Abstract
The phylogenetic diversity and species richness of ammonia-oxidizing archaea (AOA) and bacteria (AOB) were examined with aquarium biofiltration systems. Species richness, deduced from rarefaction analysis, and diversity indices indicated that the phylogenetic diversity and species richness of AOA are greater than those of AOB; the diversity of AOA and of AOB is minimized in cold-water aquaria. This finding implies that temperature is a key factor influencing the population structure and diversity of AOA and AOB in aquarium biofiltration systems.
In aquaria and recirculating aquaculture systems, the accumulation of ammonia, an end product of protein metabolism in aquatic life, must be prevented because of its toxicity to fish and other aquatic organisms (6, 24, 27). Nitrifying bacteria are thought to be largely responsible for the oxidization of ammonia to nitrate via nitrite in these systems. Ammonia oxidation, the rate-limiting process in nitrification, is driven by chemolithotrophic ammonia-oxidizing bacteria (AOB) belonging to the Betaproteobacteria and Gammaproteobacteria classes. Recently, it was discovered that the autotrophic oxidation of ammonia is not restricted to the domain Bacteria. Könneke and colleagues (13) isolated an ammonia-oxidizing archaeon, Nitrosopumilus maritimus, from a tropical marine fish tank, representing the first cultivated isolate of the ubiquitous marine group 1 of the Crenarchaeota phylum. These organisms contain putative genes for all three subunits (amoA, amoB, and amoC) of ammonia monooxygenase, the key enzyme for ammonia oxidation and a specific biomarker for the molecular detection of ammonia oxidizers. To date, it has been reported that nitrification in aquarium biofiltration is performed mainly by organisms of the Nitrosomonas and Nitrosospira genera belonging to the Betaproteobacteria class (3), while the relevance of ammonia-oxidizing archaea in aquarium biofiltration remains unknown. It is crucial to determine which ammonia oxidizers are mainly responsible for biofiltration systems in aquaria, as seeding of nitrifiers is commonly used; seeding greatly accelerates the nitrification process in aquaria and recirculating aquaculture systems (10). This study provides a genetic clarification of the ubiquity, diversity, and abundance of ammonia-oxidizing archaea (AOA) and AOB in aquarium biofiltration systems.
Sample collection.
Water and surficial gravity filtration materials (sand) were collected from three seawater fish tanks of a public aquarium (Aqua World Ibaraki Prefectural Oarai Aquarium, Ibaraki, Japan) in September 2004 (Table 1). In brief, the first fish tank, named “coastal fish tank,” keeps temperate coastal fish at 19.9°C. A second fish tank, called “cold-water tank,” keeps deep-sea fish at a lower temperature (5.5°C). A third fish tank, named “ocean sunfish tank,” keeps large ocean sunfish (Mola mola) at 19.0°C. Physicochemical data for the water were monitored using a water quality checker (U-10; Horiba Instruments Ltd., Kyoto, Japan). Nutrient samples were filtered through cellulose acetate syringe filters (pore size, 0.8 μm) and analyzed by using an autoanalyzer (TRAACS-800; Bran+Luebbe GmbH, Tokyo, Japan). Ammonium ions were detected only in the cold-water tank (0.01 mg liter−1); no NO2−-N was detected (Table 1). Nearly all inorganic nitrogen was in the NO3−-N form. These results confirmed that the biofiltration systems were operated adequately in all fish tanks. Direct microscopic counting was performed as described previously (25). The mean total bacterial counts ± standard deviations for the tank water were (1.0 ± 0.10) × 105 (n = 3) cells ml−1 in the coastal fish tank, (2.6 ± 0.48) × 105 cells ml−1 in the cold-water tank, and (2.6 ± 0.75) × 105 cells ml−1 in the ocean sunfish tank, indicating that the cell numbers in the aquaria were lower than those in coastal waters, where cell counts generally reach 106 cells ml−1.
TABLE 1.
Characteristics of the aquaria analyzed in this studya
| Tank name | Temp (°C) | pH | DOb (mg liter−1) | Salinity (psuc) | NH4+-N (mg liter−1) | NO3−-N (mg liter−1) | Vol (m3) | Organism reared in the fish tank |
|---|---|---|---|---|---|---|---|---|
| Coastal fish tank | 19.9 | 8.2 | 8.4 | 34 | 0.00 | 1.25 | 4.5 | Crimson seabream (Evynnis japonica) |
| Red gurnard (Chelidonichthys spinosus) | ||||||||
| Knobsnout parrotfish (Scarus ovifrons) | ||||||||
| Cold-water tank | 5.5 | 8.4 | 12.4 | 33 | 0.01 | 1.53 | 4.5 | Sailfin poacher (Podothecus sachi) |
| Pink snailfish (Careproctus rastrinus) | ||||||||
| Queen crab (Chionoecetes opilio) | ||||||||
| Ocean sunfish tank | 19.0 | 8.4 | 8.3 | 34 | 0.00 | 0.30 | 289 | Ocean sunfish (Mola mola) |
NO2−-N was not detected in water from any of the fish tanks.
DO, dissolved oxygen.
psu, practical salinity units.
DNA extraction, PCR, cloning, and DNA sequencing.
DNA was extracted from approximately 0.6 g of wet sand, using bead-beating disruption (FastPrep 120; Qbiogene, Inc., Carlsbad, CA) and an ISOIL Beads for Beating kit (Nippon Gene Co. Ltd., Tokyo, Japan) according to the manufacturer's protocol. The primers Arch-amoAF and Arch-amoAR were used for PCR, as described previously (8), to amplify the archaeal amoA gene fragments (ca. 635 bp). The bacterial amoA gene fragments (ca. 491 bp) were amplified using the primers amoA-1F and amoA-2R, as described previously (25). The amplified archaeal and bacterial amoA genes were cloned into a TOPO TA cloning kit (Invitrogen Corp., San Diego, CA). Nucleotide sequences were determined for both strands by cycle sequencing reaction, with a sequencing kit (BigDye Terminator cycle version 3.1; Applied Biosystems, Foster City, CA) and a capillary DNA sequencer (ABI 3100; Applied Biosystems).
Phylogenetic analyses.
The archaeal and bacterial amoA sequences that were determined were aligned manually and compared with those available from databases, using nucleotide-nucleotide BLAST software. Alignment editing and phylogenetic analyses (neighbor-joining and maximum parsimony methods) based on the amino acid sequences were implemented using MEGA version 3.1 software (14).
Diversity indices and statistical analyses.
To compare the relative amoA gene-based operational taxonomic unit (OTU) richness among fish tanks, rarefaction analysis was performed using Analytic Rarefaction version 1.3 software (http://www.uga.edu/∼strata/software/Software.html). Even though appropriate sequence identity thresholds have yet to be carefully considered for defining OTUs for various functional genes (28), these thresholds might vary depending on the evolutionary history of the targeted gene and the population size. Although thresholds ranges of 2 to 5% were considered to be adequate for grouping closely related amoA phylotypes, the OTUs were defined as groups of sequences that differed by one amino acid residue because relatively restricted phylotypes were detected in the samples. The Shannon-Weiner index (H′) and the Simpson index (D) were calculated as the diversity indices. The Shannon-Weiner index, which is the negative sum of each OTU's proportional abundance multiplied by the log of its proportional abundance, is a measure of the amount of information (entropy) in the system. The Simpson index assigns a strong weighting to dominant OTUs. Value D gives the probability that two clones chosen at random will be from the same OTU (11). Coverage was calculated as described previously (9). An Arlequin population genetics software package was used to compare the levels of genetic variation within and between fish tank samples (5). The mean pairwise genetic distances (π) within (πw) and between (πB) sampled populations were calculated from sequence data to estimate the levels of genetic variation within and between fish tanks (17). Dissimilarity indices (Fst) were calculated for pairs of fish tanks (16, 21).
Quantitative real-time PCR assays.
All quantitative real-time PCR (Q-PCR) assays targeting the archaeal and bacterial amoA genes were carried out with three replications per sample on a capillary system of LightCycler (Roche Applied Science, Indianapolis, IN). We designed a primer set, Arch-amoA-79F (5′- ATTAAYGCAGGWGAYTAYAT-3′) and Arch-amoA-479R (5′-TATGGTGGYAAYGTDGGTC-3′), based on the archaeal amoA clone sequences obtained in this study because previously published archaeal Q-PCR primers contained significant numbers of mismatches for our clone sequences (15, 29). The bacterial amoA gene Q-PCR detection was carried out using the previously described primer set amoA-1F and amoA-2R (25). The PCR mixture consisted of 10 μl of 2× SYBR Premix Ex Taq (Perfect Real Time; TaKaRa), 0.6 μM final concentration of the archaeal amoA primer or 0.2 μM of the bacterial amoA primer, and 20 ng of DNA template in a final volume of 20 μl. The real-time PCR steps were as follows: an initial denaturation at 95.0°C for 20 s; 40 cycles of 95.0°C for 7 s, 50.0°C for 20 s (archaea), or 55.0°C for 20 s (bacteria); and 72.0°C for 20 s, followed by a melting curve analysis (65°C to 95°C with a heating rate of 0.1°C/s). The standard curves used for quantification were from plasmid DNA prepared from OA-SA8-117 for archaeal amoA and OA-SA10-28 for bacterial amoA. Data were analyzed with the second-derivative-maximum method using Light Cycler software (version 3.5.3; Roche Applied Science).
Phylogenetic diversity of ammonia-oxidizing archaea.
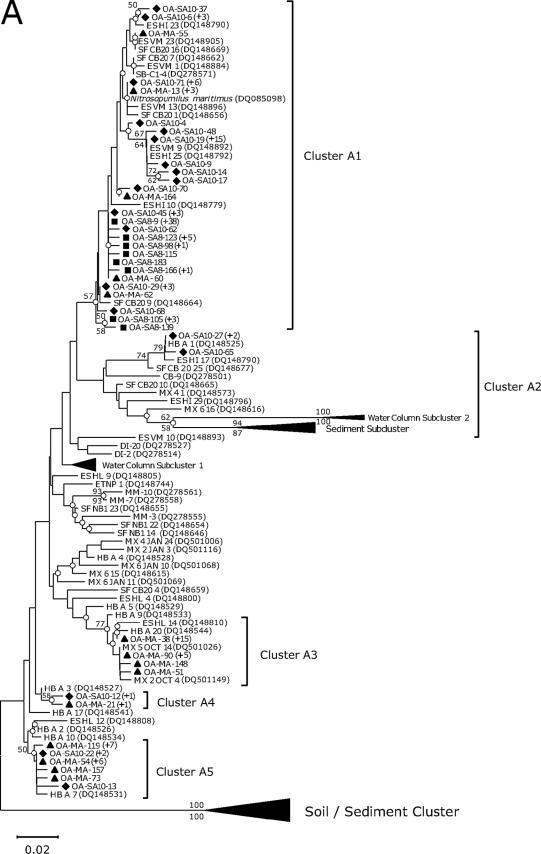
Three archaeal amoA clone libraries were prepared, and a total of 157 archaeal amoA sequences were analyzed. As a result, 41 OTUs were recovered based on one amino acid residue cutoff (Table 2). The number of OTUs recovered from individual libraries ranged from 8 OTUs from the cold-water tank to 19 OTUs from the coastal fish tank. Phylogenetic analyses revealed that all sequences fell into five clusters (Fig. 1A). The archaeal amoA sequences belonging to cluster A1 were closely related to that of Nitrosopumilus maritimus, which shared 93 to 100% amino acid sequence similarity. N. maritimus-related sequences consisted of 66% OTU and were detected in all fish tanks examined. Our result suggested that N. maritimus is ubiquitously distributed in aquarium biofiltration systems and might play an important role in aquarium biofiltration systems. Cluster A2 consisted solely of coastal fish tank clones and is grouped with the water column subcluster 2 and the sediment subcluster, which were proposed by Francis and colleagues (1, 8) (Fig. 1A). Cluster A3 consisted mainly of ocean sunfish tank clones, and a majority of clones in this aquarium fell into this cluster. Clusters A4 and A5 consisted of clones from the coastal fish and ocean sunfish tanks and did not contain clones from the cold-water tank. All archaeal amoA sequences analyzed in this study showed low similarity to water column subcluster 1 (83 to 89% amino acid sequence similarity) and 86 to 91% with the sediment subcluster. In contrast, none of the 157 clones recovered from the three fish tanks fell into the soil/sediment cluster, which shared 79 to 89% amino acid sequence similarity and contained archaeal amoA clone sequences recovered from wastewater treatment plants (19). Overall, archaeal sequences obtained from the biofiltration systems resembled environmental clones recovered from marine sediments of the San Francisco Central Bay, the Elkhorn Slough estuary Vierra Marsh, and the Hunting Beach aquifer, which shared 92 to 100% amino acid sequence similarity, and water columns of the Black Sea, Monterey Bay, and the Eastern Tropical North Pacific, which shared 93 to 98% amino acid sequence similarity (8).
TABLE 2.
Properties of the distribution of phylotypes in clone libraries from aquarium biofiltration sands
| Tank name | Archaeal amoA
|
Bacterial amoA
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| na | No. of OTUs | H′ | D | πw | n | No. of OTUs | H′ | D | πw | |
| Coastal fish tank | 54 | 19 | 2.36 | 0.14 | 49.50 | 62 | 14 | 1.86 | 0.27 | 13.67 |
| Cold-water tank | 52 | 8 | 1.16 | 0.48 | 16.36 | 80 | 5 | 0.43 | 0.81 | 4.09 |
| Ocean sunfish tank | 51 | 14 | 2.07 | 0.17 | 67.56 | 70 | 14 | 1.69 | 0.28 | 24.69 |
n, number of clones sequenced.
FIG. 1.
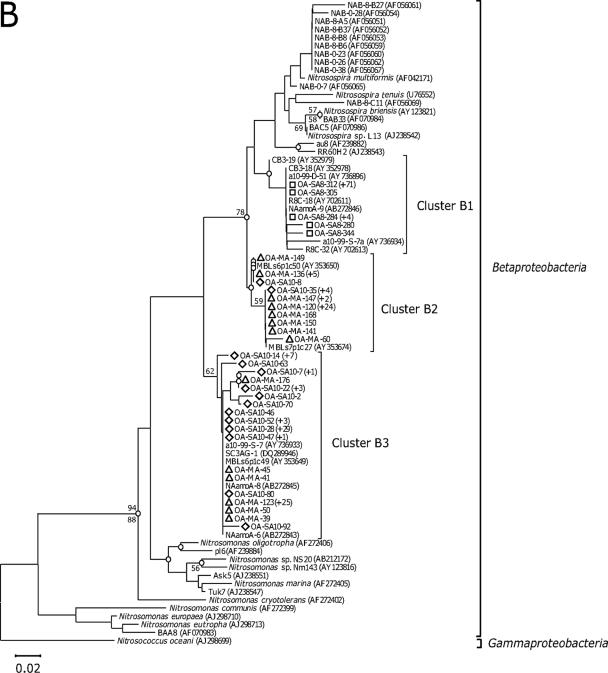
Neighbor-joining trees show phylogenetic relationships among archaeal amoA (A) and bacterial amoA (B) sequences recovered from aquarium biofiltration systems. The clone sequences reported in this study are coded by symbol according to the fish tank sampled (i.e., diamond, coastal fish tank; square, cold-water tank; and triangle, ocean sunfish tank). The number of sequences showing identical amino acid sequences is indicated by values in parentheses. The branch nodes supported by two phylogenetic analyses (neighbor joining [NJ] and maximum parsimony [MP]) are indicated as open circles. Numbers at branch nodes are bootstrap values obtained from NJ (above the branch) and MP analyses (below the branch); only value nodes supported by bootstrap values greater than 50% and those which are important for defining the phylogenetic major groups are indicated. For outgroups, a soil/sediment cluster, proposed by Beman and Francis (1), and Nitrosococcus oceani are used, respectively, for the AOA and AOB trees. The bar represents 0.01 estimated substitution per site. The archaeal amoA gene, which corresponds to 198 amino acids, and the bacterial amoA gene, which corresponds to 150 amino acids, were used for phylogenetic analyses. A distance matrix was inferred by using a pairwise difference method to construct a phylogenetic tree based on the amino acid alignments. NJ trees based on the Jones-Taylor-Thornton matrix as an amino acid replacement model and MP trees were constructed based on alignments of amino acid sequences. Distance-based and parsimony-based bootstrap analyses were conducted and used to estimate the reliability of phylogenetic reconstructions with 500 replicates for both archaeal and bacterial amoA sequences.
Phylogenetic diversity of ammonia-oxidizing bacteria.
Three bacterial amoA clone libraries were prepared, and a total of 212 bacterial amoA sequences were analyzed. In all, 33 OTUs were recovered, based on one amino acid residue cutoff (Table 2). The number of OTUs recovered from individual libraries ranged from 5 OTUs from the cold-water tank to 14 OTUs each from the coastal fish tank and the ocean sunfish tank. Phylogenetic analyses revealed that all sequences fell into three clusters (Fig. 1B). The bacterial amoA sequences affiliated with cluster B1, sharing 88 to 95% amino acid similarity, contained all sequences recovered from the cold-water tank. These 80 sequences related to the Nitrosospira lineage were most similar to those of environmental clones (>98% amino acid sequence similarity) reported from various marine environments, such as the Chesapeake Bay (7), the Monterey Bay (18), Plum Island Sound (2), and the deep-sea sediments from the Pacific Ocean (23). Cluster B2 was dominated by sequences recovered from the ocean sunfish tank and included several coastal fish tank sequences, whereas cluster B3 was represented mainly by coastal fish tank clones, several ocean sunfish tank clones, and environmental clones (12). Interestingly, these two clusters contained no previously known isolates of AOB species and were not affiliated with either the Nitrosomonas lineage or the Nitrosospira lineage. In previous studies, these sequences have been treated as Nitrosospira-like sequences (2, 7) or described as sequences from a sister clade to Nitrosospira (12), but little is known about the ecophysiology of these ammonia oxidizers. It is noteworthy that all Nitrosospira-like sequences identified as environmental clones were reported from marine environments, suggesting that these Nitrosospira-like AOB are adapted to marine environments and might be potentially important in aquarium biofiltration systems and permeable marine sediments, which are similar to aquarium biofiltration systems and are impacted by advectional pore water flow (12).
Archaeal and bacterial amoA diversity/richness.
Interestingly, more archaeal and bacterial amoA sequences recovered from the cold-water tank fell into restricted clusters than those recovered from the coastal fish tank and the ocean sunfish tank (Fig. 1). Rarefaction analysis indicated that the lowest archaeal and bacterial amoA diversity/richness indices were from the cold-water tank (see Fig. S1 in the supplemental material). Coverage values for archaeal and bacterial amoA sequences recovered from the cold-water tank were higher than those of the other tanks, suggesting that the presence of low OTU numbers is expected in clone libraries (see Fig. S1 in the supplemental material). The diversity indices of AOA and AOB from the cold-water tank were estimated using the Shannon-Weiner index (H′) and the Simpson index (D). The result showed that the diversity indices of AOA and AOB from the cold-water tank were the smallest among the fish tanks examined (Table 2). The mean number of pairwise differences (π) within a sample of DNA sequences (πw) is an estimate of the amount of genetic variation within the sample. The archaeal and bacterial amoA sequences from the ocean sunfish tank contained the highest πw values, and the cold-water tank yielded the lowest πw value (Table 2). The π value between tanks (πB) is a meaningful estimate of the genetic variation between two tanks. Regarding genetic variation among the archaeal amoA genes between tanks, the πB value for the coastal fish tank and the cold-water tank pair indicates that the communities in these two tanks are the most similar, i.e., they exhibit the least genetic variation between any two tanks (see Table S1 in the supplemental material). For genetic variation among the bacterial amoA genes between the tanks, the πB value for the coastal fish tank and ocean sunfish tank pair indicates that the communities of these two tanks are the most similar (see Table S2 in the supplemental material). Dissimilarity (Fst) indices are useful for comparing the average genetic variation within a group with the genetic variation between groups. This analysis evaluates the degree to which randomly generated groups of the sequences partition variation compared with the observed sequence distribution. The Fst values of all pairwise comparisons indicated that all populations differed significantly (Fst > 0.05; P < 0.05). Based on phylogenetic and population diversity data, it is apparent that the archaeal and bacterial amoA sequences recovered from the cold-water tank have the lowest diversity levels among the aquaria examined.
Comparison of archaeal and bacterial diversity indices.
Recently, the ecological importance of AOA has been acknowledged in various environments in terms of abundance (15). However, a comparison of AOA and AOB diversity has been reported only from coastal environments (1). Rarefaction analysis and diversity indices clearly showed that the AOA diversity indices in aquarium biofiltration systems were greater than the AOB diversity indices in all the aquaria examined (Table 2 and see Fig. S1 in the supplemental material).
Temperature as a potential factor influencing diversity of AOA and AOB.
The factors that influence the diversity of AOA and AOB in aquarium biofiltration systems remain unknown. Salinity has been considered a key factor influencing the distribution and diversity of AOB in coastal marine environments (2, 4). Ammonium concentration and the aquatic organisms reared in fish tank might influence the community structures of AOA and AOB because ammonium is a primary energy source for ammonia oxidizers. As reported previously, the archaeal amoA library from Huntington Beach, which is characterized by a hydrologic connection with groundwater and which is highly enriched in NH4+, showed high genetic diversity (8). In the present study, salinity, pH, and ammonium concentration, which have been reported as factors important to the determination of the community structures of AOB (2, 20), were almost identical among the aquaria (Table 1). Species richness deduced from polyphasic analyses clearly indicates that the diversity indices of both AOA and AOB are low in the cold-water aquarium, which implies that temperature might be a key factor influencing the population structure and diversity of AOA and AOB in aquaria biofiltration systems.
Quantification of AOA and AOB.
Quantification of AOA and AOB was performed by using real-time PCR assays. The copy number of AOA and AOB ranged from (2.0 ± 0.14) × 103 (n = 3) to (3.0 ± 0.10) × 103 per ng of DNA and from (2.2 ± 0.17) × 103 to (3.8 ± 0.04) × 103 per ng of DNA, respectively (Table 3). The gene copy number was converted to cell numbers by using a formula based on the total bacterial counts and the total amount of extracted DNA (26). The population sizes of AOA and AOB were between (5.3 ± 0.11) × 106 and (7.4 ± 0.25) × 106 cells per g of dry sand and (2.5 ± 0.02) × 106 and (3.0 ± 0.23) × 106 cells per g of dry sand, respectively. The estimated numbers of AOA and AOB corresponded to between 2.8% ± 0.06% and 3.9% ± 0.13% and to between 1.3% ± 0.01% and 1.6% ± 0.12% of total cell counts, respectively. The archaeal amoA/bacterial amoA ratio ranged from 1.8 to 3.0, suggesting that AOA numbers are slightly larger than the AOB numbers in biological filtration systems.
TABLE 3.
Quantitative analyses of AOA and AOB by real-time PCRa
| Tank name | AOA
|
AOB
|
Ratio of AOA to AOB cells | ||||
|---|---|---|---|---|---|---|---|
| No. of copies (±SD) (103) ng−1 DNA | No. of cells (±SD) (106) g−1 of dry sand | % of AOA cells g−1 of sand (±SD) | No. of copies (±SD) (103) ng−1 DNA | No. of cells (±SD) (106) g−1 of dry sand | % of AOB cells g−1 of sand (±SD) | ||
| Coastal fish tank | 3.0 (±0.10) | 7.4 (±0.25) | 3.9 (±0.13) | 2.5 (±0.03) | 2.5 (±0.02) | 1.3 (±0.01) | 3.0 |
| Cold-water tank | 2.8 (±0.06) | 5.3 (±0.11) | 2.8 (±0.06) | 3.8 (±0.04) | 2.9 (±0.03) | 1.5 (±0.02) | 1.8 |
| Ocean sunfish tank | 2.0 (±0.14) | 6.8 (±0.46) | 3.6 (±0.24) | 2.2 (±0.17) | 3.0 (±0.23) | 1.6 (±0.12) | 2.3 |
Real time PCR assays were carried out with the newly designed primers (Arch-amoA-79F and Arch-amoA-479R), which target clusters A1, A3, A4, and A5, as shown in Fig. 1A, but not cluster A2 and the soil/sediment cluster proposed by Beman et al. (1). AOA and AOB data are means ± standard deviations (SD) for n = 3. The numbers of cells g−1 of dry sand assume a single amoA copy per AOA genome and 2.5 amoA copies per AOB genome (15). Percentages of AOA and AOB cell counts from aquarium biofiltration sand were derived from the total cell counts, assumed to be 1.9 × 108 cells g−1 of sand (22).
Application of new insights into fortification of seed ammonia oxidizers.
Although we have not yet examined the activity of AOA, the results of this study suggest that the diversity indices of AOA and AOB can be high and can vary with the temperature in aquarium biofiltration systems. We also found that the abundance levels of AOA in seawater aquarium biofiltration systems are slightly larger than those of AOB. Our findings will strengthen development strategies for seeding ammonia oxidizers into aquarium biofiltration systems.
Nucleotide sequence accession numbers.
The archaeal and bacterial amoA sequences reported in this study have been deposited in the GenBank database under accession numbers AB373235 to AB373391 (AOA) and AB373392 to AB373603 (AOB).
Supplementary Material
Acknowledgments
We thank M. Kosaka (Director of Aqua World Ibaraki Prefectural Oarai Aquarium) for providing the sampling opportunity. We also thank Sumiko Kawabata, Masahiro Takahashi, Toshifumi Osaka, and Hiromi Watanabe for their technical assistance. We also appreciate Martin Könneke, Oldenburg University, Germany, for critical reading of the manuscript. Helpful comments and suggestions by three anonymous reviewers are gratefully acknowledged.
This research was partially supported by a Grant-in-Aid for Scientists (no. 18201003) and a Grant-in-Aid for Young Scientists (no. 17688009) from the Ministry of Education, Culture, Sports, Science and Technology of Japan to H.U.
Footnotes
Published ahead of print on 7 December 2007.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Beman, J. M., and C. A. Francis. 2006. Diversity of ammonia-oxidizing archaea and bacteria in the sediments of a hypernutrified subtropical estuary: Bahía del Tóbari, Mexico. Appl. Environ. Microbiol. 72:7767-7777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bernhard, A. E., T. Donn, A. E. Giblin, and D. A. Stahl. 2005. Loss of diversity of ammonia-oxidizing bacteria correlates with increasing salinity in an estuary system. Environ. Microbiol. 7:1289-1297. [DOI] [PubMed] [Google Scholar]
- 3.Burrell, P. C., C. M. Phalen, and T. A. Hovanec. 2001. Identification of bacteria responsible for ammonia oxidation in freshwater aquaria. Appl. Environ. Microbiol. 67:5791-5800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.de Bie, M. J. M., A. Speksnijder, G. A. Kowalchuk, T. Schuurman, G. Zwart, J. R. Stephen, O. E. Diekmann, and H. J. Laanbroek. 2001. Shifts in the dominant populations of ammonia-oxidizing β-subclass Proteobacteria along the eutrophic Schelde estuary. Aquat. Microb. Ecol. 23:225-236. [Google Scholar]
- 5.Excoffier, L., G. Laval, and S. Schneider. 2005. Arlequin ver. 3.0: an integrated software package for population genetics data analysis. Evol. Bioinform. 1:47-50. [PMC free article] [PubMed] [Google Scholar]
- 6.Frances, J., B. F. Nowak, and G. L. Allan. 2000. Effects of ammonia on juvenile silver perch (Bidyanus bidyanus). Aquaculture 183:95-103. [Google Scholar]
- 7.Francis, C. A., G. D. O'Mullan, and B. B. Ward. 2003. Diversity of ammonia monooxygenase (amoA) genes across environmental gradients in Chesapeake Bay sediments. Geobiology 1:129-140. [Google Scholar]
- 8.Francis, C. A., K. J. Roberts, J. M. Beman, A. E. Santoro, and B. B. Oakley. 2005. Ubiquity and diversity of ammonia-oxidizing archaea in water columns and sediments of the ocean. Proc. Natl. Acad. Sci. USA 102:14683-14688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Good, I. J. 1953. The population frequencies of species and the estimation of population parameters. Biometrika 40:237-264. [Google Scholar]
- 10.Gross, A., A. Nemirovsky, D. Zilberg, A. Khaimov, A. Brenner, E. Snir, Z. Ronen, and A. Nejidat. 2003. Soil nitrifying enrichments as biofilter starters in intensive recirculating saline water aquaculture. Aquaculture 223:51-62. [Google Scholar]
- 11.Hill, T. C. J., K. A. Walsh, J. A. Harris, and B. F. Moffett. 2003. Using ecological diversity measures with bacterial communities. FEMS Microbiol. Ecol. 43:1-11. [DOI] [PubMed] [Google Scholar]
- 12.Hunter, E. M., H. J. Mills, and J. E. Kostka. 2006. Microbial community diversity associated with carbon and nitrogen cycling in permeable shelf sediments. Appl. Environ. Microbiol. 72:5689-5701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Könneke, M., A. E. Bernhard, J. R. de la Torre, C. B. Walker, J. B. Waterbury, and D. A. Stahl. 2005. Isolation of an autotrophic ammonia-oxidizing marine archaeon. Nature 437:543-546. [DOI] [PubMed] [Google Scholar]
- 14.Kumar, S., K. Tamura, and M. Nei. 2004. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinform. 5:150-163. [DOI] [PubMed] [Google Scholar]
- 15.Leininger, S., T. Urich, M. Schloter, L. Schwark, J. Qi, G. W. Nicol, J. I. Prosser, S. C. Schuster, and C. Schleper. 2006. Archaea predominate among ammonia-oxidizing prokaryotes in soils. Nature 442:806-809. [DOI] [PubMed] [Google Scholar]
- 16.Martin, A. P. 2002. Phylogenetic approaches for describing and comparing the diversity of microbial communities. Appl. Environ. Microbiol. 68:3673-3682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nei, M., and W. H. Li. 1979. Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc. Natl. Acad. Sci. USA 76:5269-5273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.O'Mullan, G. D., and B. B. Ward. 2005. Relationship of temporal and spatial variabilities of ammonia-oxidizing bacteria to nitrification rates in Monterey Bay, California. Appl. Environ. Microbiol. 71:697-705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Park, H. D., G. F. Wells, H. Bae, C. S. Criddle, and C. A. Francis. 2006. Occurrence of ammonia-oxidizing archaea in wastewater treatment plant bioreactors. Appl. Environ. Microbiol. 72:5643-5647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Prinčič, A., I. Mahne, F. Megusar, E. A. Paul, and J. M. Tiedje. 1998. Effects of pH and oxygen and ammonium concentrations on the community structure of nitrifying bacteria from wastewater. Appl. Environ. Microbiol. 64:3584-3590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reynolds, J., B. S. Weir, and C. C. Cockerham. 1983. Estimation of the coancestry coefficient: basis for a short-term genetic distance. Genetics 105:767-779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sugita, H., H. Nakamura, and T. Shimada. 2005. Microbial communities associated with filter materials in recirculating aquaculture systems of freshwater fish. Aquaculture 243:403-409. [Google Scholar]
- 23.Tamegai, H., R. Aoki, S. Arakawa, and C. Kato. 2007. Molecular analysis of the nitrogen cycle in deep-sea microorganisms from the Nankai Trough: genes for nitrification and denitrification from deep-sea environmental DNA. Extremophiles 11:269-275. [DOI] [PubMed] [Google Scholar]
- 24.Thurston, R. V., R. C. Russo, and G. A. Vinogradov. 1981. Ammonia toxicity to fishes. Effect of pH on the toxicity of the un-ionized ammonia species. Environ. Sci. Technol. 15:837-840. [Google Scholar]
- 25.Urakawa, H., S. Kurata, T. Fujiwara, D. Kuroiwa, H. Maki, S. Kawabata, T. Hiwatari, H. Ando, T. Kawai, M. Watanabe, and K. Kohata. 2006. Characterization and quantification of ammonia-oxidizing bacteria in eutrophic coastal marine sediments using polyphasic molecular approaches and immunofluorescence staining. Environ. Microbiol. 8:787-803. [DOI] [PubMed] [Google Scholar]
- 26.Urakawa, H., H. Maki, S. Kawabata, T. Fujiwara, H. Ando, T. Kawai, T. Hiwatari, K. Kohata, and M. Watanabe. 2006. Abundance and population structure of ammonia-oxidizing bacteria that inhabit canal sediments receiving effluents from municipal wastewater treatment plants. Appl. Environ. Microbiol. 72:6845-6850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wajsbrot, N., A. Gasith, A. Diamant, and D. M. Popper. 1993. Chronic toxicity of ammonia to juvenile gilthead seabream Sparus aurata and related histopathological effects. J. Fish Biol. 42:321-328. [Google Scholar]
- 28.Ward, B. B. 2002. How many species of prokaryotes are there? Proc. Natl. Acad. Sci. USA 99:10234-10236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wuchter, C., B. Abbas, M. J. L. Coolen, L. Herfort, J. van Bleijswijk, P. Timmers, M. Strous, E. Teira, G. J. Herndl, J. J. Middelburg, S. Schouten, and J. S. S. Damste. 2006. Archaeal nitrification in the ocean. Proc. Natl. Acad. Sci. USA 103:12317-12322. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.