Abstract
Symbiotic bacteria residing in the hindgut chambers of scarab beetle larvae may be useful in paratransgenic approaches to reduce larval root-feeding activities on agricultural crops. We compared the bacterial community profiles associated with the hindgut walls of individual Dermolepida albohirtum third-instar larvae over 2 years and those associated with their plant root food source among different geographic regions. Denaturing gradient gel electrophoresis analysis was used with universal and Actinobacteria-specific 16S rRNA primers to reveal a number of taxa that were found consistently in all D. albohirtum larvae but not in samples from their food source, sugarcane roots. These taxa included representatives from the “Endomicrobia,” Firmicutes, Proteobacteria, and Actinobacteria and were related to previously described bacteria from the intestines of other scarab larvae and termites. These universally distributed taxa have the potential to form vertically transmitted symbiotic associations with these insects.
There has been growing research interest in developing novel approaches to manage insect pests and the pathogens they transmit through the genetic manipulation of microorganisms that intimately associate with these insects (3, 19). Initially, this work was focused on the genetic manipulation of insect symbionts to block the transmission of pathogens vectored by insects (4). This approach has since widened to target insect pests directly (12, 17, 22, 25). As a first step toward the development of such an approach with Australian scarab pests, we have characterized a subset of the bacterial community that naturally associates with the scarab hindgut, in order to identify potential candidates for genetic manipulation strategies targeting the feeding activity of these beetles.
The hindgut chambers of scarab larvae are known to house dense microbial communities that participate in the fermentation of plant material (1, 13). Molecular characterization of the bacterial communities in the midguts, hindguts, and food sources of humivorous larvae of the East African scarab Pachnoda ephippiata showed significant differences across all three environments (6), although some gut taxa matched close relatives found in rhizosphere communities, suggesting that transient associations occur in the larval gut environment. However, in the case of root-feeding larvae of the European cockchafer Melolontha melolontha, terminal restriction fragment length polymorphism profiles compared across regions revealed a diverse and consistent bacterial community on the hindgut walls of larvae (5). Many of the taxa were related to gut bacteria identified in other beetles, termites, and the vertebrate rumen, suggesting a specific and functional role in cellulose digestion.
In Australia, scarab beetles are major pests in sugarcane production through the feeding damage that larvae cause to plant root systems. Nineteen species of endemic melolonthine beetles cause significant damage to Australian sugarcane (14). The most important species is Dermolepida albohirtum, a species with a 1-year life cycle in which the third-instar larvae feed on the roots of the crop in late summer and autumn. Despite the considerable economic importance of these beetles, relatively few control strategies exist for their management and virtually nothing is known about the bacteria that associate with their digestive tracts. Here, we used denaturing gradient gel electrophoresis (DGGE) analysis of 16S rRNA amplicons to identify specific bacterial taxa that are associated with the hindgut walls of D. albohirtum larvae across the geographic range of the species. The results of this analysis provide a better understanding of nontransient microorganisms that are associated with this pest and are potentially amenable to future manipulation.
MATERIALS AND METHODS
Sample collection.
Third-instar larvae of the melolonthine scarab D. albohirtum were collected from commercial sugarcane fields in April 2005 and April 2006. Larvae were collected from four regions approximately 200 to 300 km apart near Cairns, Tully, Ayr, and Mackay in northern Queensland, Australia. Each region is geographically distinct and contains different soil types. Within each region, larvae were collected from two sites approximately 10 km apart. Larvae of the humus-feeding dynastine Dasygnathus sp. were opportunistically collected from the same sugarcane root bowls as D. albohirtum larvae in 2005. All larvae were maintained separately in soil-free sterile containers and kept from food and water for up to 7 days before processing. Larvae were placed at −80°C for 30 min, surface sterilized by immersion in 5% (wt/vol) sodium hypochlorite for 2 min, and rinsed with 70% ethanol. Larvae were then dissected, the complete intestine of each larva was removed, and the hindgut paunch was severed from the midgut and colon, as described by Lemke et al. (13). The hindgut paunch was then cut open, and the hindgut wall was rinsed three times in sterilized water to remove lumen contents. Hindgut wall tissue was then stored at −80°C until further analysis.
In 2006, we also collected plant roots and soil samples from the immediate surroundings of the larvae at one site each in the Cairns, Ayr, and Mackay regions. Six plant and six soil samples were collected from each site at locations approximately 10 m apart. Loose soil was shaken off plant roots and collected in clean trays. The soil samples collected from each site were thoroughly mixed. Plant roots were washed three times in sterilized water. Plant roots collected from each site were combined and ground to a pulp using a mortar and pestle. Soil samples and plant roots were stored separately at −30°C within 6 h of field collection until further analysis.
All DNA extractions were carried out using the PowerSoil DNA isolation kit by following the protocol of the manufacturer (MO BIO Laboratories, Carlsbad, CA). For the larvae collected in 2005, the entire hindgut walls of individual larvae were used in separate DNA extractions. DNA was extracted from separate samples of approximately 2 g of plant root pulp or soil from each site. For larvae collected in 2006, the hindgut walls of six individual larvae from each site were pooled before DNA extraction.
PCR amplification.
Fragments of 16S rRNA genes were amplified from DNA extracted from the gut, plant, and soil samples by using the following primer sets and conditions. The general DGGE primer set targeting eubacterial 16S rRNA genes, comprising F-968-GC (5′-CGC CCG GGG CGC GCC CCG GGC GGG GCG GGG GCA CGG GGG GAA CGC GAA GAA CCT TAC-3′) and R-1401 (5′-CGG TGT GTA CAA GAC CC-3′) (16), was used to amplify DNA extracted from the hindgut walls of individual larvae collected in 2005. The reaction mixture contained, in a total volume of 20 μl, 1× PCR buffer (New England BioLabs [NEB], Beverly, MA), 1 U of Taq DNA polymerase (NEB), 50 μM deoxynucleoside triphosphates (Promega, Madison, WI), 0.5 μM (each) primers (Invitrogen, Carlsberg, CA), and 1 μl of a 1:10 dilution of gut DNA extract. PCR amplification was undertaken using the hot-start technique (2) and the following reaction conditions: initial denaturation at 94°C for 5 min; 30 cycles of 94°C for 30 s, annealing at 55°C for 30 s, and extension at 72°C for 90 s; and terminal extension at 72°C for 10 min. Visual analysis of PCR products (4-μl aliquots) on 1% agarose gels was accomplished by gel electrophoresis with ethidium bromide staining.
We used a seminested PCR method to enrich DNA extracts from plant, soil, and hindgut wall samples with 16S rRNA amplicons from the Actinobacteria phylum (20). Gut samples from six individuals from each site of collection were pooled for amplification. First-round amplification used the Actinobacteria-specific forward primer F243 (5′-GGA TGA GCC CGC GGC CTA-3′) (7) and R-1401. Second-round amplification used primers F-968-GC and R-1401. All amplification conditions were the same as those described above, except that the volume used in first-round reactions contained 1 μl of plant or 5 μl of soil DNA extract and the second-round reaction mixtures contained 1 μl of a 1:20 dilution of first-round PCR mixtures.
“Endomicrobium”-specific primers TG1-209F (5′-AAT GCG TTT TGA GAT GGT CCT G-3′) and TG1-1325R (GAT TCC TAC TTC ATG TTG-3′) (23) were used to amplify 16S rRNA genes from hindgut wall samples collected in 2006. DNA extracts from the hindgut walls of larvae from three regions were pooled, and 1-μl aliquots were used under standard PCR conditions. Ten-microliter aliquots were loaded onto 1% agarose gels, and PCR products of the correct size were cut out and purified using the MinElute gel extraction kit (QIAGEN, Hilden, Germany).
DGGE.
Heterogeneous 16S rRNA amplicons were separated using a DGGE system (Bio-Rad Laboratories, Hercules, California). Samples (2 to 6 μl) were loaded onto 7% polyacrylamide gels (30 to 60% gradient [100% denaturant corresponds to 7 M urea and 40% formamide]) and run for 16 h at 80 V using 1× Tris-acetate-EDTA buffer. Gels were silver stained (21) and viewed on a light box. Bands of interest, including dominant bands that upon visual inspection appeared to be broadly distributed among larval hindgut samples and to be of high intensity, were cut from gels, and DNA was eluted overnight at 4°C in 50 μl of water. Eluted DNA (5-μl aliquots) was then reamplified using primers F-968-GC and R-1401 and purified from agarose gels for subsequent cloning and sequencing. Sequences extracted from DGGE bands that had >99% similarity were considered to represent the same taxa.
Cloning and sequencing.
PCR products were ligated into pGEM-T Easy vector (Promega) and cloned into Escherichia coli DH5α. Plasmids were purified from randomly selected clones using a QIAprep spin miniprep kit (QIAGEN), checked for correct insert size by PCR amplification, and sequenced. Two clones from each reamplified DGGE band were sequenced. Three clones from the “Endomicrobium”-specific PCR products were sequenced in both forward and reverse directions. 16S rRNA gene sequences were labeled Da-1 to Da-9, respectively. The nomenclature Da corresponds to the first letters of the name of the scarab D. albohirtum, from which the sequences were first identified.
Phylogenetic analysis.
DNA sequences were assembled using Lazergene 5 (DNASTAR, Madison, WI), and sequence similarity searches were performed using Seqmatch in the Ribosomal Database Project II (RDP II) (http://rdp.cme.msu.edu/). For phylogenetic analysis, 16S rRNA sequences were aligned with the nearest neighbors found in the RDP II using ClustalX version 1.81 (26) and manually checked. Trees were constructed using maximum likelihood and Bayesian inference. Maximum-likelihood trees were constructed in PAUP* version 4.0b10 (24), and construction included the preselection of nucleotide substitution models in Modeltest version 3.5 utilizing the Akaike information criterion (18). Model TrN + I + G was identified for both alignments and incorporated in the Bayesian trees constructed using MrBayes 3.1.2 (10). Bayesian analyses were carried out using 106 generations and a sample frequency of 100. The first 25 to 30% of trees were discarded, and consensus trees were computed according to 50% majority rule. The 16S rRNA fragments used to construct trees that were derived from DGGE gels and “Endomicrobium”-specific primers were 433 bp (E. coli numbering, bp 968 to 1401) and 1,116 bp (E. coli numbering, bp 209 to 1325) in length, respectively.
Nucleotide sequence accession numbers.
Nucleotide sequences were deposited in the GenBank database under accession numbers EU073940 to EU073948.
RESULTS
Hindgut wall DGGE analysis.
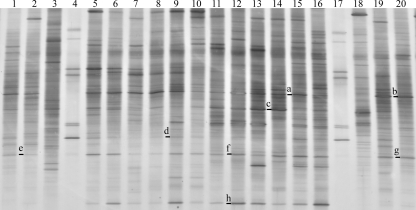
The bacterial community profiles associated with the hindgut walls of individual D. albohirtum larvae from the four regions (Cairns, Tully, Ayr, and Mackay) were compared. DGGE analysis using general eubacterial primers targeting 16S rRNA genes showed that a number of bands were shared among all D. albohirtum larvae. A representative subset of amplifications from individuals is shown in Fig. 1. There was also considerable variation among profiles of individual D. albohirtum larvae and between these profiles and those of larvae of Dasygnathus sp. from the same location. Dominant bands (Fig. 1) belonged to taxa from the Clostridiales (16S rRNA gene sequence Da-2 was isolated from DGGE bands a and b), Deltaproteobacteria (16S rRNA gene sequence Da-5 was isolated from DGGE bands e to g), and Actinobacteria (16S rRNA gene sequence Da-3 was isolated from DGGE band h). Less common bands matched taxa from the Betaproteobacteria (16S rRNA gene sequences Da-6 to Da-8 were isolated from DGGE band d) and Bacteroides (16S rRNA gene sequence Da-9 was isolated from DGGE band c).
FIG. 1.
DGGE profiles of bacteria present on the hindgut walls of D. albohirtum third-instar larvae collected in 2005 from regions approximately 200 km apart by using general primers targeting the 16S rRNA gene. Sampling areas were the Cairns region (lanes 1 and 2), Tully region site 1 (lanes 5 to 7) and site 2 (lanes 8 to 10), Ayr region site 1 (lanes 11 to 13) and site 2 (lanes 14 to 16), and the Mackay region (lanes 19 and 20). Lanes 4 and 17 are ladders. Lanes 3 and 18 contain samples from hindguts of humus-feeding Dasygnathus sp. larvae found in the same root bowls as the larva samples in lanes 2 and 19, respectively. Letters to the left of the lanes indicate bands that were selected for sequencing. Sequence Da-5 was isolated from bands a and b. Sequence Da-2 was isolated from bands e to g, and Da-3 was isolated from band h. Dominant bands are presented in the phylogenetic tree (Fig. 2).
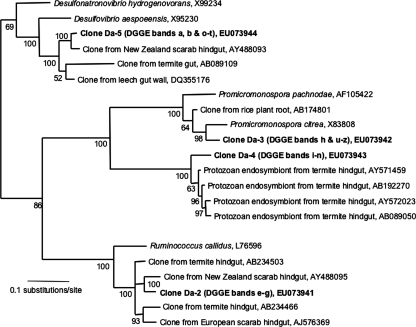
The dominant bands isolated from the DGGE gel shown in Fig. 1 are presented in the phylogenetic tree in Fig. 2, along with the closest relatives found in the RDP II database. The phylogenetic tree (Fig. 2) shows close relationships to noncultured taxa detected in samples from the hindguts of larvae of Pachnoda ephippiata (6), the New Zealand scarab Costelytra zealandica (H. Zhang and T. Jackson, personal communication), and termites (8, 9).
FIG. 2.
Phylogenetic relationship of sequences isolated from dominant bands in DGGE profiles (Fig. 1 and 3) of bacteria from the hindgut walls of D. albohirtum third-instar larvae (in bold) and the nearest neighbors found using the RDP II. Accession numbers of reference sequences are indicated. Tree construction used Bayesian analysis of 433 bp of the 16S rRNA gene, and Bayesian posterior probabilities are indicated at each node. Sequence Da-2 was present in DGGE bands e to g. Sequence Da-3 was present in DGGE bands h and u to z. Sequence Da-4 was present in DGGE bands i to n. Sequence Da-5 was present in DGGE bands a, b, and o to t.
However, the large diversity of taxa corresponding to the sequences amplified using general primers made it difficult to isolate bands belonging to a single bacterial species. Consequently, phylum-specific primers were used to focus on the Actinobacteria phylum in a second DGGE analysis that also included plant roots and soil samples. The Actinobacteria phylum was chosen because the dominant band corresponding to Da-3 in Fig. 1 and 2 matched closely with a sequence from Promicromonospora pachnodae, which was originally isolated from the rose chafer Pachnoda marginata (1). In addition, we needed to reduce the diversity among DGGE profiles to identify specific bacterial species when comparing diverse soil and plant root environments to D. albohirtum larval gut profiles.
Plant-soil-gut DGGE analysis.
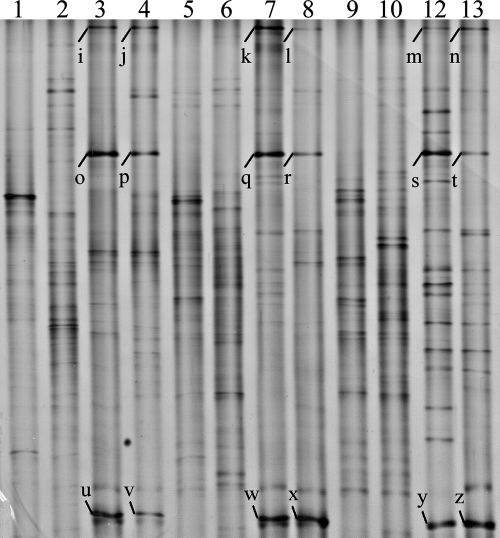
Actinobacteria-specific primers were used in a seminested PCR to enrich 16S rRNA genes from the Actinobacteria phylum for use in DGGE analysis. The community profiles for the hindgut walls of D. albohirtum larvae, the larvae's food source (sugarcane roots), and the surrounding soil were compared. DGGE analysis utilizing Actinobacteria-specific primers showed three bands (Fig. 3) that were found consistently in the three geographic regions and over two generations of larvae (2005 and 2006). These three common bands were not detected in plant and soil samples (Fig. 3). Two of these bands corresponded to the Actinobacteria representative Da-3 (DGGE bands u to z) and the Clostridiales representative Da-2 (DGGE bands o to t) from Fig. 1 and 2, while the upper band in Fig. 3 (16S rRNA gene sequence Da-4 was isolated from DGGE bands i to n) matched the recently described “Endomicrobia” candidate phylum (23). Taxa corresponding to these three bands are presented in the phylogenetic analysis (Fig. 2).
FIG. 3.
DGGE profiles of the bacteria present on plant roots, in rhizosphere soil, and on the hindgut walls of D. albohirtum third-instar larvae collected from regions approximately 200 km apart in 2005 (lanes 4, 8, and 12) and 2006 (lanes 3, 7, and 11) as determined by using Actinobacteria-specific primers targeting the 16S rRNA gene. Plant roots (lanes 1, 5, and 9) and soil samples (lanes 2, 6, and 10) from the immediate surroundings of the larvae collected in 2006 were collected at the same time as the larvae. The sampling regions were Cairns (lanes 1 to 4), Ayr (lanes 5 to 8), and Mackay (lanes 9 to 12). Letters to the left of lanes indicate dominant bands that were sequenced (Da-4 was isolated from bands i to n, Da-5 was isolated from bands o to t, and Da-3 was isolated from bands u to z) and presented in the phylogenetic tree (Fig. 2).
We chose to use the forward primer F243 in order to subsequently enrich the representation of taxa from the Actinobacteria phylum. This primer also matches a number of taxa from other phyla, giving it limited specificity. Using this primer, we detected representatives from the candidate phylum “Endomicrobia” and Da-2 from the Clostridiales. There were other DGGE bands identified by visual analysis that did not occur in all regions or in all plant and soil communities and bands that were unique to gut, plant, and soil samples within a region.
“Endomicrobium”-specific primers.
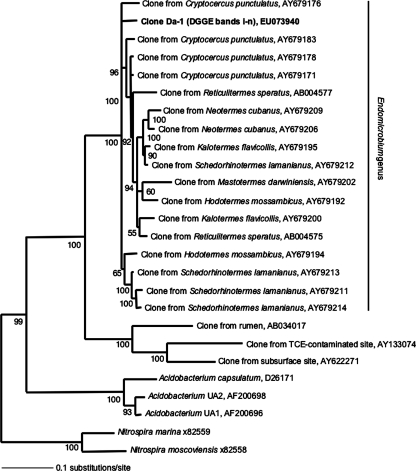
Upon the discovery of Da-4 from the “Endomicrobia” phylum (Fig. 3), specific primers for this phylum were used to sequence a 1,116-bp fragment of the 16S rRNA gene from D. albohirtum hindgut wall samples. All three amplicons that were sequenced had a 100% match to sequence Da-1. Both Bayesian (Fig. 4) and maximum-likelihood (data not shown) trees placed Da-1 within the “Endomicrobium” genus among the endosymbionts of cellulose-fermenting protists. These protists develop an obligate symbiosis with the wood-feeding cockroach Cryptocercus punctulatus and the lower termite group (23).
FIG. 4.
Phylogenetic relationship of the 16S rRNA gene Da-1 sequence, listed in bold, recovered from the hindgut of D. albohirtum third-instar larvae and the nearest neighbors found using the RDP II. “Endomicrobium” clones are represented by the host species; Cryptocercus punctulatus is the wood-feeding cockroach, and all others are lower termites. Reference organisms chosen for tree construction were the same as those in reference 23. Accession numbers of reference sequences are indicated. Tree construction used Bayesian analysis of 1,116 bp of the 16S rRNA gene, and Bayesian posterior probabilities of >50% are indicated at each node. TCE, trichloroethene.
DISCUSSION
The larval intestine of root-feeding scarab beetles is exposed to an enormous diversity of microorganisms within the rhizosphere. This diversity creates difficulties in discerning between bacteria that are transiently acquired and species that may be intimately associated with the host. Using DGGE analysis, we have identified species that are associated with the larval host over a broad geographic distribution and across generations. Potentially, these species could be isolated in pure culture and genetically transformed to express compounds that inhibit the feeding activity of larvae on commercial sugarcane crops.
We identified species from numerous phyla that are associated closely with the hindgut walls of root-feeding D. albohirtum larvae. The 16S rRNA gene fragments Da-2, Da-3, and Da-4 were found in all larvae over two generations and were not readily detected in the samples from the larval food source or the local soil environment of the larvae. Unfortunately, definitive conclusions on the detection of rare bacteria within the rhizosphere are beyond the scope of this work, as limitations are imposed by the competitive nature of mixed-template PCR amplification and DGGE detection limits. However, it is possible that these bacteria are not transiently acquired while the larvae feed in the rhizosphere. Alternatively, these commonly found bacteria may be inherited from the previous generation via an unknown mechanism.
The phylogenetic positioning of the commonly found gut bacteria showed interesting correlations to results from other studies. Taxa closely matched hindgut bacteria found in larvae of the New Zealand scarab beetle C. zealandica (Zhang and Jackson, personal communication), which is an endemic pasture pest (11). Costelytra zealandica gut profiles analyzed with DGGE had gut bacteria from the Clostridiales, Deltaproteobacteria, and Bacteroides, with close identity to those in D. albohirtum. Other significant correlations were found with gut bacteria from larvae of the European scarab Pachnoda ephippiata (6) and with a dominant (hemi)cellulolytic Actinobacteria isolate, Promicromonospora pachnodae (1), from Pachnoda marginata larvae. Unfortunately, the molecular characterization of the guts of root-feeding M. melolontha larvae used a different region of the 16S rRNA gene, preventing direct sequence comparisons to the findings in our study (5). However, the phylogenetic placement of taxa from the Clostridiales, Deltaproteobacteria, and Actinobacteria phyla correlates with that of D. albohirtum hindgut taxa. These similarities suggest that applying phylum-specific DGGE analyses to other phyla will provide rapid identification of other gut bacteria associated with the hindguts of scarab larvae.
A significant finding from this study is the discovery of a representative from the “Endomicrobium” genus present in D. albohirtum larval samples from different regions and years (Fig. 3). This finding provides strong support for the occurrence of digestive symbiosis in the hindguts of root-feeding scarab beetle larvae. Previously, members of the “Endomicrobium” genus had been detected only in the hindguts of wood-feeding cockroaches and the lower termites, where these bacteria form an obligate endosymbiotic relationship with hindgut protozoa (23). A brief microscopic examination of D. albohirtum larval hindgut contents showed that protozoa were commonly found in all three larvae inspected (data not shown).
Assuming that the Da-1 species forms an endosymbiotic association with protozoa in the gut of D. albohirtum, our discovery adds to the debate on the ancestral transfer of symbionts between cockroaches and termites (15). The phylogenetic placement of the Da-1 species within the “Endomicrobium” genus suggests that a recent horizontal acquisition of symbiotic protozoa has occurred between these deeply divergent insect groups.
Visual inspection of the DGGE gels revealed bands that were unique to individual gut profiles (Fig. 1) and bands that were common among gut, soil, and plant root environments (Fig. 3). If these bands represent bacteria that are transiently acquired by larvae and are capable of surviving in both rhizosphere and hindgut environments for prolonged periods, then these bacteria may also be potential candidates for the development of transgenic strategies aimed at reducing the feeding activities of scarab pests of sugarcane.
Acknowledgments
We thank Bill Harris, Mohamed Sallam, and Peter Samson for assistance in locating and collecting scarab larvae. We also thank members of the O'Neill Laboratory, particularly Inaki Iturbe-Ormaetxe and Jeremy Brownlie for technical support, Beth McGraw for phylogenetic advice, and Hongyu Zhang and Trevor Jackson for advice on DGGE analysis.
This work was supported by an Australian Research Council grant.
Footnotes
Published ahead of print on 14 December 2007.
REFERENCES
- 1.Cazemier, A. E., J. C. Verdoes, F. A. G. Reubsaet, J. H. P. Hackstein, C. van der Drift, and H. J. M. Op den Camp. 2003. Promicromonospora pachnodae sp. nov., a member of the (hemi)cellulolytic hindgut flora of larvae of the scarab beetle Pachnoda marginata. Antonie Leeuwenhoek 83:135-148. [DOI] [PubMed] [Google Scholar]
- 2.Chou, Q., M. Russel, D. E. Birch, J. Raymond, and W. Bloch. 1992. Prevention of pre-PCR mis-priming and primer dimerization improves low-copy-number amplifications. Nucleic Acids Res. 20:1717-1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Douglas, A. E. 2007. Symbiotic microorganisms: untapped resources for insect pest control. Trends Biotechnol. 25:338-342. [DOI] [PubMed] [Google Scholar]
- 4.Durvasula, R. V., A. Gumbs, A. Panackal, O. Kruglov, S. Aksoy, R. B. Merrifield, F. F. Richards, and C. B. Beard. 1997. Prevention of insect-borne disease: an approach using transgenic symbiotic bacteria. Proc. Natl. Acad. Sci. USA 94:3274-3278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Egert, M., U. Stingl, L. D. Bruun, B. Pommerenke, A. Brune, and M. W. Freidrich. 2005. Structure and topology of microbial communities in the major gut compartments of Melolontha melolontha larvae (Coleoptera: Scarabaeidae). Appl. Environ. Microbiol. 71:4556-4566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Egert, M., B. Wagner, T. Lemke, A. Brune, and M. W. Friedrich. 2003. Microbial community structure in midgut and hindgut of the humus-feeding larva of Pachnoda ephippiata (Coleoptera: Scarabaeidae). Appl. Environ. Microbiol. 69:6659-6668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Heuer, H., M. Krsek, P. Baker, K. Smalla, and E. M. H. Wellington. 1997. Analysis of actinomycete communities by specific amplification of genes encoding 16S rRNA and gel-electrophoretic separation in denaturing gradients. Appl. Environ. Microbiol. 63:3233-3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hongoh, Y., L. Ekpornprasit, T. Inoue, S. Moriya, S. Trakulnaleamsai, M. Ohkuma, N. Noparatnaraporn, and T. Kudo. 2006. Intracolony variation of bacterial gut microbiota among castes and ages in the fungus-growing termite Macrotermes gilvus. Mol. Ecol. 15:505-516. [DOI] [PubMed] [Google Scholar]
- 9.Hongoh, Y., M. Ohkuma, and T. Kudo. 2003. Molecular analysis of bacterial microbiota in the gut of the termite Reticulitermes speratus (Isoptera; Rhinotermitidae). FEMS Microbiol. Ecol. 44:231-242. [DOI] [PubMed] [Google Scholar]
- 10.Huelsenbeck, J. P., and F. Ronquist. 2001. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17:754-755. [DOI] [PubMed] [Google Scholar]
- 11.Jackson, T. A., and T. R. Glare (ed.). 1992. Use of pathogens in scarab pest management. Intercept, Andover, United Kingdom.
- 12.Kuzina, L. V., E. D. Miller, B. Ge, and T. A. Miller. 2002. Transformation of Enterobacter gergoviae isolated from pink bollworm (Lepidoptera: Gelechiidae) gut with Bacillus thuringiensis toxin. Curr. Microbiol. 44:1-4. [DOI] [PubMed] [Google Scholar]
- 13.Lemke, T., U. Stingl, M. Egert, M. W. Friedrich, and A. Brune. 2003. Physiochemical conditions and microbial activities in the highly alkaline gut of the humus-feeding larva of Pachnoda ephippiata (Coleoptera: Scarabaeidae). Appl. Environ. Microbiol. 69:6650-6658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miller, L. J., and P. G. Allsopp. 2000. Identification of Australian canegrubs (Coleoptera: Scarabaeidae: Melolonthini). Invertebr. Taxon. 14:377-409. [Google Scholar]
- 15.Nalepa, C. A. 1991. Ancestral transfer of symbionts between cockroaches and termites: an unlikely scenario. Proc. R. Soc. Lond. B 246:185-189. [DOI] [PubMed] [Google Scholar]
- 16.Nubel, U., B. Engelen, A. Felske, J. Snaidr, A. Wieshuber, R. I. Amann, W. Ludwig, and H. Backhaus. 1996. Sequence heterogeneities of genes encoding 16S rRNAs in Paenibacillus polymyxa detected by temperature gradient gel electrophoresis. J. Bacteriol. 178:5636-5643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Peloquin, J., C. R. Lauzon, S. Potter, and T. A. Miller. 2002. Transformed bacterial symbionts re-introduced to and detected in host gut. Curr. Microbiol. 45:41-45. [DOI] [PubMed] [Google Scholar]
- 18.Posada, D., and K. A. Crandall. 1998. MODELTEST: testing the model of DNA substitution. Bioinformatics 14:817-818. [DOI] [PubMed] [Google Scholar]
- 19.Riehle, M. A., and M. Jacobs-Lorena. 2005. Using bacteria to express and display anti-parasite molecules in mosquitoes: current and future strategies. Insect Biochem. Mol. Biol. 35:699-707. [DOI] [PubMed] [Google Scholar]
- 20.Salles, J. F., F. A. De Souza, and J. D. van Elsas. 2002. Molecular method to assess the diversity of Burkholderia species in environmental samples. Appl. Environ. Microbiol. 68:1595-1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sanguinetti, C. J., E. D. Neto, and A. J. G. Simpson. 1994. Rapid silver staining and recovery of PCR products separated on polyacrylamide gels. BioTechniques 17:914-921. [PubMed] [Google Scholar]
- 22.Schloss, P. D., I. Delalibera, Jr., J. Handelsman, and K. F. Raffa. 2006. Bacteria associated with the guts of two wood-boring beetles: Anoplophora glabripennis and Saperda vestita (Cerambycidae). Environ. Entomol. 35:625-629. [Google Scholar]
- 23.Stingl, U., R. Radek, H. Yang, and A. Brune. 2005. “Endomicrobia”: cytoplasmic symbionts of termite gut protozoa form a separate phylum of prokaryotes. Appl. Environ. Microbiol. 71:1473-1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Swofford, D. L. 2000. PAUP*: phylogenetic analysis using parsimony (*and other methods). Sinauer Associates, Sunderland, MA.
- 25.Tang, C., F. Sun, X. Zhang, T. Zhao, and J. Qi. 2004. Transgenic ice nucleation-active Enterobacter cloacae reduces cold hardiness of corn borer and cotton bollworm larvae. FEMS Microbiol. Ecol. 51:79-86. [DOI] [PubMed] [Google Scholar]
- 26.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 24:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]