 185 | A/B/C | 0.25 |
eBurst analysis.
Population structure of the B. cereus group was investigated by using the eBurst algorithm (18, 59).
Initially, the eBurst algorithm subdivides STs into groups. eBurst analysis was performed by either (i) using the default, very stringent parameters (http://eburst.mlst.net/), in which STs are assigned to the same group only if six of seven alleles in the MLST loci are identical or (ii) relaxing the default parameters to allow groups of STs sharing five or more identical alleles. In our analyses, we considered 449 isolates belonging to 297 distinct MLST profiles. The eBurst analyzed data set was produced, combining ST profiles determined in present study together with those available at the B. cereus group MLST database (http://pubmlst.org/bcereus/), which implements the MLSTdbnet software (33).
HGT detection.
HGT was investigated by applying multiple approaches, which limits the risk of identifying false events of recombination or overlooking the occurrence of true recombinations (50, 51). We performed recombination analyses by applying the following algorithms: BooTScan (56), CHIMAERA (50), GENECONV (48), MAXIMUM χ2 (44), RDP (42), and SiScan (21), as implemented in the RDP2 program (43).
The MLST data set was used in all of the analyses performed with the RDP2 program.
RESULTS
MLST sequence analysis.
Newly determined sequences of glpF, gmk, ilvD, pta, pur, pycA, and tpi housekeeping genes were compared to allelic sequences available at the MLST B. cereus database. Comparisons identified 22 new alleles that were confirmed with a second, independent round of amplification and sequencing. The new alleles were distributed among each of the MLST loci with three of glpF, one of gmk, four of ilvD, three of pta, five of pur, four of pycA, and two of tpi. MLST profile analysis of food-borne isolates proved that the majority belonged to new ST profiles. Indeed, 34 of 47 MLST profiles (72.3% of the studied samples) were included in the 12 new ST profiles, while the remaining profiles had a counterpart in the MLST B. cereus database (Table 1).
Isolates, including the strains studied here, listed as food-borne in the MLST database corresponded to 18.7% of the whole currently available set (84 of 449). Food-borne strains were included in 50 different ST profiles, 45 of which were isolated exclusively from foodstuff, while 5 were collected from other sources.
Phylogenetic analysis.
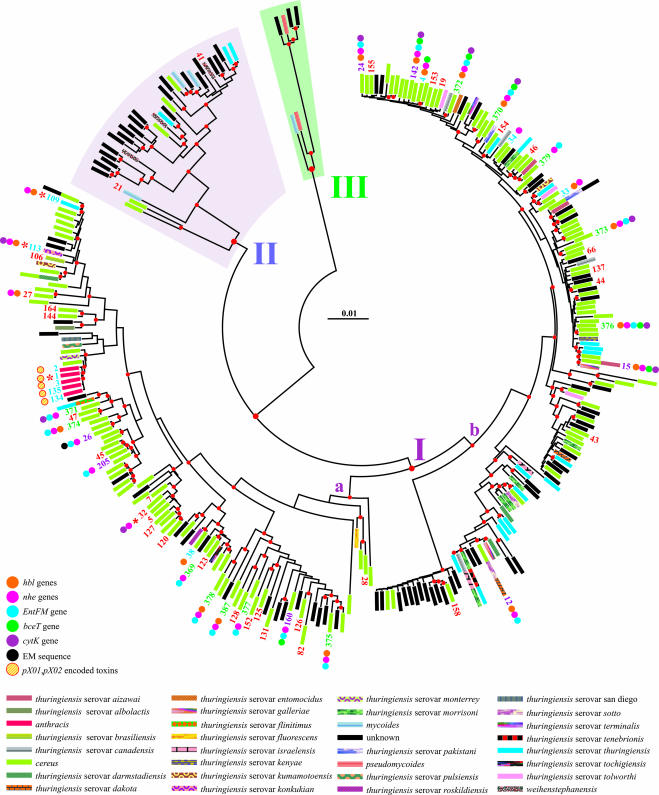
MLST multiple alignment was 2,829 bases long and contained 296 unique MLST sequences. The phylogenetic tree obtained from the analysis performed on the MLST data set is presented in Fig. 1. Three main lineages, hereafter named I, II, and III, were recognized as all receiving strong BT support. Lineage I included B. anthracis, many B. cereus strains, and most of the variously named B. thuringiensis serovars. Lineage I was the largest cluster and was subdivided in the two major groups, Ia and Ib, both receiving statistical support. Group Ia included 102 STs mostly belonging to B. cereus and also contained all STs used to define clade 1 (B. cereus) of Priest and coworkers (52). Clade 1 was named B. cereus clade by Priest et al. since that was the predominant organism of the cluster (52). Group Ia contained all B. anthracis isolates that were tightly linked and formed a monophyletic taxon. Finally, group Ia included ST profiles identifying isolates for which full-length genome sequences are available in GenBank (Fig. 1). Group Ib included STs belonging to diverse B. thuringiensis serovars and unknown B. cereus complex isolates, as well as B. cereus strains. Group Ib also contained the STs used to delimit the B. thuringiensis clade 2 by Priest et al. (52). Lineage I included all new food-borne isolates investigated in the present study and nearly all of the food-borne isolates contained in the MLST database. The only noteworthy exceptions were ST 21 and ST 41, which were placed within lineage II. New food-borne profiles identified in the present study were scattered all over lineage I. Among the 50 known food-borne ST profiles, 58% were included within group Ia. Lineage II contained B. mycoides, B. weihenstephanensis, and B. cereus isolates, as well as unknown isolates. Lineage III included only six isolates. Notably, different serovars of B. thuringiensis and B. cereus isolates showed identical MLST sequence (e.g., ST 12) (Fig. 1).
FIG. 1.
Phylogeny of the B. cereus group. An NJ tree depicts the phylogenetic relationships among unique MLST isolates. The scale bar shows the distance calculated according to the Tamura and Nei evolutionary model. Major clusters are numbered arbitrarily I, II, and III. Nodes supported by bootstrap values of >50% are marked with a red dot. Different toxin genes and species are labeled according to the self-explanatory color scheme listed below the tree. ST labels defining isolates were omitted to avoid overcrowding. Exceptions are represented by newly determined unique isolates (green), new isolates with a counterpart in the public database (purple), food-borne isolates (red), and public isolates (blue), for which toxin data are available in the literature. The distribution of toxins and toxin genes is not known for most of the food-borne isolates labeled here. The distribution of toxins and toxin genes is also currently unknown for the unlabeled STs depicted in this figure. An ST marked with a red asterisk indicates that at least a full-length genome sequence is available in GenBank for an isolate with the same ST profile.
Toxin genes analysis and mapping.
The results of multiplex amplifications performed to test the presence of enterotoxin genes in the bacterial chromosome and em plasmid sequence of 47 food-borne isolates are summarized on Table 1. Occurrences of various toxins in different STs was mapped on the NJ tree produced in the phylogenetic analysis (Fig. 1), which also combined data derived from previous studies with the current results. All strains analyzed in the present study were tested for the presence or absence of toxin genes (Table 1). Current data on toxins are available only for a limited number of strains listed in Fig. 1. Thus, the absence of toxins and/or toxin genes in many strains depicted in Fig. 1 reflects lack of knowledge rather than the true absence of the toxins.
Toxin genes exhibited a scattered distribution over all of lineage I (Fig. 1). However, distribution patterns varied among different genes. The nhe genes were present in almost all STs sampled with the exclusion of ST 38. The hbl and entF genes also exhibited a broad distribution. Conversely, the bceT gene was limited mainly to the cluster ranging from ST 24 to ST 15, with the only exception represented by ST 160. A similar behavior was observed for cytK gene with the exception represented, in this case, by ST 371. The em sequence showed the narrowest distribution, being restricted only to ST 26. The maximum number of co-occurring toxin genes was 5. Toxin genes shared among different STs varied from 0 to 5 with the most spread combination represented by hbl plus nhe plus entFM genes. Current limits on the knowledge of toxin distribution notwithstanding, some patterns can be identified. It is plausible to imagine that hbl, nhe, entFM, bceT, and cytK genes were already present in the common ancestor of lineage I. The loss of genes followed a different pattern in groups Ia and Ib. In fact, all five types of genes have been retained (coinherited) in several STs belonging to the Ib group (e.g., ST 142 and ST 376), while a more marked reduction occurred in the Ia group, where the loss particularly affected bceT and cytK genes. Note that gene loss is not linked to a particular species. The pX01- and pX02-encoded toxins appeared during evolution just once in the B. anthracis taxon. The more widespread coinheritance occurred for the three groups of genes: hbl and nhe; hbl, nhe, and entFM; and nhe and entFM. Nothing can be said for lineages II and III because no data on toxin distribution in these lineages are currently available.
eBurst analysis.
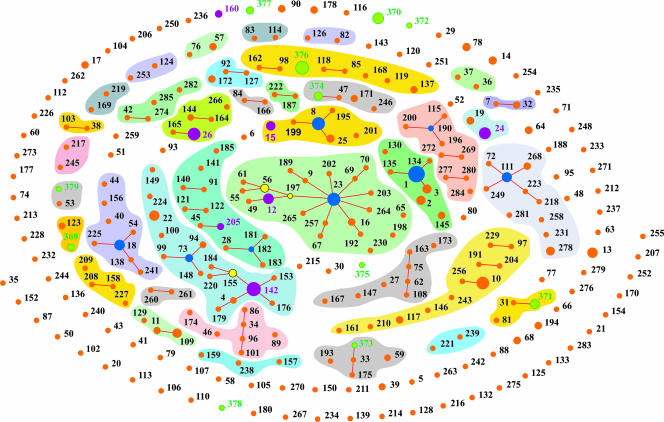
Population structure analysis performed using the eBurst algorithm identified 35 potential clonal complexes (CCs) (Fig. 2 and Fig. 3). These complexes included 210 isolates belonging to 115 ST allelic profiles. The remaining 239 isolates, included in 181 ST allelic profiles, did not cluster with any other ST profile and were taken as singletons (18). Only four complexes included five or more STs, seven contained four STs, one had three STs, and each of the remaining 23 complexes included only two STs (see additional information in the supplemental material).
FIG. 2.
Population snapshot of the B. cereus group. Numbers refer to the ST allelic profiles (newly determined or available in the MLST database). Clusters of linked isolates correspond to potential CCs (i.e., STs sharing at least six identical alleles) (18). Primary founders, colored blue except for 142, which is purple (see below), are positioned centrally in the cluster, and subgroup founders are shown in yellow. Newly determined unique isolates are green, while new isolates with a counterpart in the public database are shown in purple. ST profiles sharing at least five identical alleles are grouped according to a common color background. STs present only in a public B. cereus MLST database are shown in orange. Dot sizes are proportional to the number of isolates with the same ST allelic profile.
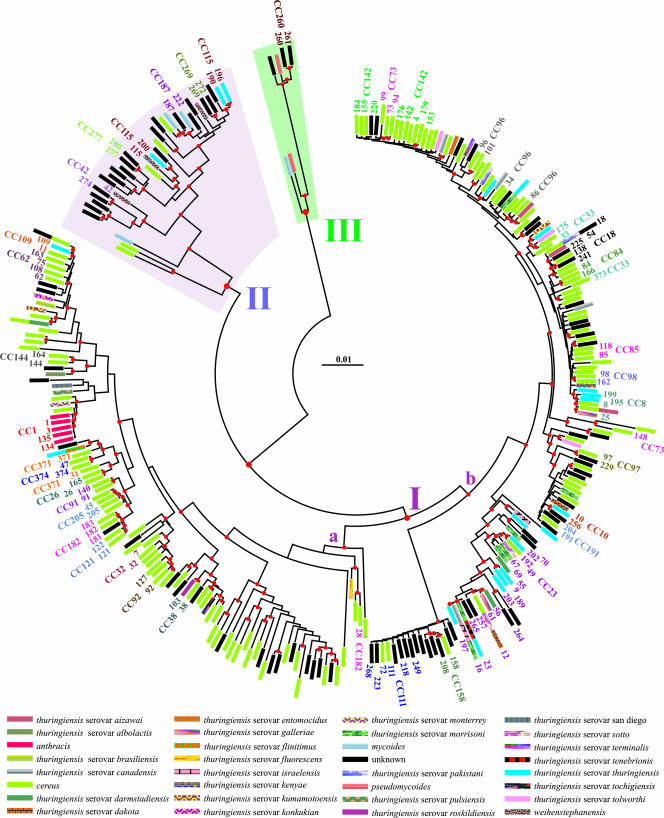
FIG. 3.
Distribution of potential CCs, identified by eBurst analysis, on the NJ tree. The scale bar indicates distance calculated according to the Tamura and Nei evolutionary model. The 35 complexes of ST profiles obtained through eBurst analysis were mapped on the phylogenetic tree previously produced with MEGA program. The species legend listed below the tree is identical to that provided in Fig. 1. Numbers at the tip of the tree terminal branches refer to the corresponding ST profiles labeled according to the scheme provided in the MLST database dedicated to B. cereus (http://pubmlst.org/bcereus/). CCs are labeled according to the eBurst results (see the supplemental material).
The 35 potential CCs were formed by STs distributed throughout the NJ tree (see Fig. 3). The most numerous complex recovered by eBurst (CC23 in Fig. 3 and see also the supplemental material) included 36 isolates representing 19 STs belonging to B. cereus, unknown B. cereus complex species, and 9 different serovars of B. thuringiensis. Within CC23, ST 23 was identified as the primary founder (89% bootstrap support) and ST 197 and ST 56 were predicted to be secondary founders but without statistical support (BT < 49%) (Fig. 2). CC1 proved to be the cluster encompassing most of ST attributed to B. anthracis (with exclusion of ST 2). ST 1 was identified as the founder of complex CC1.
Several CCs identified by eBurst analysis resulted in a paraphyletic (e.g., CC111) or polyphyletic (e.g., CC73 and CC182) in the phylogenetic tree (Fig. 3). This discrepancy between the phylogenetic reconstruction and eBurst output suggests an extensive occurrence of HGT among various isolates (see also paragraph below).
HGT detection.
The main outputs of the HGT analyses performed by using the RDP2 package are summarized in Table 3 and the HGT figure (see the supplemental material). Only HGT events with both major and minor sequences parents identified in the MLST data set were considered. Major and minor sequences parents (43) are defined as the two sequences that produced each recombined ST allelic profile through the HGT process. The occurrence of a potential HGT event was accepted only if validated by at least three distinct methods and sustained by strong statistical support. HGT events with only one potential parent identified were not further investigated even if statistically supported. As a consequence, Table 3 provides a conservative estimate of HGT events and does not fully cover the whole range of HGT events affecting the MLST data set. Nevertheless, this conservative approach revealed at least 25 HGT events, involving 85 STs. Some of the HGTs shared the same major and minor sequence parents and this applied both to HGTs spanning two or more genes (see, for example, references 14 to 17), as well as to HGT restricted to a single gene (15, 21). All seven gene fragments concatenated in MLST profiles were affected by HGT events. In 14 HGTs, event breakpoints were located within a single gene, and the pur locus was particularly affected by this phenomenon (Table 3). In three HGT events, breakpoints were limited to pairs of genes arranged consecutively, but not adjacent to one another, in genomes sequenced thus far (ilvD versus pur, pycA versus tpi). The remaining eight HGT breakpoints involved nonconsecutive genes (glpF versus gmk, ilvD versus pta, and pur versus pta). Each identified HGT spanning more than a single gene could actually be the result of two independent events that were not detectable with currently available algorithms. STs involved in HGT events are scattered all over the tree (see the HGT figure in the supplemental material), and no clear bias was detected that linked peculiar species and/or serovars to HGT occurrences. An analysis of alleles involved in single HGT events did not identify alleles clearly favoring HGT occurrence. For example, the five HGTs (HGTs 3, 4, 5, 6, and 7) restricted to the ilvD gene showed that 11 different ilvD alleles were involved. Most of them were restricted to the single HGT event, with a partial overlapping in HGTs 5 and 6. Five of the newly described food-borne strains (ST 369, ST 371, ST 373, ST 374, and ST 378) were also involved in HGT events. For most STs involved in HGT events the toxin distribution is not known (Table 3). Thus, it was impossible to investigate toxin transmission patterns in relation to HGT occurrence.
TABLE 3.
Detection of horizontal gene transfer events in MLST data seta
| HGT event | ST(s) involved in HGT event | Nb | PPSTc
|
Ad | Breakpoint
|
Gene(s)e | Pf | ||
|---|---|---|---|---|---|---|---|---|---|
| M | m | Start | End | ||||||
| 1 | 82 | 1 | 217 | 274 | 6 | 255 | 876 | glpF-gmk | ≤4.072 × 10−12 |
| 2 | 21 | 1 | 222 | 59 | 5 | 381 | 876 | gmk | ≤1.707 × 10−14 |
| 3 | 207 | 1 | 275 | 112 | 6 | 885 | 1245 | ilvD | ≤4.656 × 10−7 |
| 4 | 125 | 1 | 250 | 127 | 5 | 933 | 1245 | ilvD | ≤8.296 × 10−5 |
| 5 | 271, 277, 279, 280, 283, 284 | 6 | 196 | 208 | 6 | 882 | 1251 | ilvD | ≤9.370 × 10−15 |
| 6 | 273, 282, 285 | 3 | 196 | 208 | 6 | 882 | 1251 | ilvD | ≤1.660 × 10−9 |
| 7 | 124, 253 | 2 | 153 | 102 | 4 | 1101 | 1266 | ilvD | ≤4.179 × 10−6 |
| 8 | 80 | 1 | 59 | 140 | 6 | 879 | 2031 | ilvD-pur | ≤1.890 × 10−13 |
| 9 | 12, 16, 23, 56, 61, 197, 257, 203, 265 | 9 | 204 | 102 | 3 | 885 | 1749 | ilvD-pur | ≤2.125 × 10−3 |
| 10 | 148 | 1 | 17 | 11 | 6 | 879 | 1302 | ilvD-pta | ≤2.613 × 10−17 |
| 11 | 36 | 1 | 272 | 5 | 6 | 885 | 1302 | ilvD-pta | ≤1.203 × 10−12 |
| 12 | 173 | 1 | 76 | 232 | 3 | 1302 | 1674 | pta | ≤8.100 × 10−8 |
| 13 | 38, 78, 103, 123, 132, 369 | 6 | 266 | 84 | 5 | 1531 | 2031 | pta-pur | ≤8.880 × 10−3 |
| 14 | 47, 171, 371, 374 | 4 | 266 | 84 | 5 | 1551 | 2031 | pta-pur | ≤1.843 × 10−2 |
| 15 | 28, 79, 188 | 3 | 266 | 84 | 5 | 1551 | 2031 | pta-pur | ≤1.453 × 10−4 |
| 16 | 6 | 1 | 266 | 84 | 1551 | 2031 | pta-pur | ≤7.535 × 10−3 | |
| 17 | 45, 91, 121, 122, 140, 141, 181, 182, 183, 185, 205 | 11 | 266 | 84 | 5 | 1767 | 1914 | pur | ≤2.304 × 10−5 |
| 18 | 1, 2, 3, 107, 110, 112, 130, 134, 135, 145 | 10 | 62 | 229 | 6 | 1693 | 1890 | pur | ≤1.535 × 10−9 |
| 19 | 5, 7, 32, 90, 92, 120, 127, 169, 172, 180, 211, 219, 234 | 13 | 144 | 373 | 6 | 1710 | 2031 | pur | ≤4.752 × 10−4 |
| 20 | 26, 31, 165 | 3 | 144 | 373 | 6 | 1710 | 2031 | pur | ≤6.820 × 10−8 |
| 21 | 105 | 1 | 164 | 210 | 4 | 1740 | 1911 | pur | ≤1.028 × 10−4 |
| 22 | 230 | 1 | 203 | 207 | 6 | 1687 | 2031 | pur | ≤1.933 × 10−11 |
| 23 | 264 | 1 | 16 | 235 | 5 | 1693 | 2011 | pur | ≤1.640 × 10−13 |
| 24 | 193 | 1 | 213 | 248 | 6 | 2034 | 2391 | pycA | ≤3.809 × 10−12 |
| 25 | 110, 214 | 2 | 106 | 378 | 6 | 2061 | 2697 | pycA-tpi | ≤2.557 × 10−3 |
Underlined values indicate possible misidentifications of breakpoint positions. Boldface values indicate STs for which the toxin distribution is available. Only HGT events detected by at least three methods and having both potential ST parents present in the MLST data set are listed.
N, number of STs subject to HGT.
PPST, potential parent ST; M, major potential parent; m, minor potential parent.
A, number of different algorithms supporting the HGT event.
That is, the gene fragments involved in the HGT event.
The P value was obtained after multiple-comparison correction. Because each HGT event was supported by at least three algorithms, Table 3 lists only the highest P value obtained for the specific HGT (i.e., the least statistically significant). Values listed below the breakpoint's start and end refer to the positions of the MLST data set of the first ST recorded for the specific HGT event.
DISCUSSION
Phylogeny of B. cereus group.
The phylogenetic analysis provided in the present study was based on the largest data set thus far studied for the B. cereus group. Our main evolutionary outputs agree with previous analyses (see, for example, references 28-31, 52, 58, and 60) in identifying three major lineages within the B. cereus complex (Fig. 1). Lineage I is the richest of STs and includes mostly B. cereus and B. thuringiensis serovar thuringiensis strains. Many other B. thuringiensis serovars are included within this cluster and intermixed with B. cereus and B. thuringiensis serovar thuringiensis isolates. This finding is consistent with previous molecular analyses (7, 8, 28, 29, 31, 36, 37, 52, 58, 60).
The majority of STs associated with food-borne strains (58%) were assigned to group Ia, which comprised 34.4% of the total ST profiles. Group Ia contains mostly B. cereus isolates and can be viewed as an expansion of the B. cereus clade 1 defined by Priest et al. (52). The presence of food-borne isolates mainly within group Ia suggests that strains belonging to this group could preferentially grow on some foodstuff matrices. The B. cereus species has long been recognized as a food-borne microbe that plays a key role in food poisoning. The inclusion of food-borne isolates within a group dominated by B. cereus strains is in agreement with microbiological evidence. However, in the present study, a food matrix preference (i.e., egg product, milk products, or other) for different B. cereus isolates was not found, in contrast to a previous report (22). However, the limited isolate sampling suggests the need for further investigation of this phenomenon. Thus, additional studies on a larger number of food-borne isolates are needed to better address this issue. In addition, experiments based on artificial contaminations of food and analysis of growth will also be necessary in order to elucidate the features of different strains characterized through MLST or other genomic approaches.
As a final remark to this paragraph, it must be noted that the broad occurrence of HGT within the B. cereus complex may affect the phylogenetic relationships described in here, as well as in previous analyses (7, 8, 28, 29, 31, 36, 37, 52, 58, 60) (see below for further discussion of this point).
Evolution of toxin genes in B. cereus group.
All toxin genes are contained in group I, which includes most of the strains considered in the present study (Fig. 1). Lineages II and III contain the B. mycoides and B. weihenstephanensis species, which are currently not known as pathogens. Thus, the absence of toxin genes could explain why these species are not isolated from human clinical samples and are not associated with disease. The broad distribution of nhe, hbl, and entF genes makes the possibility that they are widespread or even ubiquitous within lineage I plausible. A narrower distribution characterizes the gene bceT, since it seems restricted mostly to a subgroup of lineage I, with the only exception being represented by ST 160. This latter occurrence could be due to a recent event of HGT (see below), or it could be the result of a generalized loss of the bceT gene in other STs. Current evidence does not help to discriminate between these opposing hypotheses, and further research is needed to settle this point. We used the same set of primers for all sampled strains; thus, the lack of a specific toxin gene should be evidence of the true absence of that toxin in the analyzed strain. However, we cannot exclude the possibility that the regions on which PCR primers were designed underwent some rearrangement, thus preventing us from amplifying the specific toxin gene. Distribution of the em sequence is restricted solely to the ST 26. Location of the em sequence is on a plasmid; thus, its sporadic occurrence appears to be tightly linked to the presence of this plasmid (14, 15). Genes encoding lethal toxins carried by pX01 and pX02 plasmids are currently known only for B. anthracis strains (25, 34, 35). Both phylogeny and eBurst analysis recovered B. anthracis as a monophyletic taxon; thus, it is plausible to suggest that pX01 and pX02 plasmids were acquired just once in this species.
The presence of toxin-encoding genes does not automatically equate with pathogenicity. Regulation of enterotoxin expression is complex, and the current available knowledge is incomplete. Furthermore, the enterotoxin production (12, 47, 62) alone does not fully explain the food poisoning phenomenon that is regulated by other key factors, such as bacterial concentration. Identification of a toxin gene in the bacterial genome indicates only the potential pathogenicity of the strain, while the true poisoning activity may be absent due to the lack of genes regulating toxin expression. All of the food-borne isolates described here are potential causes of food poisoning, since all possess at least one enterotoxin gene. Quite remarkably, isolates with the highest number of enterotoxin genes (ST 142, ST 370, ST 372, and ST 376) are all located in the Ib group.
A possible mechanism of transmission of toxin genes between two bacterial strains was recently identified by Han et al. (25). These researchers found transposable elements in the genomic portion encoding for toxin genes in pathogenic strains of B. cereus and B. thuringiensis, which could be a potential mechanism for gene transmission. Thus, HGT of toxin-encoding genes mediated by transposable elements could be the general mechanism shaping the evolution and distribution of toxin genes within the B. cereus complex. The present analysis supports the view that HGT is a major player in the evolution of B. cereus complex.
Population structure of B. cereus group and HGT.
The population structure of B. cereus cannot be discussed without considering the effect of HGT. This genomic mechanism is pervasive in B. cereus strains. Our analyses identified at least 25 independent events of HGT involving more than one-fourth (28.72%) of all analyzed ST profiles. Our estimate of HGT was very conservative (see above), suggesting that HGT is a very widespread phenomenon in the B. cereus group. This finding was corroborated by several independent methods developed to detect HGT; therefore, we believe that misidentification of HGTs is unlikely. The occurrence of HGT in the B. cereus complex has not been seen consistently in the literature, since some reports cite strong evidence of HGT (28, 37), whereas other analyses showed evidence for clonal behavior (29-31, 52, 58, 60).
In the present study we benefit from an unparalleled number of ST profiles (n = 296) which can be used to check for the occurrence of HGT events. The large size of MLST profiles drastically improved the statistical power of detection by the suite of methods applied here (50, 51). We were able to provide evidence that HGT is a general phenomenon characterizing the evolution of the B. cereus group.
The broad occurrence of HGT is reflected in the marked discrepancies observed between phylogenetic analysis and eBurst population outputs. Several of the potential CCs did not appear monophyletic in the phylogenetic tree, and even the relaxation of the number of identical alleles shared among members of same complex did not modify the pattern of clear differences between phylogeny and eBurst analysis. HGT occurrence, however, acts both on eBurst and phylogenetic outputs. Thus, an integrated approach is necessary to fully investigate the evolutionary history of the B. cereus complex. A better instantaneous representation of the relationships among strains of B. cereus complex is provided by eBurst analysis, which was specifically developed for this task (18). The algorithm has proven to be efficient with other bacterial species exhibiting high levels of recombination (e.g., Campylobacter jejuni) (18). Conversely, when investigation of species relationships is extended throughout their evolutionary history, a phylogenetic approach is preferable (46). Routinely applied phylogenetic algorithms are not able to cope with the HGT occurrence, and thus new algorithms are necessary to correctly represent the partly reticulated and partly dichotomous phylogenetic relationships among B. cereus complex species. HGT may also explain the current difficulty in assigning a single strain to a monophyletic group based on the serovar concept (e.g., see the scatter distribution of B. thuringiensis serovar tolworthi in lineage Ib). Another important effect of HGT is that formally defining subspecies or species within the B. cereus complex becomes a very difficult task. In fact, the use of different names for identifying serovars with a peculiar pathogenic behavior or restricted to a particular host and/or geographic area is, in most cases, not supported by biological evidence, even though it may have a practical utility. More generally, our study provides evidence that the evolution of B. cereus complex is characterized by reticulate evolution with continuous and extensive exchange of genomic portions among different strains.
In conclusion, traditional phylogenetic analysis of MLST sequences might not fully resolve the taxonomy of B. cereus isolates due to the presence of extensive HGT. An eBurst analysis and HGT detection are also necessary to deal with this topic. Prediction of pathogenicity in particular strains is not a trivial task with the currently available evidence, since isolates bearing enterotoxin genes do not belong to a single phylogenetic group.
The development of phylogenetic algorithms that take HGT into account, as well as provide a more accurate identification of HGT events, will greatly improve our ability to understand the evolutionary history of the B. cereus complex. Limits of currently available analytical methods notwithstanding, the MLST approach proved to be a highly reproducible, fast, and accurate method for strain genetic typing. International food trade requires methods suitable for tracking the spread of disease-causing organisms and MLST analysis of B. cereus isolates might become the method of choice for the epidemiology of food-borne diseases produced by this bacterium. Our analysis demonstrated the presence of toxin genes in many strains of the B. cereus complex that are potentially pathogenic. However, further studies are required to characterize the key factors that regulate toxin expression and to determine the different levels of pathogenicity among the various strains.
Supplementary Material
Acknowledgments
We thank three anonymous reviewers for very helpful and constructive criticisms on the manuscript.
Footnotes
Published ahead of print on 14 December 2007.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Agata, N., M. Ohta, Y. Arakawa, and M. Mori. 1995. The bceT gene of Bacillus cereus encodes an enterotoxin protein. Microbiology 141:983-988. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 3.Anonymous. 2004. Microbiology of food and animal feeding stuffs—horizontal method for enumeration of presumptive Bacillus cereus—colony-count technique at 30°C. Document ISO 7932. International Organization for Standardization, Geneva, Switzerland.
- 4.Asano, S. I., Y. Nukumizu, H. Bando, T. Iizuka, and T. Yamamoto. 1997. Cloning of novel enterotoxin genes from Bacillus cereus and Bacillus thuringiensis. Appl. Environ. Microbiol. 63:1054-1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barker, M., B. Thakker, and F. G. Priest. 2005. Multilocus sequence typing reveals that Bacillus cereus strains isolated from clinical infections have distinct phylogenetic origins. FEMS Microbiol. Lett. 245:179-184. [DOI] [PubMed] [Google Scholar]
- 6.Beecher, D. J., J. L. Schoeni, and A. C. Wong. 1995. Enterotoxic activity of hemolysin BL from Bacillus cereus. Infect. Immun. 63:4423-4428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carlson, C. R., D. A. Caugant, and A.-B. Kolstø. 1994. Genotypic diversity among Bacillus cereus and Bacillus thuringiensis strains. Appl. Environ. Microbiol. 60:1719-1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carlson, C. R., T. Johansen, and A.-B. Kolstø. 1996. The chromosome map of Bacillus thuringiensis subsp. canadensis HD224 is highly similar to that of Bacillus cereus type strain ATCC 14579. FEMS Microbiol. Lett. 141:163-167. [DOI] [PubMed] [Google Scholar]
- 9.Damgaard, P. H., P. E. Granum, J. Bresciani, M. V. Torregrossa, J. Eilenberg, and L. Vealentino. 1997. Characterization of Bacillus thuringiensis isolated from infections in burn wounds. FEMS Immunol. Med. Microbiol. 18:47-53. [DOI] [PubMed] [Google Scholar]
- 10.Dietrich, R., C. Fella, S. Strich, and E. Märtlbauer. 1999. Production and characterization of monoclonal antibodies against the hemolysin BL enterotoxin complex produced by Bacillus cereus. Appl. Environ. Microbiol. 65:4470-4474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dietrich, R., M. Moravek, C. Burk, P. E. Granum, and E. Märtlbauer. 2005. Production and characterization of antibodies against each of the three subunits of the Bacillus cereus nonhemolytic enterotoxin complex. Appl. Environ. Microbiol. 71:8214-8220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Duport, C., A. Zigha, E. Rosenfeld, and P. Schmitt. 2006. Control of enterotoxin gene expression in Bacillus cereus F4430/73 involves the redox-sensitive ResDE signal transduction system. J. Bacteriol. 188:6640-6651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ehling-Schulz, M., M. Fricker, and S. Scherer. 2004. Bacillus cereus the causative agent of an emetic type of food-borne illness. Mol. Nutr. Food Res. 48:479-487. [DOI] [PubMed] [Google Scholar]
- 14.Ehling-Schulz, M., M. Fricker, and S. Scherer. 2004. Identification of emetic toxin producing Bacillus cereus strains by a novel molecular assay. FEMS Microbiol. Lett. 232:189-195. [DOI] [PubMed] [Google Scholar]
- 15.Ehling-Schulz, M., B. Svensson, M. H. Guinebretiere, T. Lindback, M. Andersson, A. Schulz, M. Fricker, A. Christiansson, P. E. Granum, E. Martlbauer, C. Nguyen-The, M. Salkinoja-Salonen, and S. Scherer. 2005. Emetic toxin formation of Bacillus cereus is restricted to a single evolutionary lineage of closely related strains. Microbiology 151:183-197. [DOI] [PubMed] [Google Scholar]
- 16.Ehling-Schulz, M., N. Vukov, A. Schulz, R. Shaheen, M. Andersson, E. Märtlbauer, and S. Scherer. 2005. Identification and partial characterization of the nonribosomal peptide synthetase gene responsible for cereulide production in emetic Bacillus cereus. Appl. Environ. Microbiol. 71:105-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ehling-Schulz, M., M. Fricker, P. Grallert, H. Riek, M. Wagner, and S. Scherer. 2006. Cereulide synthetase gene cluster from emetic Bacillus cereus: structure and location on a mega virulence plasmid related to Bacillus anthracis toxin plasmid pXO1. BMC Microbiol. 6:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Feil, E. J., B. C. Li, D. M. Aanensen, W. P. Hanage, and B. G. Spratt. 2004. eBURST: inferring patterns of evolutionary descent among clusters of related bacterial genotypes from multilocus sequence typing data. J. Bacteriol. 186:1518-1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Felsenstein, J. 2004. Inferring phylogenies. Sinauer, Sunderland, MA.
- 20.From, C., R. Pukall, P. Schumann, V. Hormazábal, and P. E. Granum. 2005. Toxin-producing ability among Bacillus spp. outside the Bacillus cereus group. Appl. Environ. Microbiol. 71:1178-1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gibbs, M. J., J. S. Armstrong, and A. J. Gibbs. 2000. Sister-Scanning: a Monte Carlo procedure for assessing signals in recombinant sequences. Bioinformatics 16:573-582. [DOI] [PubMed] [Google Scholar]
- 22.Granum, P. E., and T. Lund. 1997. Bacillus cereus and its food poisoning toxins. FEMS Microbiol. Lett. 157:223-228. [DOI] [PubMed] [Google Scholar]
- 23.Granum, P. E., K. O'Sullivan, and T. Lund. 1999. The sequence of the non-hemolytic enterotoxin operon from Bacillus cereus. FEMS Microbiol. Lett. 177:225-229. [DOI] [PubMed] [Google Scholar]
- 24.Guinebretiere, M. H., V. Broussolle, and C. Nguyen-The. 2002. Enterotoxigenic profiles of food-poisoning and food-borne Bacillus cereus strains. J. Clin. Microbiol. 40:3053-3056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Han, C. S., G. Xie, J. F. Challacombe, M. R. Altherr, S. S. Bhotika, D. Bruce, C. S. Campbell, M. L. Campbell, J. Chen, O. Chertkov, C. Cleland, M. Dimitrijevic, N. A. Doggett, J. J. Fawcett, T. Glavina, L. A. Goodwin, K. K. Hill, P. Hitchcock, P. J. Jackson, P. Keim, A. R. Kewalramani, J. Longmire, S. Lucas, S. Malfatti, K. McMurry, L. J. Meincke, M. Misra, B. L. Moseman, M. Mundt, A. C. Munk, R. T. Okinaka, B. Parson-Quintana, L. P. Reilly, P. Richardson, D. L. Robinson, E. Rubin, E. Saunders, R. Tapia, J. G. Tesmer, N. Thayer, L. S. Thompson, H. Tice, L. O. Ticknor, P. L. Wills, T. S. Brettin, and P. Gilna. 2006. Pathogenomic sequence analysis of Bacillus cereus and Bacillus thuringiensis isolates closely related to Bacillus anthracis. J. Bacteriol. 188:3382-3390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hansen, B. M., and N. B. Hendriksen. 2001. Detection of enterotoxic Bacillus cereus and Bacillus thuringiensis strains by PCR analysis. Appl. Environ. Microbiol. 67:185-189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hansen, B. M., P. E. Høiby, G. B. Jensen, and N. B. Hendriksen. 2003. The Bacillus cereus bceT enterotoxin sequence reappraised. FEMS Microbiol. Lett. 223:21-24. [DOI] [PubMed] [Google Scholar]
- 28.Helgason, E., D. A. Caugant, M.-M. Lecadet, Y. Chen, J. Mahillon, A. Lovgren, I. Hegna, K. Kvaloy, and A.-B. Kolstø. 1998. Genetic diversity of Bacillus cereus/B. thuringiensis isolates from natural sources. Curr. Microbiol. 37:80-87. [DOI] [PubMed] [Google Scholar]
- 29.Helgason, E., O. A. Økstad, D. A. Caugant, H. A. Johansen, A. Fouet, M. Mock, I. Hegna, and A.-B. Kolstø. 2000. Bacillus anthracis, Bacillus cereus, and Bacillus thuringiensis—one species on the basis of genetic evidence. Appl. Environ. Microbiol. 66:2627-2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Helgason, E., H. A. Caugant, I. Olsen, and A. B. Kolstø. 2000. Genetic structure of population of Bacillus cereus and Bacillus thuringiensis isolates associated with periodontitis and other human infections. J. Clin. Microbiol. 38:1615-1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Helgason, E., N. J. Tourasse, R. Meisal, D. A. Caugant, and A.-B. Kolstø. 2004. Multilocus sequence typing for bacteria of the Bacillus cereus group. Appl. Environ. Microbiol. 70:191-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jackson, S. G., R. B. Goodbrand, R. Ahmed, and S. Kasatiya. 1995. Bacillus cereus and Bacillus thuringiensis isolated in gastroenteritis outbreak investigations. Lett. Appl. Microbiol. 21:103-105. [DOI] [PubMed] [Google Scholar]
- 33.Jolley, K. A., M.-S. Chan, and M. C. J. Maiden. 2004. mlstdbNet: distributed multi-locus sequence typing (MLST) databases. BMC Bioinformatics. 5:86-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Keim, P., A. Kalif, J. M. Schupp, K. K. Hill, S. E. Travis, K. Richmond, D. M. Adair, M. E. Hugh-Jones, C. R. Kuske, and P. Jackson. 1997. Molecular evolution and diversity in Bacillus anthracis as detected by amplified fragment length polymorphism markers. J. Bacteriol. 179:818-824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Keim, P., L. B. Price, A. M. Klevytska, K. L. Smith, J. M. Schupp, R. Okinaka, P. J. Jackson, and M. E. Hugh-Jones. 2000. Multiple-locus variable-number tandem repeat analysis reveals genetic relationships within Bacillus anthracis. Appl. Environ. Microbiol. 182:2928-2936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim, W., Y.-P. Hong, J.-H. Yoo, W.-B. Lee, C.-S. Choi, and S.-I. Chung. 2001. Genetic relationships of Bacillus anthracis and closely related species based on variable-number tandem repeat analysis and BOX-PCR genomic fingerprinting. FEMS Microbiol. Lett. 207:21-27. [DOI] [PubMed] [Google Scholar]
- 37.Ko, K. S., J. W. Kim, J. M. Kim, W. Kim, S. I. Chung, I. J. Kim, and Y. H. Kook. 2004. Population structure of the Bacillus cereus group as determined by sequence analysis of six housekeeping genes and the plcR gene. Infect. Immun. 72:5253-5261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kumar, S., K. Tamura, and M. Nei. 2004. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinform. 5:150-163. [DOI] [PubMed] [Google Scholar]
- 39.Lecadet, M.-M., E. Frachon, V. C. Dumanoir, H. Ripouteau, S. Hamon, P. Laurent, and I. Thiéry. 1999. Updating the H-antigen classification of Bacillus thuringiensis. J. Appl. Microbiol. 86:660-672. [DOI] [PubMed] [Google Scholar]
- 40.Lechner, S., R. Mayr, K. P. Francis, B. M. Pruss, T. Kaplan, E. Wiessner-Gunkel, G. S. Stewart, and S. Scherer. 1998. Bacillus weihenstephanensis sp. nov. is a new psychrotolerant species of the Bacillus cereus group. Int. J. Syst. Bacteriol. 48:1373-1382. [DOI] [PubMed] [Google Scholar]
- 41.Lund, T., M. L. De Buyser, and P. E. Granum. 2000. A new cytotoxin from Bacillus cereus that may cause necrotic enteritis. Mol. Microbiol. 38:254-261. [DOI] [PubMed] [Google Scholar]
- 42.Martin, D., and E. Rybicki. 2000. RDP: detection of recombination amongst aligned sequences. Bioinformatics 16:562-563. [DOI] [PubMed] [Google Scholar]
- 43.Martin, D. P., C. Williamson, and D. Posada. 2005. RDP2: recombination detection and analysis from sequence alignments. Bioinformatics 21:260-262. [DOI] [PubMed] [Google Scholar]
- 44.Maynard-Smith, J. 1992. Analyzing the mosaic structure of genes. J. Mol. Evol. 34:126-129. [DOI] [PubMed] [Google Scholar]
- 45.Mock, M., and A. Fouet. 2001. Anthrax. Annu. Rev. Microbiol. 55:647-671. [DOI] [PubMed] [Google Scholar]
- 46.Nei, M., and S. Kumar. 2000. Molecular evolution and phylogenetics. Oxford University Press, New York, NY.
- 47.Ouhib, O., T. Clavel, and P. Schmitt. 2006. The production of Bacillus cereus enterotoxins is influenced by carbohydrate and growth rate. Curr. Microbiol. 53:222-226. [DOI] [PubMed] [Google Scholar]
- 48.Padidam, M., S. Sawyer, and C. M. Fauquet. 1999. Possible emergence of new geminiviruses by frequent recombination. Virology 265:218-225. [DOI] [PubMed] [Google Scholar]
- 49.Phelps, R. J., and J. L. McKillip. 2002. Enterotoxin production in natural isolates of Bacillaceae outside the Bacillus cereus group. Appl. Environ. Microbiol. 68:3147-3151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Posada, D., and K. A. Crandall. 2001. Evaluation of methods for detecting recombination from DNA sequences: computer simulations. Proc. Natl. Acad. Sci. USA 98:13757-13762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Posada, D. 2002. Evaluation of methods for detecting recombination from DNA sequences: empirical data. Mol. Biol. Evol. 19:708-717. [DOI] [PubMed] [Google Scholar]
- 52.Priest, F. G., M. Barker, L. W. Baillie, E. C. Holmes, and M. C. Maiden. 2004. Population structure and evolution of the Bacillus cereus group. J. Bacteriol. 186:7959-7970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Prüss, B. M., R. Dietrich, B. Nibler, E. Martlbauer, and S. Scherer. 1999. The hemolytic enterotoxin HBL is broadly distributed among species of the Bacillus cereus group. Appl. Environ. Microbiol. 65:5436-5442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Reyes-Ramirez, A., and J. E. Ibarra. 2005. Fingerprinting of Bacillus thuringiensis type strains and isolates by using Bacillus cereus group-specific repetitive extragenic palindromic sequence-based PCR analysis. Appl. Environ. Microbiol. 71:1346-1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 56.Salminen, M. O., J. K. Carr, D. S. Burke, and F. E. McCutchan. 1995. Identification of breakpoints in intergenotypic recombinants of HIV type 1 by bootscanning. AIDS Res. Hum. Retrovir. 11:1423-1425. [DOI] [PubMed] [Google Scholar]
- 57.Schoeni, J. L., and A. C. Wong. 2005. Bacillus cereus food poisoning and its toxins. J. Food Prot. 68:636-648. [DOI] [PubMed] [Google Scholar]
- 58.Sorokin, A., B. Candelon, K. Guilloux, N. Galleron, N. Wackerow-Kouzova, S. D. Ehrlich, D. Bourguet, and V. Sanchis. 2006. Multiple-locus sequence typing analysis of Bacillus cereus and Bacillus thuringiensis reveals separate clustering and a distinct population structure of psychrotrophic strains. Appl. Environ. Microbiol. 72:1569-1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Spratt, B. G., W. P. Hanage, B. Li, D. M. Aanensen, and E. J. Feil. 2004. Displaying the relatedness among isolates of bacterial species: the eBURST approach. FEMS Microbiol. Lett. 241:129-134. [DOI] [PubMed] [Google Scholar]
- 60.Ticknor, L. O., A. B. Kolstø, K. K. Hill, P. Keim, M. T. Laker, M. Tonks, and P. J. Jackson. 2001. Fluorescent amplified fragment length polymorphism analysis of Norwegian Bacillus cereus and Bacillus thuringiensis soil isolates. Appl. Environ. Microbiol. 67:4863-4873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yang, I. C., D. Y. Shih, T. P. Huang, Y. P. Huang, J. Y. Wang, and T. M. Pan. 2005. Establishment of a novel multiplex PCR assay and detection of toxigenic strains of the species in the Bacillus cereus group. J. Food Prot. 68:2123-2130. [DOI] [PubMed] [Google Scholar]
- 62.Zigha, A., E. Rosenfeld, P. Schmitt, and C. Duport. 2007. The redox regulator Fnr is required for fermentative growth and enterotoxin synthesis in Bacillus cereus F4430/73. J. Bacteriol. 189:2813-2824. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.