Abstract
Saccharomyces cerevisiae strains from different regions of Minas Gerais, Brazil, were isolated and characterized aiming at the selection of starter yeasts to be used in the production of cachaça, the Brazilian sugar cane spirit. The methodology established took into account the screening for biochemical traits desirable in a yeast cachaça producer, such as no H2S production, high tolerance to ethanol and high temperatures, high fermentative capacity, and the abilities to flocculate and to produce mycocins. Furthermore, the yeasts were exposed to drugs such as 5,5′,5"-trifluor-d,l-leucine and cerulenin to isolate those that potentially overproduce higher alcohols and esters. The utilization of a random amplified polymorphic DNA-PCR method with primers based on intron splicing sites flanking regions of the COX1 gene, as well as microsatellite analysis, was not sufficient to achieve good differentiation among selected strains. In contrast, karyotype analysis allowed a clear distinction among all strains. Two selected strains were experimentally evaluated as cachaça producers. The results suggest that the selection of strains as fermentation starters requires the combined use of biochemical and molecular criteria to ensure the isolation and identification of strains with potential characteristics to produce cachaça with a higher quality standard.
Cachaça (pronounced “kha-sha-ssa”), the sugar cane spirit, is the most popular distilled beverage produced in Brazil. The annual production reaches 1.3 billion liters, with 15% being produced in more than 8,500 distilleries in the state of Minas Gerais.
Traditional cachaça production relies on a spontaneous fermentative process that is mediated by the microbiota present in the cane juice wort and on the surface of equipments used in the productive process. It has been already demonstrated that in such systems there occurs a succession of yeasts, with Saccharomyces cerevisiae being the predominant species. Cachaça quality depends on the ecology of the microbial populations during an initial spontaneous fermentation (18, 29, 31, 32, 39). The fermentative process occurs through a continuously open fermentative process which is completed within 24 h and generally takes place from May to November, corresponding to the sugar cane harvesting period.
Considering the conditions of production usually found in the cachaça distilleries, fermenting yeast populations have to face different types of stress (osmotic, high temperature, and high ethanol concentration). Besides, they might also present some characteristics such as a good fermentative power, no H2S production, killer activity, flocculation ability, and production of flavoring compounds. Taking all of these factors into account, we have developed a strategy to select yeast strains with appropriated characteristics to produce cachaça with potentially higher-quality standards (52).
Parallel to the selection and development of S. cerevisiae strains toward ethanolic fermentations, molecular methods were developed and validated to study the evolution of yeast flora in spontaneous but also in inoculated fermentations. Chromosomal karyotyping by pulsed-field gel electrophoresis is a complex and time-consuming process but is very efficient for the delimitation of S. cerevisiae strains since it makes it possible to distinguish strains at both the intra- and the interspecies levels. Numerous other methods of typing based on DNA polymorphism allow differentiating closely related yeast strains (8, 36, 43, 50). Restriction fragment length polymorphism (RFLP) analysis of mitochondrial DNA (mtDNA) is faster and easier than other methods (23, 31). Digestion of mtDNA with restriction enzymes such as HinfI or RsaI is associated with a high rate of polymorphism and has been used to study the authenticity of commercial wine yeast strains (14, 40, 41). A PCR method based on variations found on the mitochondrial COX1 (a gene coding for cytochrome oxidase) intron number and position has been validated to distinguish S. cerevisiae strains allowing researchers to monitor the evolution of wine fermentations conducted by commercial active dry yeast (27). Moreover, in many other studies, RAPD [random(ly) amplified polymorphic DNA]-PCR with different primers such as EL1 and LA1 has been successfully used to discriminate between wine yeast strains (2) and for differentiation of Saccharomyces species isolated in Brazil (18). In the last few years, fingerprinting of microsatellite or simple sequence repeat loci, short (1 to 10 nucleotides) DNA tandem repeats dispersed throughout the genome and with a high degree of variability, has proven to be useful for distinguishing S. cerevisiae strains (16, 20, 34, 40, 45). These loci exhibit a substantial level of polymorphism and have been used in humans for paternity tests and forensic medicine but also for the demonstration of population structures among indigenous S. cerevisiae strains (24, 42, 43).
We describe here the isolation and characterization of S. cerevisiae strains from cachaça and the use of two of these strains in cachaça production. The strains were isolated from local producers and characterized by growth in high-alcohol, high-sugar environments and in the presence of 5,5′,5"-trifluor-d,l-leucine (TFL) and cerulenin to detect overproducers of flavor compounds. We also used molecular methods to evaluate the polymorphisms of yeast strains in the fermentative process. Our results demonstrate that some methodologies based on DNA polymorphism are insufficient to evaluate the diversity of S. cerevisiae population during the cachaça production. The utilization of specific biochemical tests is necessary in order to permit a more precise characterization of the dynamics of the selected strains during the fermentative process. Only by combining biochemical and molecular methods were we able to select strains that showed a suitable profile to be used as starters. The cachaça produced with these strains was also evaluated, and the results demonstrate that the quality of the final product is better than that obtained with a commercial strain also used to produce cachaça.
MATERIALS AND METHODS
Sample collection and yeast isolation.
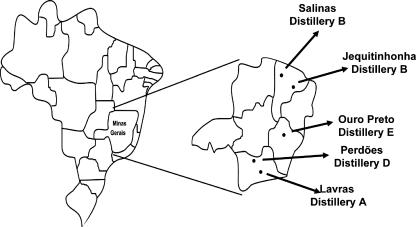
The yeast strains described here were isolated from cachaça distilleries in 2001 (Ouro Preto) and 2003 (Ouro Preto, Lavras, Perdìoes, Jequitinhonha, and Salinas) localized in different subregions of Minas Gerais, Brazil, with differences in temperature, soil composition, and sugar cane variety used to produce cachaça (Fig. 1).
FIG. 1.
Geographical location of the five distilleries in Minas Gerais state (Brazil).
Samples were collected from fermentation vats in previously sterilized 250-ml flasks. They were kept and transported in an ice-cold water bath and processed as fast as possible. Triplicates of decimal dilutions (1:10, 1:100, and 1:1,000) in sterilized water were inoculated on YP medium containing peptone (2% [wt/vol]), yeast extract (1% [wt/vol]), and agar (2% [wt/vol]) supplemented with sucrose (20% [wt/vol]) and chloramphenicol (0.1% [wt/vol]) or with sucrose (20% [wt/vol]), ethanol (8% [wt/vol]), and chloramphenicol (0.1% [wt/vol]). Plates were incubated at 30 or 37°C for 3 days. These special conditions (high sugar concentration and higher temperatures) were chosen because cachaça yeast strains are normally subjected to similar conditions during the fermentative process (18 to 22% sucrose, vats without temperature control, and a high ethanol concentration).
The isolates were first classified on the basis of morphological characteristics and then tested for their ability to grow at higher temperatures (37°C) in the presence of 8% (wt/vol) ethanol, conditions similar to the stressing environmental conditions that occur during sugar cane fermentation. Isolated colonies were replicated on YP medium containing sucrose (20% [wt/vol]), ethanol (8% [wt/vol]), and chloramphenicol (0.1% [wt/vol]). Plates were then incubated at 37°C for 3 days.
To perform the taxonomic classification of the isolated strains, all colonies were subjected to biochemical tests (based on the utilization of carbon and nitrogen sources) as previously described (23, 49).
Physiological characteristics of the Saccharomyces strains.
The ability to produce H2S was verified by inoculating the strains on bismuth sulfite agar medium (Difco Laboratories), followed by incubation at 30°C for 3 days. H2S-producing colonies showed a brown or black color (22).
Flocculating strains were selected by their growth in liquid medium. Cells from each colony were inoculated in tubes containing 4 ml of YP medium with glucose (2% [wt/vol]) and grown in a gyratory shaker (New Brunswick, model G25) at 200 rpm at 30°C until the absorbance at 600 nm reached a value of 1. Flocculation was observed as described previously (51).
In order to evaluate the capacity of the isolated strains to produce killer toxins, each colony obtained from growth in solid YPD medium was suspended in 0.9% (wt/vol) NaCl solution. Approximately 5 μl of yeast cellular suspension was spotted onto YEPD agar plates containing 0.003% methylene blue (pH 4.2), previously inoculated with 100 μl of saline suspensions of Candida glabrata NCYC 388 or S. cerevisiae NCYC 1006 as sensitive reference strains. Plates were then incubated at 25°C for 7 days. Cachaça yeast strains that produced killer toxins were identified by the formation of a blue dark halo around the spotted strains (31).
Selected yeast strains were further subjected to ethanol tolerance tests, and their growth at higher temperatures was also evaluated. Selected colonies were replicated to YP medium supplemented with glucose (2% [wt/vol]) and different ethanol concentrations (10, 15, or 20% [wt/vol]), followed by incubation at 30°C for 3 or 4 days. Ethanol-tolerant cells were then replicated onto plates containing YP medium with glucose (2% [wt/vol]) and ethanol (15% [wt/vol]), followed by incubation at different temperatures (30, 37, and 42°C).
Determination of invertase activity.
Measurement of specific invertase activity was performed as described earlier (11, 17), with slight modifications, since the assay was carried out at pH 5.1 and 37°C. The yeast strains were grown on YP medium containing raffinose as the carbon source, because the derepression of the invertase encoding SUC2 gene is usually higher in the presence of this sugar than in that of other carbon sources. One unit of invertase activity was defined as the amount of enzyme able to hydrolyze 1 μmol of sucrose per min per mg of protein. The protein concentration was determined by using a classical method (28).
Isolation of TFL- and/or cerulenin-resistant strains.
Selected yeast strains were transferred to plates containing minimal medium (SD) supplemented with glucose (2% [wt/vol]) in combination with TFL (1 mM) (1) or cerulenin (25 μM) (21), followed by incubation at 30°C for 3 days. Colonies that developed on the plates containing the inhibitors were considered resistant strains.
DNA extraction.
Strains were grown in 10 ml of YP medium supplemented with glucose (2% [wt/vol]) at 28°C for 12 h at 200 rpm on an orbital shaker. Yeast cells were then centrifuged, washed with distilled water, resuspended in a 400 μl of extraction buffer (1 M sorbitol, 100 mM sodium citrate, 60 mM EDTA [pH 7.0]) containing 0.3 mg of lyticase and 8 μl of β-mercaptoethanol/ml, followed by incubation for 3 h at 37°C. The same volume of lysis buffer (2% sodium dodecyl sulfate in 50 mM Tris-10 mM EDTA [pH 8.0]) was added, and the mixture was then gently shaken and incubated at room temperature for 10 min. After the addition of 200 μl of 5 M NaCl, the suspension was maintained in ice for 2 h. The pellet was harvested by centrifugation at 13,000 rpm for 10 min and then suspended in 200 μl of TE, and the DNA was deproteinated with a phenol-chloroform-isoamyl alcohol mixture (25:24:1). DNA from the aqueous layer was precipitated with ethanol (2 volumes), collected by centrifugation (13,000 rpm, 15 min), washed in ice-cold 70% (vol/vol) ethanol, and dissolved in 60 μl of water. Enriched mtDNA preparations were obtained essentially as previously described (36). For mtDNA analysis, total DNA isolation was carried out as previously described (32) with some minor modifications (40).
Amplification of the ribosomal DNA internal transcribed spacer region and RFLP analyses.
The primers used to amplify the ribosomal DNA internal transcribed spacer region were ITS1 (GTAGGTGAACCTGCGG) and ITS4 (TCCGCTTATTGATATGC) (19, 53). Aliquots (10 μl) of PCR products were digested with 5 U of restriction enzyme (CfoI, HaeIII, and HinfI) in a 20-μl reaction volume. The resulting fragments were analyzed by polyacrylamide gel electrophoresis, followed by silver staining for improved fragment visualization. The restriction patterns obtained from the ITS1-ITS4 region were compared to the expected fragment lengths of the corresponding sequences from the Saccharomyces Genome Database (http://genome-www2.stanford.edu).
For mtDNA RFLP analysis we performed an overnight digestion reaction containing 60 to 120 μg of mtDNA, isolated as described previously (35, 36). The mtDNA restriction fragments were separated on a 1.5% agarose gel containing ethidium bromide and photographed.
Chromosomal polymorphisms.
The cachaça strains, Saccharomyces paradoxus and Lachancea kluyveri, and the commercial Saccharomyces boulardii strain (an S. cerevisiae strain was used as a probiotic) were selected for a comparative electrophoretic karyotype analysis. The preparation of intact chromosomal DNA containing plugs and their separation by transverse alternating field electrophoresis were performed as previously described (8, 40).
Microsatellite amplification.
The amplification of six trinucleotide microsatellite loci as molecular markers for S. cerevisiae occurred in two multiplex reactions containing primer pairs for the loci ScAAT1, ScAAT6, and ScAAT4 or the loci ScAAT2, ScAAT3, and ScAAT5. The forward primer of each pair was labeled with fluorescent dyes (FAM, HEX, or TET; MWG Biotech, Germany). The PCR products were amplified and analyzed in an ABI Prism 310 DNA sequencer (Applied Biosystems, Foster City, CA) and analyzed by using the corresponding Genescan software, as previously described (33, 40).
PCR amplification for RAPD-PCR and COX1-PCR analysis.
For RAPD-PCR assays, the following primers were used: EI1 (CTGGCTTGGTGTATGT) and LA1 (GCGATCGGTGTACTAAC) complementary to the intron splice site sequences (2). The amplification reaction was performed in a 50-μl volume containing 20 pmol of each primer, approximately 300 ng of DNA template, a 0.25 mM concentration of each deoxynucleoside triphosphate, 1.5 mM MgCl2, and 0.5 U of Taq polymerase. The reactions were run as previously described (15).
For COX1-PCR assays, the primers 3L (GCTTTAATTGGWGGWTTTGG), 3R (ATTGTCATACCATTTGTYCTYAT), 4L (GAAGTAGCAGGWGGWGGWGA), and 5R (GTTAGCTAAGGCWACWCCWGT) were used (27). PCR was performed in a 50-μl volume containing 50 pmol of each primer, approximately 300 ng of DNA as a template, 0.25 mM concentrations of each deoxynucleoside triphosphate, 1.5 mM MgCl2, and 0.5 U of Taq polymerase. Initial denaturation (3 min at 94°C) was followed by 35 cycles of denaturation (94°C for 1 min), annealing (51°C for 1 min), and extension (72°C for 2 min), with a final 5-min extension at 72°C. Amplification products were resolved on polyacrylamide gels (6% [vol/vol]). The gels were stained with silver nitrate and scanned.
Fermentation conditions.
Cachaça was produced by laboratory-scale fermentations (15 liters; room temperature, without agitation), using two selected strains (A4 and E70) that were previously characterized by molecular and biochemical methods, and a commercial strain (Maurivin; Australian Wine Research Institute, Adelaide, Australia) that is sometimes used as a starter for cachaça production. Sugar cane (variety RB855156) was freshly harvested from the experimental agricultural unit of the Universidade Federal de Viçosa, MG, Brazil, and subjected to mechanical juice extraction, followed by immediate dilution to 15% (wt/vol) sucrose, measured by refractometry. Strains were propagated from the slants in sugar cane juice by increasing culture volumes until the biomass necessary to obtain an initial cellular density of 12 × 106 cells/ml in each of the 15-liter vessels was achieved. Sterilized sugar cane juice (121°C, 15 min) was used only for the first propagation phase in order to guarantee propagation of the inoculated strains without the eventually occurring indigenous flora originating from sugar cane juice. Thereafter, similar conditions of industrial fermentation were maintained, without previous sterilization of the grape juice in the 15-liter vessels. At 8-h intervals fermenting cane juice samples were collected and subsequently analyzed for methanol, acetaldehyde, and volatile acidity by using the official methods adopted by the Ministry of Agriculture of Brazil. For the acidity level, the samples were distilled in an analytical distiller (Gibertini Vade 3) before analysis in order to eliminate density interferences and to evaluate only volatile acidity.
In order to evaluate the impact of the yeast strains over higher alcohol concentrations, the procedure generally used by distilleries in the cachaça production has been adopted for the distillation of cachaça produced in the laboratory. Immediately after the end of fermentation, fermented cane juice from each fermentation vessel was subjected to distillation in a copper pot still. The total isoamyl alcohol content was evaluated by gas chromatography, and the concentration was calculated from a standard curve, obtained in the same methodological conditions with 0 to 300 mg of isoamyl alcohols.
Implantation of the inoculated yeast strains throughout the fermentative process was monitored by plating diluted sugar cane from different stages on YPD medium. After incubation (2 days, 30°C), 30 colonies were randomly withdrawn and genotyped (mtDNA RFLP).
Sensory evaluation of cachaça samples.
The sensorial quality of the cachaça produced experimentally with the three different yeast strains was evaluated for overall impressions in a trial with 19 trained or consuming panelists using a hedonic scale of nine points (from grade 1 [extremely disliked] to grade 9 [extremely pleased]) (44).
Reproducibility of results.
All experiments were performed at least three times and showed consistent results. Representative results are presented.
RESULTS
Isolation and physiological characteristics of cachaça yeast strains.
From the fermenting sugar cane juice in each of the five distilleries mentioned in Fig. 1, approximately 1,000 yeast isolates were obtained and preliminarily identified based on morphological characteristics. According to standard methods for identification of the yeast species S. cerevisiae (23, 49), the number of initially isolated strains was slightly reduced. For convenience, we decided to continue with only part of the isolated S. cerevisiae strains, using 500 colonies to perform subsequent tests.
The yeast isolates were tested for various characteristics, taking into account the particularities related to the cachaça production process. The 500 isolates were tested for the capacity to produce H2S, an undesirable compound associated with an off-flavor and unpleasant taste, that must be absent from fermented beverages (37). From the initial set of isolates, H2S-producing strains were excluded, and the remaining yeasts were tested for cellular growth at different ethanol concentrations (10, 15, and 20% [wt/vol]) at 37°C. Combining both selection criteria (i.e., no H2S production and high tolerance to temperature and ethanol), we chose an average number of 140 isolates for each distillery.
These isolates were further screened for TFL and cerulenin resistance. As suggested by the data in Table 1, there is a clear variation in the percentage of strains that present resistance to such drugs. The isolated strains are more sensitive to TFL (27 to 59% of resistance) than to cerulenin (81 to 96% of resistance); moreover, 26 to 53% of the strains were resistant to both compounds. Double-resistant strains (36 from distillery A, 40 from distillery B, 70 from distillery C, and 45 from distillery D) were tested for their potential fermentative capacity by measuring invertase activity since sucrose is the principal sugar found in sugar cane juice (13, 31). A total of 21 isolates (5 from distillery A, 5 from distillery B, 6 from distillery C, and 5 from distillery D [data not shown]) presented an invertase activity higher than 10,000 U, which were selected and tested for their capacity to flocculate. Except for distillery C, where no flocculent yeast was found, one flocculent isolate from each distillery was selected for further tests. From the remaining isolates, we selected four (one from each distillery) that were able to produce killer toxins. To perform this analysis, the sensitive reference yeast strains Candida glabrata NCYC 388 and S. cerevisiae NCYC 1006 were used (data not shown).
TABLE 1.
Resistance of S. cerevisiae strains isolated from different distilleries in Minas Gerais state to TFL and cerulenin
| Distillery | No. of tested strains | Drug(s) | No. of resistant strains | % of resistant strains |
|---|---|---|---|---|
| A | 140 | TFL | 38 | 27 |
| Cerulenin | 133 | 95 | ||
| TFL and cerulenin | 36 | 26 | ||
| B | 140 | TFL | 41 | 29 |
| Cerulenin | 129 | 92 | ||
| TFL and cerulenin | 40 | 28 | ||
| C | 140 | TFL | 85 | 58 |
| Cerulenin | 135 | 96 | ||
| TFL and cerulenin | 70 | 53 | ||
| D | 140 | TFL | 67 | 47 |
| Cerulenin | 114 | 81 | ||
| TFL and cerulenin | 45 | 37 | ||
| Total strains resistant to TFL and cerulenin | 115 | 20 |
Molecular genetic polymorphism characterization of cachaça yeast strains.
Restriction pattern analysis of the internal transcriber spacer sequences (25, 53) showed that all four strains belonged to the species S. cerevisiae (data not shown). We also included the laboratory strain W303 and an L. kluyveri strain as a reference.
To further characterize the four selected strains and to verify the level of genetic polymorphism, different strategies were used: analysis of chromosomal patterns, mtDNA RFLP, RAPD-PCR with the primers EI1 and LA1, analysis of six microsatellite loci (ScAAT1 to ScAAT6), and COX1-PCR analysis. For all tests, we included two strains isolated before from distillery E (E22 and E70) that differ from each other in that E70 is resistant to TFL and cerulenin and E22 is sensitive to both compounds (52). Additional Saccharomyces strains were included as controls: S. cerevisiae (genetic background W303), a commercially used probiotic S. boulardii, and S. paradoxus, as well as an L. kluyveri strain.
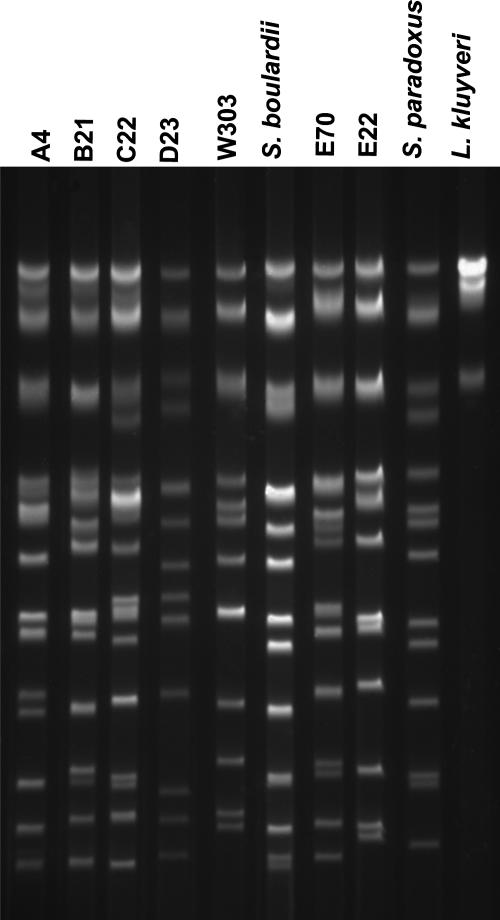
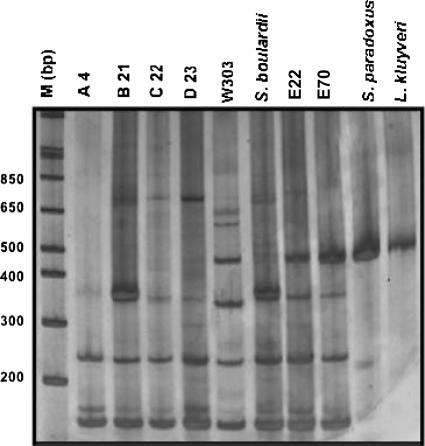
The cachaça strains (A4, B21, C22, D23, E22, and E70) isolated from different distilleries showed six clearly distinctive electrophoretic karyotype patterns (Fig. 2). Strains E22 and E70, which were isolated from the same distillery and display distinct behavior regarding drug resistance (see above), could be distinguished based on their chromosomal patterns.
FIG. 2.
Electrophoretic karyotype patterns of S. cerevisiae strains isolated from five distilleries.
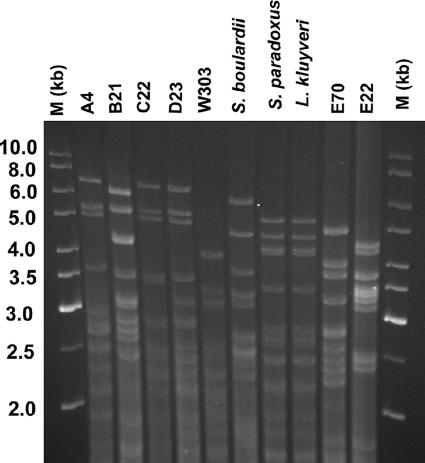
Although the mtDNA RFLP analysis (Fig. 3) showed a very high level of polymorphism, an identical pattern was found for strains E22 and E70. Strains C22 and D23 have very similar patterns and can be distinguished by a band of ∼3.0 kb that is only present in strain D23.
FIG. 3.
mtDNA RFLP analysis of S. cerevisiae strains isolated from five distilleries.
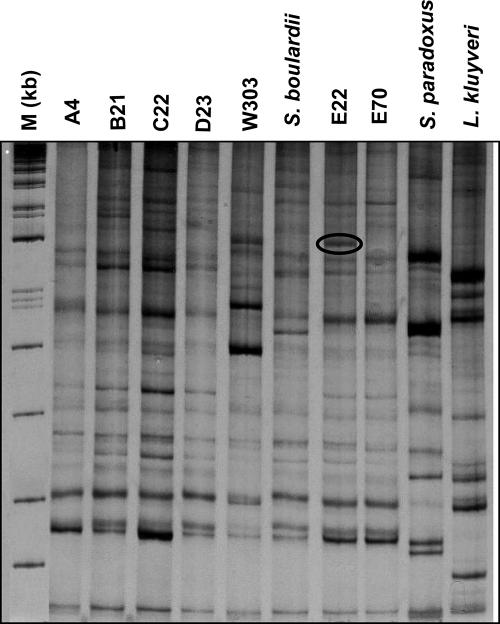
RAPD-PCR-based analysis has been used to differentiate yeast strains and to determine the level of genetic polymorphism (3, 12, 18, 33). Based on our previous results (15), the primers EI1 and LA1 were very efficient for strain delimitation; however, in the present study the profiles obtained for S. cerevisiae strains isolated from different distilleries were similar (Fig. 4), being distinguishable by minor changes of their banding profiles. Strains E22 and E70, which showed identical mtDNA RFLP patterns, could only be distinguished by an extra band in the range of 1,600 bp (indicated by a circle Fig. 2) that was not present in strain E70.
FIG. 4.
RAPD-PCR profiles prepared by using EI1 and LA1 primers of S. cerevisiae strains isolated from five distilleries. The circle around the 1,600-bp band on strain E22 indicates the unique difference found among the cachaça yeast strains.
Table 2 shows the results obtained for the analysis of six microsatellite loci ScAAT1 to ScAAT6. The number of alleles found was quite variable with the lowest polymorphism in the loci ScAAT5 and ScAAT6. Again, unique patterns were obtained for all strains, except for strains E22 and E70, which showed identical allelic combinations.
TABLE 2.
Allelic diversity of S. cerevisiae strains isolated from different distilleries compared to a laboratory wild-type strain (W303) and S. boulardii (an S. cerevisiae strain used as a probiotic)a
| Strain no. | Allelic diversity at microsatellite locus:
|
|||||
|---|---|---|---|---|---|---|
| ScAAT1 | ScAAT2 | ScAAT3 | ScAAT4 | ScAAT5 | ScAAT6 | |
| S. cerevisiae | ||||||
| A4 | 210 | 372/381 | 232/250 | 308/329 | 222 | 322/340 |
| B21 | 192/207 | 351 | 259/283 | 305 | 222 | 256 |
| C22 | 198 | 372/381 | 265 | 329 | 228 | 256 |
| D23 | 198 | 357/366 | 247 | 329 | 225 | 366 |
| W303 | 221 | 396 | 271 | 305 | 225 | 263 |
| S. boulardii | 174/198 | 363 | 247 | 302/329 | 225 | 256 |
| E22 | 195 | 381 | 232 | 305 | 231 | 256 |
| E70 | 195 | 381 | 232 | 305 | 231 | 256 |
The analysis is based on the number and length (in base pairs) of alleles for six microsatellite loci (ScAAT1 to ScAAT6).
The COX1 gene introns from the isolated yeast strains were amplified by using the primers 3L, 3R, 4L, and 5R (Fig. 5). Fragments of ca. 50, 60, and 240 bp were present in all S. cerevisiae strains and also in S. boulardii. Strain-specific additional bands (300 bp, strain D23; 360 bp, strains B21, C22, D23, E22, and E70; 720 bp, strains B21, C22, D23, and E22) were also apparent. Strains B21, C22, and D23 showed identical amplification patterns. Strains E22 and E70 that showed identical microsatellites and mtDNA restriction patterns could be distinguished by one weak band of 720 bp that was apparent in strain E22.
FIG. 5.
COX1-PCR profiles by using primers 3L, 3R, 4L, and 5R of S. cerevisiae strains isolated from five distilleries.
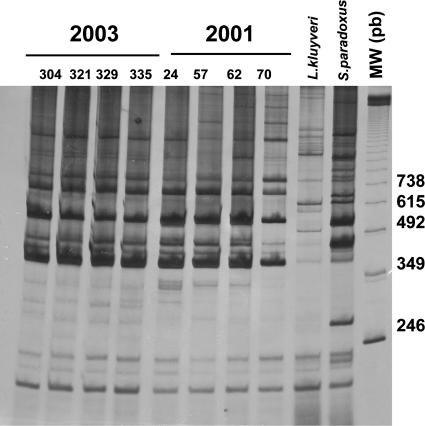
RAPD analysis revealed very similar or identical patterns among a group of strains that we consider candidate strains for cachaça production (Fig. 6). Although desirable, we could not find any correspondence between cerulenin and/or TFL resistance/sensitivity and characteristic banding patterns, since strains 70 and 321 (resistant to both drugs) showed distinctive RAPD profiles. On the contrary, strains 321 and 24, which were sensitive and resistant to both drugs, respectively, showed identical banding patterns.
FIG. 6.
RAPD-PCR profiles by using EI1 and LA1 primers of S. cerevisiae strains isolated from one distillery in two different years (2001 and 2003).
The described methods allowed us to differentiate strains with similar biochemical properties that were isolated from different distilleries. Their application to a unique distillery to follow the presence of a typical yeast strain seems to be limited. In fact, strains with similar biochemical properties (70 versus 321) can present different electrophoretic profiles, and strains with different biochemical properties (24 versus 321) show similar electrophoretic profiles.
Evaluation of the quality of cachaça produced with selected yeast strains.
Another important aspect investigated in the present study was the quality of cachaça obtained by using the isolated strains. We therefore produced cachaça by using three different strains: A4 and E70 (from distilleries A and E, respectively), as well as the commercial wine yeast strain Maurivin (Australian Wine Research Institute), a frequently used cachaça starter strain. To ensure that the selected strains were predominant during the fermentation process, 30 isolates were obtained from the fermenting sugar cane juice of the vessel inoculated with the strain and subjected to mtDNA-RFLP analysis. The results showed that strain E70 dominated the fermentative process during three fermentative cycles (data not shown).
As summarized in Table 3, strains A4 and E70 produced much less acetaldehyde than did the commercial wine strain (the legal limit is 30 mg/100 ml of anhydrous alcohol). Acidity, within certain limits, is necessary for sensory characteristics of the spirit, but excess acidity (limit, 150 mg/100 ml of anhydrous alcohol) renders cachaça unsuitable for consumption. The two selected strains produced cachaça with a seven to ten times lower volatile acidity content than the widely used commercial strain. This result clearly demonstrates the superiority of strains A4 and E70 isolated in the present study. The methanol concentration of cachaça produced with all strains was in conformity with legal standards (limit, 20 mg/100 ml of ethanol).
TABLE 3.
Some parameters obtained in the fermentative process recovered in batch distillates for three of the strains characterized
| Strain | Amt (mg/100 ml of ethanol)
|
Sensorial grade of cachaça samples (1 to 9) | ||
|---|---|---|---|---|
| Acetaldehyde | Volatile acidity | Methanol | ||
| A4 | 25.26 | 68.27 | 12.27 | 5.1 |
| E70 | 34.92 | 98.28 | 9.99 | 5.6 |
| Commercial strain | 91.42 | 671.86 | 9.64 | 3.9 |
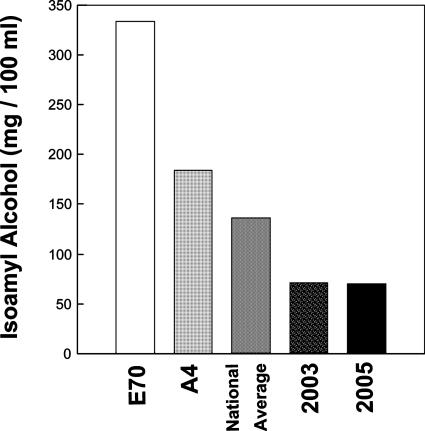
Figure 7 shows that the selected strains A4 and E70 produced a 1.4- to 2.5-fold higher amount of isoamyl alcohol than the national average, which is a value obtained for different brands of commercial cachaça (4). The highly increased isoamyl alcohol content is a characteristic of strain E70, since the isoamyl alcohol content of cachaça produced in distillery E, from which this strain originated, was five times lower in 2003 and 2005. Our data show that, among the yeast flora found in distillery E, very efficient isoamyl alcohol producers can be isolated, and the finding of strain E70 demonstrates the usefulness of our selection program rationale for cachaça production with superior aromatic characteristics. Sensory evaluation was performed by a panel with 19 trained panelists to confirm the quality of cachaça produced by strains A4, E70, and Maurivin. As shown in Table 3, the scores were considerably higher for cachaça obtained with strains A4 and E70 than for cachaça obtained strain Maurivin but were, nevertheless, similar to the score of 5.4, the mean value obtained in the same panel for traditional brands, which have been produced in Brazil for decades (data not shown).
FIG. 7.
Concentration of isoamyl alcohol in different cachaça samples obtained with selected yeast cells compared to concentrations produced by a commercial strain. The national average values in 2003 and 2005 samples of cachaça produced in Ouro Preto by spontaneous fermentation are also shown.
DISCUSSION
This study comprises two objectives: (i) to isolate and characterize yeast strains from cachaça distilleries in the state Minas Gerais (Brazil) and (ii) to examine the quality of the cachaça produced by the utilization of selected strains in the fermentative process.
Due to the special conditions of the fermentative process related to cachaça production, S. cerevisiae is the predominant species in the production of this beverage (31, 32; the present study). Our strategy to select appropriate strains to be used as starters in cachaça production (52) took into account important traits that cachaça yeasts need to possess, such as high resistance to osmotic, temperature, and ethanol stress, in order to counter the stressful fermentative conditions. Considering that sugar cane juice fermentation starts with high sucrose concentrations (ca. 20% [wt/vol]), proceeds under elevated temperatures (ca. 37°C), and terminates with high ethanol concentrations (6 to 8% [vol/vol]) at the end of each fermentative cycle, strains endowed with natural resistance to these conditions are more appropriate.
Our strategy also included strains with additional characteristics that are considered desirable, namely, a high fermentative power, here estimated “potentially” in terms of invertase activity (13, 52); the absence of H2S production, since this compound is considered an off flavor (37); killer activity, in order to guarantee the efficient elimination of competitors (6, 29, 31, 48); flocculation ability, to separate the yeast biomass from the fermented juice at the end of the process; and the production of high levels of flavoring compounds, such as higher alcohols and esters.
Screening for drug resistance such as TFL and cerulenin, which are an l-leucine analogue and a potent fatty acid synthetase (FAS) inhibitor, respectively, has been shown to be useful for isolating yeast strains with a high capacity to produce flavoring compounds (1, 5, 9, 10, 21, 38). In TFL-resistant strains there is an increase in the production of higher alcohols and their esters, particularly isoamyl alcohol and isoamyl acetate, associated with a fruity and sweet aroma. Previous studies have shown that LEU4 mutant strains yield an α-isopropylmalate synthase that is insensitive to feedback inhibition by leucine resistance to TFL. Such mutant strains are leucine overproducers and also produce higher amounts of isoamyl alcohol (1, 9, 10).
Ethyl caproate, one of the principal aroma compounds of sake, is synthesized from the precursors ethanol and caproic acid or caproyl acetate. The medium-chain fatty acid caproic acid is synthesized by the multifunctional FAS. Changing the balance of the individual FAS catalytic activities also changes the chain length of synthesized fatty acids. It seems that a specific mutation is responsible for cerulenin resistance and higher production of esters such as ethyl caproate (46). Since cerulenin is an inhibitor of FAS, it can be expected that cerulenin-resistant strains have a higher capacity to produce fatty acids (21) such as caproic acid.
In our view, the strategy of selection including TFL and or cerulenin resistance might be also applied in order to isolate cachaça yeast strains, mainly because flavoring substances such as isoamyl alcohol, isoamyl acetate, caproic acid, and ethyl caproate are found in different fermented beverages, including cachaça (4, 21, 30, 52).
Depending on the distillery from which the strains were isolated, the proportion of cerulenin- and TFL-resistant strains varied from 26 to 53%. A similar elevated variability concerning the additional characteristics tested in the present study was found among the strains isolated from each distillery. These findings show that each distillery or cachaça-producing region may possess a unique and characteristic S. cerevisiae flora, characterized by a high phenotypic variability related to traits that are of industrial importance.
It has already been demonstrated that S. cerevisiae strains isolated from different distilleries present a high genetic diversity (18). For this reason, we used different molecular approaches to characterize strains with the desired combination of characteristics for the achievement of better cachaça quality. Different molecular methods have been applied to characterize and differentiate yeast strains used in the production of fermented beverages such as wine (3, 7, 26, 27, 40) or cachaça (18) and also derived from other sources (47).
We present here results obtained by karyotyping (40), microsatellite analysis (33, 40), RAPD-PCR analysis using EI1 and LA1 primers (3, 15), and PCR analysis based on the variation in the number and position of introns in the mitochondrial gene COX1 (27). Although all of these techniques have proved useful for distinguishing the isolated strains (Fig. 3 to 6 and Table 2), they were unable to discriminate those with the appropriate characteristics for producing a better-quality beverage (at least potentially). Indeed, it has already been shown that some characteristics present in starter strains are not accessed by traditional molecular methods (12).
The isolation of strains with the same genetic profiles but with distinctive biochemical and/or physiological characteristics (H2S resistance, flocculation behavior, the production of flavoring compounds) makes it clear that an appropriate selection strategy for S. cerevisiae strains involved in cachaça fermentations should always be a combination of molecular, biochemical, and physiological methods. This approach should also be used for the monitoring of fermentations to ensure that the fermentative process is dominated by the inoculated strain(s) with appropriated characteristics so as to produce cachaça with a potentially higher and more reproducible quality standard.
Cachaça produced by the selected strains showed better characteristics compared to the sugar cane spirit produced by means of a commercial strain that is frequently used by cachaça producers. This was demonstrated by measuring the concentration of acetaldehyde, methanol, volatile acidity, and isoamyl alcohol. The cachaça produced by using strains A4 and E70 showed 1.4- and 2.6-fold increased isoamyl alcohol concentrations compared to the national average value (130 mg/100 ml of anhydrous alcohol) (4). Strain E70 from distillery E produced cachaça with a fivefold higher isoamyl alcohol content than the cachaça produced via the traditional, spontaneous fermentative process in the same distillery in 2003 and 2005. Cachaça produced with the selected strains was also evaluated by a trained sensory panel, and strain E70 scored better (sensorial grade of 5.6) than the national average (sensorial grade of 5.4), while the commercial wine strain received a rather low evaluation (sensorial grade of 3.9) Apparently, the commercial wine yeast strain (Maurivin) is not well adapted for producing a high-quality cachaça, since commercial wine strains are selected for properties that are important in winemaking, such as efficient fermentation of glucose and fructose, the two main sugars of wine must.
The findings from chemical and sensorial analysis emphasize the rationale of our selection procedure. Our results clearly show that strains selected from the flora of cachaça distilleries, where they underwent continuous adaptation to the particular cachaça environment through long-term cultivation in successive fermentation, produce cachaça with a higher quality than that of the cachaça obtained by either the traditional process or by using a commercial wine yeast strain. In our view, the procedure proposed here is a more adequate approach for isolating yeast strains to be used as starter strains in cachaça fermentations.
Acknowledgments
This study was supported by grants from the Fundação de Capacitação de Pessoal de Nível Superior from the Ministry of Education, Universidade Federal de Ouro Preto, Fundação de Amparo à Pesquisa do Estado de Minas Gerais (process EDT-2080/03), the programs POCI 2010 (FEDER/FCT, POCI/AGR/56102/2004; Fundação para a Ciência e Tecnologia) and AGRO (ENOSAFE, no. 762; Instituto Nacional de Investigação Agrária), and research fellowships from the Conselho Nacional de Desenvolvimento Científico e Tecnológico of Brazil (CNPq; process 300998/89-9 [R.L.B.]; process 490717/04-7 [CNPq/Grices], and process 301255/01-6 [L.G.F.]).
We thank Hugo Alves, a former research fellow from the Centro de Biologia Molecular e Ambiental, and Sylvie Dequin and Brigitte Cambon from the Institut Nacional de la Recherche Agronomique, UMR Sciences pour l'Oenologie, Montpellier, France, for their assistance with karyotype analysis.
Footnotes
Published ahead of print on 7 December 2007.
REFERENCES
- 1.Ashida, S., E. Ichikawa, K. Suginami, and S. Imayasu. 1987. Isolation and application of mutants producing sufficient isoamyl acetate, a sake flavor component. Agric. Biol. Chem. 51:2061-2065. [Google Scholar]
- 2.Barros-Lopes, M., A. Soden, P. A. Hebschke, and P. Langridge. 1996. PCR differentiation of commercial yeast strains using intron splice site primers. Appl. Environ. Microbiol. 62:4514-4520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barros-Lopes, M., J. R. Bellon, N. J. Shirley, and P. F. Ganter. 2002. Evidence for multiple interspecific hybridization in Saccharomyces sensu stricto species. FEMS Yeast Res. 1:323-331. [DOI] [PubMed] [Google Scholar]
- 4.Boscolo, M., C. W. B. Bezerra, D. R. Cardoso, B. S. L. Neto, and D. F. Franco. 2000. Identification and dosage by HRGC of minor alcohols and esters in sugar-cane spirit. Chem. Soc. 11:86-90. [Google Scholar]
- 5.Bussey, H., and H. E. Umbarger. 1970. Biosynthesis of branched-chain amino acids in yeast: a trifluoroleucine-resistant mutant with altered regulation of leucine uptake. J. Bacteriol. 103:286-294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cansado, J., E. Longo, P. Calo, C. Siero, J. B. Velazques, and T. G. Villa. 1991. Role of a killer character in spontaneous fermentation from NW Spain: ecology, distribution and significance. Appl. Microbiol. Biotechnol. 34:643-647. [Google Scholar]
- 7.Cappello, M. S., G. Bleve, F. Grieco, F. Dellaglio, and G. Zacheo. 2004. Characterization of Saccharomyces cerevisiae strains isolated from must of grape grown in experimental vineyard. J. Appl. Microbiol. 10:1-7. [DOI] [PubMed] [Google Scholar]
- 8.Carle, G. F., and M. W. Olson. 1985. An electrophoretic karyotype for yeast. Proc. Natl. Acad. Sci. USA 81:2965-2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Casalone, E., G. Fia, C. Barberio, D. Cavalieri, L. Turbanti, and M. Polsinelli. 1997. Genetic and biochemical characterization of Saccharomyces cerevisiae mutants resistant to trifluoroleucine. Res. Microbiol. 148:613-623. [DOI] [PubMed] [Google Scholar]
- 10.Cavalieri, D., E. Casalone, B. Bendoni, G. Fia, C. Barberio, M. Polsinelli, and C. Barberio. 1999. Trifluoroleucine resistance and regulation of α-isopropyl malate synthase in Saccharomyces cerevisiae. Mol. Gen. Genet. 261:152-160. [DOI] [PubMed] [Google Scholar]
- 11.Celenza, J. L., and M. Carlson. 1989. Mutational analysis of the Saccharomyces cerevisiae SNF1 protein kinase and evidence for functional interaction with the SNF4 protein. Mol. Cell. Biol. 9:5045-5054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dunn, B., R. P. Levine, and G. Sherlock. 2005. Microarray karyotyping of commercial wine yeast strains reveals shared, as well as unique, genomic signatures. BMC Genomics 6:53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ekunsanmi, T. J., and S. A. Odunfa. 1990. Ethanol tolerance, sugar tolerance and invertase activities of same yeast strains isolated from steep water of fermenting cassava tubers. J. Appl. Bacteriol. 69:672-675. [Google Scholar]
- 14.Fernández-Espinar, M. T., V. López, D. Ramón, E. Bartra, and A. Querol. 2001. Study of the authenticity of commercial wine yeast strains by molecular techniques. Int. J. Food Microbiol. 70:1-10. [DOI] [PubMed] [Google Scholar]
- 15.Fietto, J. L., R. S. Araújo, F. N. Valadão, L. G. Fietto, R. L. Brandão, M. J. Neves, F. C. Gomes, J. R. Nicoli, and I. M. Castro. 2004. Molecular and physiological comparisons between Saccharomyces cerevisiae and Saccharomyces boulardii. Can. J. Microbiol. 50:615-621. [DOI] [PubMed] [Google Scholar]
- 16.Gallego, F. J., M. A. Perez, I. Martinez, and P. Hidalgo. 1998. Microsatellites obtained from database sequences are useful to characterize Saccharomyces cerevisiae strains. Am. J. Enol. Vitic. 49:350-351. [Google Scholar]
- 17.Goldstein, A., and J. O. Lampen. 1975. Beta-d-fructofuranoside fructohydrolase from yeast. Methods Enzymol. 42:504-511. [DOI] [PubMed] [Google Scholar]
- 18.Guerra, J. B., R. C. A. Araújo, C. Pataro, G. R. Franco, E. S. A. Moreira, L. C. Mendonça-Hagler, and C. A. Rosa. 2001. Genetic diversity of Saccharomyces cerevisiae strains during the 24 h fermentative cycle for the production of the artisanal “cachaça”. Lett. Appl. Microbiol. 33:106-111. [DOI] [PubMed] [Google Scholar]
- 19.Guillamón, J. M., J. Sabaté, E. Barrio, J. Cano, and A. Querol. 1998. Rapid identification of wine yeast species based on RFLP analysis of the ribosomal internal transcriber spacer (ITS) region. Arch. Microbiol. 169:387-392. [DOI] [PubMed] [Google Scholar]
- 20.Hennequin, C., A. Thierry, G. F. Richard, G. Lecointre, H. V. Nguyen, C. Gaillardin, and B. Dujon. 2001. Microsatellite typing as a new tool for identification of Saccharomyces cerevisiae strains. J. Clin. Microbiol. 39:551-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ichikawa, E., N. Hosokawa, Y. Hata, Y. Abe, K. Suginami, and S. Imayasu. 1991. Breeding of sake yeast with improved ethyl caproate productivity. Agric. Biol. Chem. 55:2153-2154. [Google Scholar]
- 22.Jiranek, V., P. Langridge, and P. A. Henschke. 1995. Validation of bismuth-containing indicator media for predicting H2S-producing potential of Saccharomyces cerevisiae wine yeast under enological conditions. Am. J. Enol. Vitic. 46:269-273. [Google Scholar]
- 23.Kreger-van Rij, N. J. W. (ed.). 1984. The yeasts: a taxonomic study, 3rd ed. Elsevier Science, Amsterdam, The Netherlands.
- 24.Legras, J. L., D. Merdinoglu, J. M. Cornuet, and F. Karst. 2007. Bread, beer and wine: Saccharomyces cerevisiae diversity reflects human history. Mol. Ecol. 16:2091-2102. [DOI] [PubMed] [Google Scholar]
- 25.Longo, E., J. B. Velazquez, C. Sieiro, J. Cansado, P. Calo, and T. G. Villa. 1992. Production of higher alcohols, ethyl acetate, acetaldehyde, and other compounds by 14 Saccharomyces cerevisiae wine strains isolated from the same region (Salnés, N. W. Spain). World J. Microbiol. Biotechnol. 8:539-541. [DOI] [PubMed] [Google Scholar]
- 26.López, V., M. T. Fernadez-Espinar, E. Barrio, D. Ramón, and A. Querol. 2003. A new PCR-based method for monitoring inoculated wine fermentations. Int. J. Food Microbiol. 81:63-71. [DOI] [PubMed] [Google Scholar]
- 27.López, V., A. Querol, D. Ramón, and M. T. Fernadez-Espinar. 2001. A simplified procedure to analyse mitochondrial DNA from industrial yeasts. Int. J. Food Microbiol. 68:75-81. [DOI] [PubMed] [Google Scholar]
- 28.Lowry, O. H., N. J. Rosebrough, A. L. Farr, and R. J. Randall. 1951. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 193:265-275. [PubMed] [Google Scholar]
- 29.Morais, P. B., C. A. Rosa, V. R. Linardi, C. Pataro, and A. B. R. A. Maia. 1997. Characterization and succession of yeast populations associated with spontaneous fermentations during the production of sugarcane aguardente. World J. Microbiol. Biotechnol. 13:241-243. [Google Scholar]
- 30.Nonato, E. A., E. A. Carazza, F. S. Silva, C. R. Carvalho, and Z. L. Cardeal. 2001. A headspace solid-phase micro extraction method for the determination of some secondary compounds of sugar cane spirits by gas chromatography. J. Agric. Food. Chem. 49:3533-3539. [DOI] [PubMed] [Google Scholar]
- 31.Pataro, C., A. Santos, S. R. Correa, P. B. Morais, V. R. Linardi, and C. A. Rosa. 1998. Physiological characterization of yeasts isolated from artisanal fermentation in an aguardente distillery. Rev. Microbiol. 29:104-108. [Google Scholar]
- 32.Pataro, C., J. B. Guerra, M. L. Petrillo-Peixoto, L. C. Mendonça-Hagler, V. R. Linardi, and C. A. Rosa. 2000. Yeast communities and genetic polymorphism of Saccharomyces cerevisiae strains associated with artisanal fermentation in Brazil. J. Appl. Microbiol. 88:1-9. [DOI] [PubMed] [Google Scholar]
- 33.Perez, M. A., F. J. Gallego, I. Martinez, and P. Hidalgo. 2001. Detection, distribution and selection of microsatellites (SSRs) in the genome of the yeast Saccharomyces cerevisiae as molecular markers. Lett. Appl. Microbiol. 33:461-466. [DOI] [PubMed] [Google Scholar]
- 34.Pérez, M. A., F. J. Gallego, and P. Hidalgo. 2001. Evaluation of molecular techniques for the genetic characterization of Saccharomyces cerevisiae strains. FEMS Microbiol. Lett. 205:375-378. [DOI] [PubMed] [Google Scholar]
- 35.Querol, A., E. Barrio, and D. Ramón. 1992. A comparative study of different methods of yeast strain characterization. Syst. Appl. Microbiol. 15:439-446. [Google Scholar]
- 36.Querol, A., E. Barrio, T. Huerta, and D. Ramón. 1992. Molecular monitoring of wine fermentation conducted by active dry yeast strains. Appl. Environ. Microbiol. 58:2948-2953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ribeiro, C. A. F., and J. Horii. 1999. Potencialidades de linhagens de levedura Saccharomyces cerevisiae para fermentação do caldo de cana. Sci. Agric. 56:255-263. [Google Scholar]
- 38.Satyanarayana, T., H. E. Umbarger, and G. Lindegren. 1968. Biosynthesis of branched-chain amino acids in yeast: correlation of biochemical blocks and genetic lesions in leucine auxotrophs. J. Bacteriol. 96:2012-2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schuller, D., E. Valero, S. Dequin, and M. Casal. 2004. Survey of molecular methods for the typing of wine yeast strains. FEMS Microbiol. Lett. 231:19-26. [DOI] [PubMed] [Google Scholar]
- 40.Schuller, D., H. Alves, S. Dequin, and Casal M. 2005. Ecological survey of Saccharomyces cerevisiae strains from vineyards in the Vinho Verde Region of Portugal. FEMS Microbiol. Ecol. 51:167-177. [DOI] [PubMed] [Google Scholar]
- 41.Schuller, D., and M. Casal. 2007. The genetic structure of fermentative vineyard-associated Saccharomyces cerevisiae populations revealed by microsatellite analysis. Antonie van Leeuwenhoek 91:137-150. [DOI] [PubMed] [Google Scholar]
- 42.Schuller, D., L. Pereira, H. Alves, B. Cambon, S. Dequin, and M. Casal. 2007. Genetic characterization of commercial Saccharomyces cerevisiae isolates recovered from vineyard environments. Yeast 24:625-636. [DOI] [PubMed] [Google Scholar]
- 43.Schwan, R. F., A. T. Mendonça, J. J. Silva, Jr., V. Rodrigues, and A. E. Wheals. 2001. Microbiology and physiology of cachaça (aguardente) fermentations. Antonie van Leeuwenhoek 79:89-96. [DOI] [PubMed] [Google Scholar]
- 44.Stole, H., and J. L. Sidel. 1993. Sensory evaluation practices, 2nd ed. Academic Press, Inc., San Diego, CA.
- 45.Techera, A. G., S. Jubany, F. M. Carrau, and C. Gaggero. 2001. Differentiation of industrial wine yeast strains using microsatellite markers. Lett. Appl. Microbiol. 33:71-75. [DOI] [PubMed] [Google Scholar]
- 46.Uehigashi, H., E. Nakagawa, H. Moriyama, S. Nagata, and H. Misono. 1999. Breeding of sake yeast that produces enhanced levels of flavor by transformation with a mutated FAS2 gene and their utilization in brewing rice flour shochu. J. Brew. Soc. Japan 94:63-71. [Google Scholar]
- 47.Valente, P., F. C. Gouveia, G. A. de Lemos, D. Pimentel, J. D. Van Elsas, L. C. Mendonca-Hagler, and A. N. Hagler. 1996. PCR amplification of the rDNA internal transcribed spacer region for differentiation of Saccharomyces cultures. FEMS Microbiol. Lett. 137:253-256. [DOI] [PubMed] [Google Scholar]
- 48.Van Vuuren, H. J. J., and C. J. Jacobs. 1992. Killer yeast in the wine industry: a review. Am. J. Enol. Vitic. 43:119-128. [Google Scholar]
- 49.Vaughan-Martini, A., and A. Martini. 1993. A taxonomic key the genus Saccharomyces. Syst. Appl. Microbiol. 16:113-119. [Google Scholar]
- 50.Verstrepen, K. J., G. Derdelinckx, F. R. Delvaux, J. Winderickx, J. M. Thevelein, F. F. Bauer, and I. S. Pretorius. 2001. Late fermentation expression of FLO1 in Saccharomyces cerevisiae. J. Am. Sac. Brew. Chem. 59:69-76. [Google Scholar]
- 51.Vezinhet, F., J. N. Hallet, M. Valade, and A. Poulard. 1990. Ecological survey of wine yeast strains by molecular methods of identification. Am. J. Enol. Vitic. 43:83-86. [Google Scholar]
- 52.Vicente, M. A., L. G. Fietto, I. M. Castro, A. N. G. Santos, M. X. Coutrim, and R. L. Brandão. 2006. Isolation of Saccharomyces cerevisiae strains producing higher levels of flavoring compounds for production of cachaça, the Brazilian sugarcane spirit. Int. J. Food Microbiol. 108:51-59. [DOI] [PubMed] [Google Scholar]
- 53.White, T. J., T. Bruns, S. Lee, and J. Taylor. 1990. Amplification and direct sequencing of fungal RNA genes for phylogenetics, p. 315-322. In M. A. Inns, D. H. Gelfrand, J. J. Sninsky, and T. J. With (ed.), PCR protocols: a guide to methods and applications. Academic Press, Inc., New York, NY.