Abstract
Accurate assessment of health risks associated with bovine (cattle) fecal pollution requires a reliable host-specific genetic marker and a rapid quantification method. We report the development of quantitative PCR assays for the detection of two recently described bovine feces-specific genetic markers and a method for the enumeration of these markers using a Markov chain Monte Carlo approach. Both assays exhibited a range of quantification from 25 to 2 × 106 copies of target DNA, with a coefficient of variation of <2.1%. One of these assays can be multiplexed with an internal amplification control to simultaneously detect the bovine-specific genetic target and presence of amplification inhibitors. The assays detected only cattle fecal specimens when tested against 204 fecal DNA extracts from 16 different animal species and also demonstrated a broad distribution among individual bovine samples (98 to 100%) collected from five geographically distinct locations. The abundance of each bovine-specific genetic marker was measured in 48 individual samples and compared to quantitative PCR-enumerated quantities of rRNA gene sequences representing total Bacteroidetes, Bacteroides thetaiotaomicron, and enterococci in the same specimens. Acceptable assay performance combined with the prevalence of DNA targets across different cattle populations provides experimental evidence that these quantitative assays will be useful in monitoring bovine fecal pollution in ambient waters.
Fecal pollution continues to affect the quality of environmental waters despite the development of numerous methods to enumerate fecal indicator bacteria (FIB) and discriminate among different potential sources. The U.S. Environmental Protection Agency (EPA) currently recommends the enumeration of enterococci and/or Escherichia coli as standard indicators of fecal pollution for marine water and freshwater, respectively (37). For some time researchers and water quality managers have recognized the advantages and limitations of using culture-dependent FIB for water quality monitoring. Traditional membrane filtration and most-probable-number methods such as EPA method 1600 (38) and Enterolert (Idexx Laboratories Inc., Westbrook, ME) for enterococci and EPA method 1603 and Colilert-18 (Idexx Laboratories) for E. coli provide quantitative data for fecal indicators that have been associated with human health risks. However, fecal bacterial indicators such as enterococci and E. coli can persist or even grow in various extraintestinal habitats such as soils and sediments (1, 8, 12, 40, 41) and thus reappear in the water column long after initial fecal discharge events. In addition, these FIB are common to feces produced by humans and other animals, making it difficult to determine the origins of fecal contamination. These shortcomings have led researchers to identify alternative fecal indicators that can discriminate among animal sources.
There are currently a number of microbial source tracking (MST) techniques available that can determine the presence or absence of fecal contamination from specific animal groups. Quantitative MST methods are particularly promising because they can allow for the estimation of fecal concentrations from a particular animal group and be used to develop fate and transport, mechanistic, and probabilistic models of bacterial loading over a range of conditions in water bodies. Currently available host-specific quantitative PCR (qPCR) methods designed to detect bovine fecal pollution can discriminate only between ruminant and nonruminant sources (20, 25). The development of bovine-specific qPCR assays has been restricted in part by the limited amount of genetic variation in the rRNA genes, which have been most commonly targeted by host-specific PCR methods. Several researchers have asserted that chromosomal genes directly involved in bacterium-host interactions may retain increased levels of host-specific genetic variation, making them more useful for the development of host-specific qPCR assays (16, 31, 32). A recent metagenomic survey of a bovine fecal bacterial community using genome fragment enrichment has led to the identification of several hundred candidate host-specific chromosomal sequences (30, 31). However, there is concern that these presumptively host-specific sequences may not be abundant or uniform enough within host fecal bacterial populations to be as easily detected as the more highly conserved and multiple-copy rRNA genes, particularly when they are diluted in the environment. Based on these considerations, the next step in the development of these chromosomal bacterial gene sequences for MST applications is to design qPCR assays to evaluate their abundance relative to FIB rRNA genes and to establish their range of concentrations within host populations.
Here we report the development of two qPCR assays for the enumeration of previously described bovine-specific genetic markers (31). qPCR data analysis was achieved by employing a Markov chain Monte Carlo (MCMC) approach (M. Sivaganesan, S. Seifring, M. Varma, R. A. Haugland, and O. C. Shanks, submitted for publication). qPCR performance characteristics, including specificity, range of quantification (ROQ), limit of quantification, amplification efficiency, and precision, were defined for each assay. Internal amplification controls (IACs) were designed to monitor for the presence of inhibitors that could inadvertently copurify with target DNA and confound copy number estimations. Finally, the abundance of each bovine-specific genetic marker was measured by qPCR analysis in individual fecal samples randomly collected from five different cattle populations and compared to similarly determined quantities of rRNA genes from Bacteroidetes, Bacteroides thetaiotaomicron, and enterococci fecal bacteria. The analysis of several hundred fecal samples suggests that the genes targeted by these host-specific qPCR assays are both abundant and stable within host populations.
MATERIALS AND METHODS
Sample collection.
Two hundred four individual fecal samples and five wastewater samples were collected for comparisons as previously described (31). Fecal specimens represented a total of 16 different animal species that likely affect watersheds nationwide and six bovine herds from separate geographic locations (herds 1 to 4 [various locations in Washington, summer 2005], herd 5 [Wyoming, summer 2005], and herd 6 [West Virginia, summer 2002]). Wastewater samples represent five different human populations collected from Mason, OH; Lowery, OH; Dry Creek, OH; Fairfield, OH; and Dayton, OH.
DNA extraction from fecal and wastewater samples.
All DNA extractions were performed with the FastDNA kit for soils (Q-Biogene, Carlsbad, CA) as described previously (31). DNA extract yields were quantified with a NanoDrop ND-1000 UV spectrophotometer (NanoDrop Technologies, Wilmington, DE). A general Bacteroides-Prevotella 16S rRNA gene PCR assay was used to verify the presence of fecal bacterial DNA in each extract prior to qPCR analysis (4).
Oligonucleotides and primers.
TaqMan probe and primer assays targeting the rRNA genes of the FIB groups Enterococcus (Entero1), Bacteroidetes (GB342), and B. thetaiotaomicron (Btheta) are reported elsewhere (21, 28; A. D. Blackwood and R. T. Noble, presented at the 105th General Meeting of the American Society for Microbiology, Atlanta, GA, 2005). Bovine-specific assay probe and primer sequences (Table 1) were designed with Primer Express software (Applied Biosystems, Foster City, CA) based on alignments of 62 DNA sequences isolated from PCR products amplified by previously reported bovine-specific PCR assays one and two (31) (data not shown). Amplicons were incorporated into the pCR4-TOPO plasmid vector as described by the manufacturer (Invitrogen, Carlsbad, CA). Individual clones were isolated and sequenced as described elsewhere (30). DNA sequence reads were assembled with SeqMan II (DNAstar, Inc., Madison, WI) and aligned with ClustalW (36). Briefly, sequence analysis of the CowM2 locus yielded 32 sequences. Two variant sequences were observed, each with a single polymorphism (A→G and G→A, respectively) in the probe hybridization region. Thirty sequences were aligned for the CowM3 locus, with one variant sequence observed with a single polymorphism (A→G) in the 5′ region of the CowM3R primer (Table 1). Primers and TaqMan probes were designed using the default parameters of the Primer Express software (version 1.5; Applied Biosystems). Fluorogenic probes were 5′ labeled with 6-carboxyfluorescein (FAM), VIC, or tetrachlorofluorescein (TET) and 3′ labeled with 6-carboxytetramethylrhodamine (TAMRA). Optimal primer and probe reaction concentrations were determined according to a standard Applied Biosystems protocol (2).
TABLE 1.
Oligonucleotides, primers, and probes
| Assay | Primer and probe sequences (5′→3′) | Size (bp) | Reference or source |
|---|---|---|---|
| Btheta | Forward, CGTTCCATTAGGCAGTTGGT; reverse, ACACGGTCCAAACTCCTACG; probe, FAM-CTGAGAGGAAGGTCCCCCACATTGGA-TAMRA | 110 | Blackwood and Noblea |
| GB342 | Forward, GGGGTTCTGAGAGGAAGGT; reverse, AGTAGCGTGAAGGATGACGG; probe, FAM-CAATATTCCTCACTGCTGCCTCCCGTA-TAMRA | 129 | 28 |
| Entero1 | Forward, AGAAATTCCAAACGAACTTG; reverse, AATGATGGAGGTAGAGCACTGA; probe, FAM-TGGTTCTCTCCGAAATAGCTTTAGGGCTA-TAMRA | 92 | 21 |
| CowM2 | CowM2F, CGGCCAAATACTCCTGATCGT; CowM2R, GCTTGTTGCGTTCCTTGAGATAAT; probe, FAM-AGGCACCTATGTCCTTTACCTCATCAACTACAGACA-TAMRA | 92 | This study |
| CowM3 | CowM3F, CCTCTAATGGAAAATGGATGGTATCT; CowM3R, CCATACTTCGCCTGCTAATACCTT; probe, FAM-TTATGCATTGAGCATCGAGGCC-TAMRA | 122 | This study |
| Cow IAC | Frag1, CGGCCAAATACTCCTGATCGTCCTCTAATGGAAAATGGATGGTATCTATTACTCATAGGCACTAGGAACAGGCGGCGACGAAACTACAGACAAAATTATCTCAAGAACG; Frag2, CCATACTTCGCCTGCTAATACCTTAAGGACATAGGCTTGTTGCGTTCCTTGAGATAATTTTGTCTGTAGTTTCGTCGCCGCCTGTTCCTAGTGCCTATGAGTAATAGATA; probe, VIC-TAGGAACAGGCGGCGACGA-TAMRAb | 152 | 42; this study |
Presented at the 105th General Meeting of the American Society for Microbiology.
The TaqMan probe was modified from the previously reported UT probe (42).
Putative bovine-specific qPCR CowM2 and CowM3 primer probe sets (Table 1) were tested for specificity with animal fecal and wastewater sample composites (5 ng DNA template per PCR assay). Reference fecal sample composites represented 16 animal species, including Lama pacos (alpaca; n = 2), Anser sp. (Canadian goose; n = 12), Felis catus (cat, n = 10), Gallus gallus (chicken, n = 10), Bos taurus (bovine, n = 60), Odocoileus virginianus (deer, n = 6), Canis familiaris (dog, n = 10), Anas sp. (duck, n = 12), Capra aegagrus (goat, n = 10), Equus caballus (horse, n = 5), Homo sapiens (human, n = 16), Pelecanus sp. (pelican, n = 5), Sus scrofa (pig, n = 12), Laridae (gull, n = 12), Ovis aries, (sheep, n = 10), and Meleagris sp. (turkey, n = 7), and wastewater primary effluent (n = 5).
Genomic DNA preparations from pure bacterial cultures.
American Type Culture Collection (ATCC) bacterial strains were used to prepare genomic DNA calibration standards for the general FIB qPCR assays as previously described (17; Sivaganesan et al., submitted).
Construction of IACs and plasmid DNA standards.
Two plasmid DNA constructs were developed using the composite primer technique (34). Plasmid DNA constructs were designed to function as IAC DNA targets that can be spiked into DNA extracts to monitor for PCR inhibition and also as potential plasmid DNA standards for qPCR assay calibration curves (8). Both IAC constructs were designed to generate PCR amplicons ranging from 9 to 36 bp longer than the corresponding native DNA templates to help ensure that the native DNA PCR assays maintained a competitive edge in multiplex applications. In addition, each IAC contains a single site for hybridization of a unique TaqMan VIC- or TET-labeled probe sequence flanked by multiple primer binding sequences (Table 1; Fig. 1). To build the bovine assay IAC constructs, long oligonucleotides (>100 bp) (Table 1) containing multiple primer sequences (31) were designed such that their 3′ ends overlapped. The two overlapping fragments were then combined into a single DNA molecule using overlap extension PCR (18). The IAC construct for the FIB assays was built as previously described (Sivaganesan et al., submitted). IAC constructs were then inserted into the pCR4 TOPO plasmid vector (Invitrogen), and the resulting recombinant plasmids were purified from transformed E. coli cell cultures using a Qiagen plasmid purification kit (Qiagen, Valencia, CA). IAC plasmid DNA was linearized by a NotI restriction digestion (New England BioLabs, Beverly, MA), quantified with a NanoDrop ND-1000 UV spectrophotometer (NanoDrop Technologies), and diluted in 10 mM Tris, 0.1 mM EDTA (pH 8.0) to generate samples ranging from approximately 25 to 2.5 × 106 molecules of template DNA for bovine-specific assays and from approximately 100 to 4 × 104 molecules for the FIB assays.
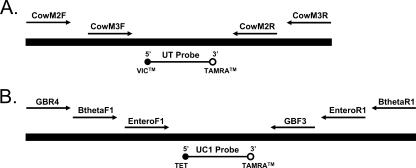
FIG. 1.
Diagram of bovine-specific (A) and fecal indicator (B) plasmid DNA composite IAC constructs. The bovine-specific IAC (152 bp) consists of a VIC-labeled universal probe binding site flanked by primer sequences for CowM2 (118 bp) and CowM3 (131 bp) qPCR assays. The fecal indicator IAC (261 bp) contains a binding site for a TET-labeled universal probe and primer sequences for GB342 (160 bp) (28), Btheta (146 bp) (Blackwood and Noble, presented at the 105th General Meeting of the American Society for Microbiology), and Entero1 (106 bp) (21) qPCR assays (Table 1).
qPCR assays and quantification.
Five qPCR assays were used in this study, including CowM2, CowM3, GB342, Btheta, and Entero1 (Table 1). Amplification was performed in either an ABI Prism 7000 or 7900 HT fast real-time sequence detector (Applied Biosystems). Reaction mixtures (25 μl) contained 1× TaqMan universal PCR master mix with AmpErase uracil-N-glycosylase (Applied Biosystems), 0.2 mg/ml bovine serum albumin (Sigma), 1 μM of each primer, 80 nM FAM- or VIC-labeled TaqMan probe (Applied Biosystems), and either 1 to 100 ng genomic DNA (fecal and wastewater samples), 25 to 2.5 × 106 target gene copies (bovine IAC plasmid DNA), or 100 to 4 × 104 target gene copies (FIB IAC plasmid or purified genomic DNA). Reaction mixtures for multiplex applications were the same as described above with the additions of both 80 nM of VIC- or TET-labeled TaqMan probes for IAC plasmid DNA and 80 nM of FAM-labeled TaqMan probe for native DNA targets and of 25 to 50 copies of IAC plasmid DNA together with the genomic DNA samples. All reactions were performed in triplicate in MicroAmp optical 96-well reaction plates with MicroAmp optical caps (Applied Biosystems). Thermal conditions were as follows: 50°C for 2 min to activate uracil-N-glycosylase, followed by 10 min of incubation at 95°C to activate AmpliTaq Gold enzyme and then 40 cycles of a short denaturation at 95°C for 15 s and a combined annealing and primer extension phase at 60°C for 1 min. Data were initially analyzed with Sequence Detector software (version 2.2.2) at threshold determinations of 0.08 for bovine-specific assays (CowM2 and CowM3) and 0.03 for FIB assays (GB342, Btheta, and Entero1). Threshold cycle (CT) values were exported to Microsoft Excel for further statistical analysis. Contaminating DNA from the laboratory environment, including other samples and amplification products, was limited by using dedicated equipment and separate laboratories for DNA extraction, PCR reagent mixing, addition of DNA template to PCR microtubes, and qPCR amplification. In addition, a minimum of three no-template amplifications with purified water substituted for template DNA were performed for each 96-well PCR.
Calculations and statistical analysis.
Master calibration curves, unknown DNA concentration estimates, and credible intervals were determined using an MCMC approach (6, 13-15; Sivaganesan et al., submitted). MCMC calculations were performed using the publicly available software WinBUGS version 1.4.1 (http://www.mrc-bsu.cam.ac.uk/bugs) (22). Run-to-run variability was incorporated using a hierarchical Bayesian model. Standard noninformative priors and the prior recommended by DuMouchel (35) were used to obtain posterior distributions of unknown parameters. See the supplemental material for lists containing the WinBUGS program code and resulting data output used to develop modified master calibration curves for CowM2 and Cow M3 plasmid DNA standards. Posterior mean, standard deviation, and lower and upper 95% credible bounds for most of the unknown parameters are also given in the supplemental material.
One-way analysis of variance (ANOVA) tests comparing simplex and multiplex CT values from reactions with a fixed amount of IAC were used to define the range where competition from target DNA did not interfere with quantification of the IAC. The precision of CT measurements determined from plasmid or genomic DNA standards was expressed as a percent coefficient of variation (%CV) (standard deviation expressed as a percentage of the mean). To estimate within-herd variance (σ2) and differences between herds, two-way nested ANOVA was used. In this analysis, bovine population was a fixed factor and fecal samples were the random factor nested within a respective bovine population. ANOVA tests were performed using SAS (SAS Institute, Cary, NC) with the procedures “PROC MIXED” and “PROC GLM” (27).
RESULTS
Calibration curves and ROQ.
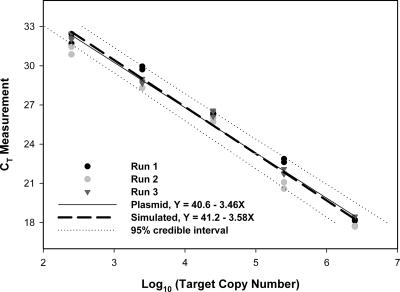
To generate calibration curve equations for FIB assays, CT values from four independent runs were plotted relative to the corresponding dilution of genomic DNA standards extracted from enumerated cell suspensions of the respective ATCC bacterial strains (Table 2). The MCMC approach was used to simultaneously estimate the mean difference in slopes and intercepts between IAC plasmid and genomic DNA standard curves reported elsewhere (Sivaganesan et al., submitted) and to adjust plasmid-derived standard curves for the CowM2 and CowM3 assays (Table 2; Fig. 2). Because the standard curves for FIB assays and bovine-specific assays are estimated simultaneously, errors in estimates for all model parameters are included in confidence bounds for calibration equations. After incorporation of these mean adjustments, the posterior mean slopes and intercepts for the bovine-specific assays were −3.58 ± 0.11 and 41.2 ± 0.60 (CowM2) and −3.67 ± 0.08 and 42.6 ± 0.42 (CowM3), respectively. Both bovine-specific assays exhibited an ROQ of 25 to 2.5 × 106 copies, while the ROQs of all FIB assays ranged from 100 to 4 × 104 copies (in both cases, the entire range of copy numbers tested). Precision across defined ROQs for all assays was less than 2.1%, and amplification efficiencies ranged from 1.87 to 2.02 (Table 2). No-template controls indicated the absence of extraneous DNA molecules in all qPCR experiments in this study.
TABLE 2.
Calibration curve equations and performance characteristics of qPCR assays
| Assay | DNA typea | Calibration equation | Amplification efficiencyc | ROQ (copies) for target DNA | %CV across ROQ | Methodd |
|---|---|---|---|---|---|---|
| CowM2 | Plasmid | y = −3.58x + 41.2b | 1.90 | 25-2.5 × 106 | 2.08 | Multiplex |
| CowM3 | Plasmid | y = −3.67x + 42.6b | 1.87 | 25-2.5 × 106 | 1.18 | Simplex |
| Btheta | Genomic | y = −3.43x + 39.1 | 1.96 | 100-4 × 104 | 0.63 | Multiplex |
| GB342 | Genomic | y = −3.27x + 38.8 | 2.02 | 100-4 × 104 | 1.11 | Multiplex |
| Entero1 | Genomic | y = −3.53x + 38.2 | 1.92 | 100-4 × 104 | 1.57 | Multiplex |
DNA standards from which the calibration equation was generated (Fig. 2).
The calibration equation was adjusted based on posterior mean differences in slope and intercept between genomic and plasmid DNA-generated standard curves from Btheta, GB342, and Entero1 assays.
Amplification efficiency = 10(1/−slope).
Either a simple approach or multiplex strategy where the target DNA was simultaneously detected with an IAC.
FIG. 2.
MCMC simulated calibration curve for the qPCR CowM2 assay, adjusted to represent mean posterior differences in genomic and IAC plasmid standard curves from three independent general fecal indicator qPCR assays (Sivaganesan et al., submitted).
Multiplex qPCR.
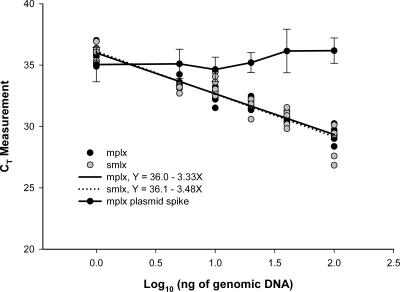
All qPCR assays were tested for potential multiplex applications to monitor accumulation of target DNA and a fixed quantity of a PCR inhibition control simultaneously. Two competitive composite IAC constructs were built where target DNA and an IAC can be coamplified with a common set of primers, under the same reaction conditions, in the same PCR tube (Fig. 1). The target DNA and IAC product were then detected with different fluorescently labeled TaqMan probes. Assays were considered suitable for multiplexing with an IAC if (i) there was no significant difference (P > 0.05) between simplex and multiplex standard curve intercepts and slopes based on analysis of variance and (ii) a fixed amount of IAC could be quantified across a range of genomic DNA standard concentrations. No significant difference was observed between simplex and multiplex curve intercepts and slopes (P > 0.05) for all assays (Fig. 3). One-way ANOVA tests comparing simplex and multiplex CT values from a fixed amount of IAC were used to define the range where competition from target DNA did not interfere with quantification of the IAC (Table 3; Fig. 3). Thresholds for the identification of PCR inhibition were established based on simplex mean CT and standard deviation values for the detection of either 25 or 50 copies of IAC (Table 3).
FIG. 3.
CowM2 multiplex qPCR with competitive IAC. The simplex (smlx) and multiplex (mplx) curves were generated from seven dilutions of bovine fecal DNA extract in the absence or presence of 50 copies of IAC plasmid DNA standard, respectively. Error bars indicate standard deviations.
TABLE 3.
IAC multiplex qPCR performance characteristics and thresholds for monitoring inhibition
| Assay | IAC spike (copies)a | Rangeb | %CV across range | Inhibition threshold range (CT)c
|
|
|---|---|---|---|---|---|
| Mean | SD | ||||
| CowM2 | 50 | 1-100 ng | 3.39 | 35.4 | 1.31 |
| CowM3 | 50 | 1 ng | |||
| Btheta | 25 | 100-4 × 103 cps | 7.00 | 35.7 | 2.37 |
| GB342 | 25 | 100-4 × 103 cps | 2.97 | 33.6 | 1.24 |
| Entero1 | 25 | 100-200 cps | 1.85 | 34.5 | 0.67 |
Number of copies of IAC plasmid control DNA added to each multiplex qPCR setup.
Span of competitor genomic DNA concentrations where accurate quantification of spiked IAC is possible.
Expected CT value from an IAC spike in the absence of PCR inhibitors.
Specificity of host-specific qPCR assays.
Primer specificity was tested for the CowM2 and CowM3 assays against DNAs from hundreds of fecal and wastewater samples, including samples collected from six different bovine populations. Host-specific assays amplified only DNA from composite bovine fecal samples, with CT values ranging from 29.5 ± 0.86 to 34.2 ± 0.32 (Table 4). Bovine IAC detection levels from CowM2 multiplex qPCR assays indicated the absence of PCR inhibitors in all composite fecal DNA extracts (expected mean CT of 35.4 ± 1.3; Table 3). All individual fecal samples used to generate composite DNA template mixtures yielded the expected PCR product when amplified with Bacteroides-Prevotella 16S rRNA gene-specific primers 32F and 708R (5), indicating the presence of amplifiable DNA from this group of fecal microorganisms.
TABLE 4.
Specificity of CowM2 and CowM3 qPCR assays
| Sample type | Localea | No. |
CT for:
|
|||||
|---|---|---|---|---|---|---|---|---|
| CowM2 multiplex assay
|
CowM3 simplex assay (FAM)
|
|||||||
| CowM2 (FAM)
|
IAC 50 copies (VIC)b
|
|||||||
| Avg | SD | Avg | SD | Avg | SD | |||
| Alpaca | WV | 2 | 34.0 | 0.9 | ||||
| Canadian Goose | GA | 12 | 34.4 | 0.24 | ||||
| Cat | WV | 10 | 36.1 | 0.79 | ||||
| Chicken | GA | 10 | 34.3 | 0.26 | ||||
| Bovine herd 1 | WA | 10 | 31.7 | 1.8 | 33.7 | 0.7 | 31.9 | 0.6 |
| Bovine herd 2 | WA | 10 | 30.8 | 0.38 | 34.7 | 1 | 29.46 | 0.86 |
| Bovine herd 3 | WA | 8 | 33.2 | 0.61 | 35.5 | 0.32 | 31.8 | 0.71 |
| Bovine herd 4 | WA | 10 | 34.0 | 0.78 | 35.8 | 0.47 | 32.8 | 0.85 |
| Bovine herd 5 | WY | 10 | 34.2 | 0.32 | 35.8 | 1.27 | 32.3 | 0.5 |
| Bovine herd 6 | WV | 12 | 31.6 | 0.58 | 34.6 | 0.55 | 30.8 | 0.34 |
| Deer | WV | 6 | 34.5 | 0.14 | ||||
| Dog | WV | 10 | 34.8 | 0.33 | ||||
| Duck | GA | 12 | 34.2 | 0.89 | ||||
| Goat | DE | 10 | 34.5 | 0.62 | ||||
| Horse | WV | 5 | 34.4 | 0.52 | ||||
| Human | WV | 16 | 34.4 | 0.17 | ||||
| Pelican | FL | 5 | 34.4 | 1.24 | ||||
| Pig | WV | 12 | 34.9 | 1.03 | ||||
| Sea gull | GA | 12 | 34.1 | 0.8 | ||||
| Sheep | DE | 10 | 34.4 | 0.76 | ||||
| Turkey | OH | 7 | 34.6 | 0.41 | ||||
| Wastewater | OH | 5 | 34.5 | 0.4 | ||||
| Total or avg | 204 | 34.7 | 0.6 | |||||
WV, West Virginia; GA, Georgia; WY, Wyoming; DE, Delaware; FL, Florida; OH, Ohio.
IAC detection levels indicate the absence of inhibition in all composites (expected CT for IAC spike, 35.4 ± 1.3 [Table 3]).
Quantification of fecal bacterial genes in bovine populations.
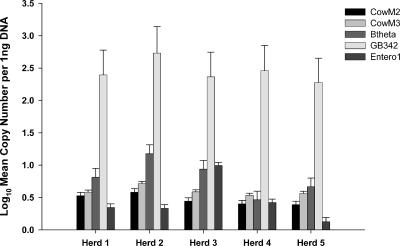
Individual bovine fecal samples were randomly collected from six herds and analyzed to estimate the host distributions of bovine-specific assays, to describe target DNA variability among and between populations, and to compare the relative abundances of each target DNA to one another. The bovine samples from West Virginia were used only to evaluate estimated host distribution because they were collected 36 months earlier and may have degraded over time. PCR inhibition was not detected in any of the bovine fecal DNA extracts based on IAC CT values from multiplex qPCR assays (data not shown). The CowM2, GB342, and Btheta assays tested positive for all individual bovine fecal samples. Estimated host distributions decreased slightly for the CowM3 (98%) and Entero1 (89.5%) assays. Within-herd variance (σ2) ranged from 0.80 to 5.95 for CowM2 and from 0.54 to 3.27 for CowM3. FIB assay variances fluctuated from 0.06 to 1.12 (GB342), from 0.36 to 2.43 (Btheta), and from 0 to 9.51 (Entero1). Of the 10 possible pairwise comparisons for five different herds, significant target copy number differences (P < 0.05) were observed in 5 to 7 pairings for the different assays. Only the pairing between herd 2 and herd 3 was significantly different (P < 0.003) for all assays. The CowM2 (six pairings showing differences) and CowM3 (five pairings showing differences) assays shared five pairings in common. A comparison of target DNA relative abundances between all assays was achieved by normalizing data sets to 1 ng of DNA template (Fig. 4). The log10 mean copy number of target DNA was measured using 5 ng DNA template for each bovine-specific assay and 2 ng DNA template for all FIB assays.
FIG. 4.
Quantification of bacterial genes from different animal herd composite DNA extracts. Error bars represent the upper 95% credible interval for each estimated measurement. Log10 mean copy number was normalized to 1 ng to compare relative abundances of each DNA target.
DISCUSSION
Bovine-specific qPCR.
We report on two qPCR assays that detected only bovine fecal DNA when tested against a panel of samples representing 16 animal species, including agriculturally important animals such as swine, poultry, horse, and sheep as well as humans and many wildlife species (Table 4). The CowM3 qPCR assay demonstrated an increased level of specificity over the previously reported end point PCR assay (Bac2F and Bac2R primers) targeting the same DNA sequence by exclusively detecting bovine fecal samples (31). In addition, host-specific qPCR assays successfully detected 98 to 100% of the individual bovine samples collected over a 36-month time span, where the host distributions of their conventional PCR counterparts ranged from 80 to 91% (31). Increases in specificity and estimated host distribution could be attributed to differences in primer design, the additional selectivity afforded by the TaqMan probe, shorter amplicons, and/or an elevated limit of detection due to a higher number of amplification thermal cycles.
qPCR is one of the most precise methods available for gene quantification, with %CV levels ranging from 2 to 20 (26). However, qPCR performance can be substantially influenced by the sample quality of DNA. DNA isolation from fecal samples may not remove all substances that can interfere with PCR, and the degree of interference may vary between samples. Therefore, internal controls designed to evaluate the suitability of isolated DNA for quantitative analysis were included for each DNA extract. In order to reduce the potential for variability in performance associated with running target and internal control assays in separate reaction tubes, we employed a multiplex strategy with the CowM2, GB342, Btheta, and Entero1 qPCR assays (Tables 2 and 3). Each fecal DNA extract was spiked with a fixed concentration of an IAC to serve as a control to monitor for the presence of amplification inhibitors. If the mean CT of the IAC standard was within the simplex detection mean value and competition from the genomic target DNA was negligible (Table 3; Fig. 3), then sample DNA quality was adequate for quantification.
Plasmid DNA standards and master calibration curves.
Quantification of the amount of initial template in bovine fecal samples requires a reliable set of DNA standards, typically prepared from genomic DNA isolated from a bacterial culture. Unfortunately, the competitive DNA hybridization method used to identify bovine-specific markers resulted in short, incomplete gene sequences that provided no definitive information regarding bacterial species of origin. Consequently, no bacterial strain was available for use as a genomic DNA standard with these assays. An alternative to bacterial genomic DNA standards is the use of plasmid-derived DNA standards to generate calibration curves. However, the assumption that plasmid and genomic DNA standards amplify with the same efficiency must be made. To account for potential differences in amplification performance between plasmid and genomic DNA approaches, an MCMC approach was used to estimate the mean difference in slope and intercept from fitted-curve equations for plasmid and genomic DNAs produced from three independent qPCR assays (Sivaganesan et al., submitted). Using the same MCMC approach, these differences were applied to the generation of bovine-specific qPCR plasmid DNA calibration curves (Fig. 2). The MCMC approach was ideal because it accounted not only for observed mean differences in plasmid and genomic DNA standards but also for intra- and interassay variation.
For many qPCR applications, calibration curves are typically generated with each assay run and quantification is only valid for that particular experiment (33). Factors such as quality of reagents, pipette calibration, and run-to-run variability can introduce uncertainty into data interpretation. However, including a standard curve with each assay run is expensive and impractical due to the limited number of reaction wells available in many qPCR instruments. Therefore, we used master calibration curves for each qPCR assay compiled from multiple independent experiments. Because calibration curves were generated from many independent runs, the intercept and slope parameters incorporate both intra- and interrun variation in the data analysis. The master calibration curves were acceptable because (i) there was no significant difference in the slopes of fitted curves between independent runs (P > 0.05), (ii) the coamplification of an IAC standard in multiplex applications provides evidence for optimal PCR performance in each reaction, (iii) the precision (%CV) over the ROQ between runs averaged less than 2.1%, and (iv) the MCMC approach accounts for run-to-run variation when creating fitted calibration curves with a 95% credible interval. Although the use of master calibration curves was adequate for this study, it remains untested whether this approach can be used over longer periods of time or between different laboratories.
Abundance of host-specific and fecal indicator genes.
Several researchers have reasoned that bacterial DNA sequences encoding proteins involved in host-bacterium interactions hold greater potential as the basis of host-specific PCR assays (16, 31, 32). However, most host-specific PCR assays target ribosomal genes which are not directly involved in bacterium-host interactions (4, 5, 9, 10). Ribosomal genes make excellent genetic targets because they are highly conserved and each bacterial genome can contain up to 15 copies (19), making it easier to detect a small number of cells at low concentrations. In contrast, most other bacterial genes, including many involved in host-bacterium interactions, contain a single copy per bacterial genome. Genes involved in bacterium-host interactions may exhibit higher levels of host specificity, but they may not be evenly distributed or abundant enough to detect in environmental matrices such as ambient surface waters. Forty-eight individual bovine fecal samples were analyzed to compare the number of bovine-specific chromosomal genes with well-described rRNA genes from Bacteroidetes, B. thetaiotaomicron, and enterococci. The general Bacteroidetes assay (GB342) yielded the highest gene concentration in all samples, which supports previous research reporting that Bacteroidetes makes up a large portion of the fecal bacterial community (23). The B. thetaiotaomicron 16S rRNA gene was the second most prevalent marker, equaling 0.07% of the detected general Bacteroidetes population (Fig. 4). The relative abundance of bovine-specific genes was greater than that of the enterococcal 23S rRNA genes in almost all samples tested. Enterococci are routinely detected in fecal polluted waters and are used for water quality monitoring (38). Although bovine-specific genes are not as abundant as the 16S rRNA gene Bacteroidetes markers, they are clearly present in sufficient quantities to be used as water quality indicators.
The ideal host-specific genetic marker should be consistently detected within host populations. However, factors such as host age and health could impact the stability of the marker within a population, while diet, climate, and geographic location may differ significantly between host populations. qPCR analysis of fecal samples collected from five bovine populations followed by pairwise comparisons of target DNA concentrations indicated that there are minimal differences within a population but significant differences between most of the herds tested. All qPCR assays exhibited less than 2.6% dispersion from a particular population mean [(two-way ANOVA standard deviation/mean) × 100] regardless of population or qPCR gene target. The GB342 assay displayed the smallest dispersion (range, 0.6 to 1.6%), while the Entero1 assay demonstrated the broadest (range, 0.5 to 2.4%). Fluctuations in dispersion, particularly for these latter assays, could result from sporadic target DNA concentrations among individuals but most likely reflect increased uncertainty associated with the amplification of lower levels of starting template. Regardless of the reason, low dispersion percentages suggest that the bovine-specific chromosomal DNA genes can be detected with the same level of confidence as the FIB rRNA genes within the tested cattle populations. Two patterns emerged based on pairwise comparisons of each bovine population. First, bovine herds 2 and 3 were significantly different (P < 0.003) regardless of the qPCR assay. Herd 2 originated from a small animal feeding operation in Washington, while herd 3 resides in a high-altitude open range in Wyoming. Differences in elevation, climate, and diet may account for this incongruity across FIB and host-specific gene concentrations. Second, the bovine-specific assays resulted in almost identical herd pairings. Based on functional annotation of each DNA target, the CowM2 sequence is involved in energy metabolism and electron transport and the CowM3 sequence in degradation of surface polysaccharides and lipopolysaccharides (31). Similar pairings of different bovine populations may have occurred by chance, or these genes may participate in similar host-bacterium-related metabolic pathways.
Quantitative MST.
Fecal pollution of ambient waters originating from both humans and other animals poses a threat to human health (3, 7, 11, 24, 39). MST studies are initiated to identify fecal pollution from different animal groups in order to prioritize polluted areas for restoration. PCR-based methods have become popular due to high levels of specificity, expedient sample processing, no requirement for culturing indicator fecal bacteria, and the potential to detect single bacterial cells. The majority of the available PCR-based MST methods rely on end point amplification, where the PCR product is detected after completion of a fixed number of thermal cycles. End point PCR methods yield qualitative data suggesting that a host-specific DNA target is either present or absent. There is no information regarding the concentration of the target DNA. Qualitative data can alert a watershed manager to the presence of fecal pollution in a sampling area and help determine which animal source(s) contributes to the total fecal load. They can also be used to make statistical inferences by collecting samples from the same location over a period of time and calculating a frequency of detection (29). However, this approach has limited statistical power and requires a large number of samples.
Researchers are now beginning to explore the application of qPCR approaches for MST. qPCR methods can extend the utility of MST applications by supplying data regarding the concentration of source-specific fecal pollution. However, in order to make inferences about the concentration of fecal pollution in a water sample, an assumption must be made about the uniformity of target DNA concentration within a host population. Our study suggests that target DNA concentrations remain relatively consistent within the tested cattle populations for all five qPCR assays but can fluctuate between groups. Based on these initial findings, it is evident that the implementation of the CowM2 and CowM3 assays will require a priori characterization of DNA target abundance in local cattle populations to establish a concentration range that can be used to determine the fecal load from a particular source. It will also be necessary to determine the stability of the DNA target range of concentration over the course of the water sampling time period. This could be accomplished by periodically testing randomly selected fecal samples from host populations impacting the watershed of interest.
Our qPCR assays are selective for bovine fecal pollution and can quantify as few as 25 copies of target DNA per reaction with a high degree of precision. DNA targets are widely distributed among host animals and more abundant than fecal enterococci in almost all fecal samples tested. It is clear that these bovine-specific qPCR assays merit further evaluation in other regions and may prove useful for quantitative MST applications.
Supplementary Material
Acknowledgments
We are grateful to James E. Graham and Mark Rodgers for critical review of the manuscript. We also thank Sam Hayes for the use of laboratory space. In addition, special thanks go to Stephanie Harris for fecal sample collection efforts.
Any opinions expressed in this paper are those of the authors and do not necessarily reflect the official positions and policies of the U.S. EPA, and any mention of products or trade names does not constitute recommendation for use.
Footnotes
Published ahead of print on 7 December 2007.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Anderson, K. L., J. E. Whitlock, and V. J. Harwood. 2005. Persistence and differential survival of fecal indicator bacteria in subtropical waters and sediments. Appl. Environ. Microbiol. 71:3041-3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Applied Biosystems. 2002. TaqMan universal PCR master mix protocol. Applied Biosystems, Foster City, CA.
- 3.Balarajan, R., V. Soni Raleigh, P. Yuen, D. Wheeler, D. Machin, and R. Caftwright. 1991. Health risks associated with bathing in sea water. Br. Med. J. 303:1444-1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bernhard, A. E., and K. G. Field. 2000. Identification of nonpoint sources of fecal pollution in coastal waters by using host-specific 16S ribosomal DNA genetic markers from fecal anaerobes. Appl. Environ. Microbiol. 66:1587-1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bernhard, A. E., and K. G. Field. 2000. A PCR assay to discriminate human and ruminant feces on the basis of host differences in Bacteroides-Prevotella genes encoding for 16S rRNA. Appl. Environ. Microbiol. 66:4571-4574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brooks, S. P. 1998. Markov chain Monte Carlo method and its application. Statistician 47:69-100. [Google Scholar]
- 7.Covert, T. C. 1999. Salmonella, waterborne pathogens. AWWA manual M48. American Water Works Association, Denver, CO.
- 8.Desmarais, T. R., H. M. Solo-Babriele, and C. J. Palmer. 2002. Influence of soil on fecal indicator organisms in a tidally influenced subtropical environment. Appl. Environ. Microbiol. 68:1165-1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dick, L. K., A. E. Bernhard, T. J. Brodeur, J. W. Santo Domingo, J. M. Simpson, S. P. Walters, and K. G. Field. 2005. Host distributions of uncultivated fecal Bacteroidales bacteria reveal genetic markers for fecal source identifcation. Appl. Environ. Microbiol. 71:3184-3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dick, L. K., M. T. Simonich, and K. G. Field. 2005. Microplate subtractive hybridization to enrich for Bacteroidales genetic markers for fecal source identification. Appl. Environ. Microbiol. 71:3179-3183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dufour, A. P. 1984. Health effects criteria for fresh recreational waters. U.S. Environmental Protection Agency, Washington, DC.
- 12.Fujioka, R., C. Sian-Denton, M. Borja, J. Castro, and K. Morphew. 1999. Soil: the environmental source of Escherichia coli and enterococci in Guam's streams. J. Appl. Microbiol. 85:83S-89S. [DOI] [PubMed] [Google Scholar]
- 13.Gelfand, A. E., and A. F. M. Smith. 1990. Sampling based approaches to calculating marginal densities. J. Am. Stat. Assoc. 85:398-409. [Google Scholar]
- 14.Gelman, A., J. C. Carlin, H. Stern, and D. B. Rubin. 1995. Bayesian data analysis. Chapman & Hall, New York, NY.
- 15.Gilks, W. R., S. Richardson, and D. J. Spiegelhalter. 1995. Markov chain Monte Carlo in practice. Chapman & Hall, New York, NY.
- 16.Hamilton, M. J., T. Yan, and M. J. Sadowsky. 2006. Development of goose- and duck-specific DNA markers to determine sources of Escherichia coli in waterways. Appl. Environ. Microbiol. 72:4012-4019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haugland, R. A., S. C. Siefring, L. J. Wymer, K. P. Brenner, and A. P. Dufour. 2005. Comparison of Enterococcus measurements in freshwater at two recreational beaches by quantitative polymerase chain reaction and membrane filter culture analysis. Water Res. 39:559-568. [DOI] [PubMed] [Google Scholar]
- 18.Higuchi, R., B. Krummel, and R. K. Saiki. 1988. A general method of in vitro preparation and specific mutagenesis of DNA fragments: study of protein and DNA interactions. Nucleic Acids Res. 16:7351-7367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Klappenbach, J. A., P. R. Saxman, J. R. Cole, and T. M. Schmidt. 2001. rrndb: the ribosomal RNA operon copy number database. Nucleic Acids Res. 29:181-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Layton, A., L. McKay, D. Williams, V. Garrett, R. Gentry, and G. Sayler. 2006. Development of Bacteroides 16S rRNA gene TaqMan-based real-time PCR assays for estimation of total, human, and bovine fecal pollution in water. Appl. Environ. Microbiol. 72:4214-4224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ludwig, W., and K.-H. Schleifer. 2000. How quantitative is quantitative PCR with respect to cell counts? Syst. Appl. Microbiol. 23:556-562. [DOI] [PubMed] [Google Scholar]
- 22.Lunn, D. J., A. Thomas, N. Best, and D. Spiegelhalter. 2000. WinBUGS—a Bayesian modeling framework: concepts, structure, and extensibility. Stat. Comput. 10:325-337. [Google Scholar]
- 23.McBride, G. B., C. E. Salmond, D. R. Bandaranayake, S. J. Turner, G. D. Lewis, and D. G. Till. 1998. Health effects of marine bathing in New Zealand. Int. J. Environ. Health Res. 8:173-189. [Google Scholar]
- 24.Okhuysen, P. C., J. H. Chappell, J. H. Crabb, C. R. Sterling, and H. L. DuPont. 1999. Virulence of three distinct Cryptosporidium parvum isolates for healthy adults. J. Infect. Dis. 180:1275-1278. [DOI] [PubMed] [Google Scholar]
- 25.Reischer, G. H., D. C. Kasper, R. Steinborn, R. L. Mach, and A. H. Farnleitner. 2006. Quantitative PCR method for sensitive detection of ruminant fecal pollution in freshwater and evaluation of this method in alpine karstic regions. Appl. Environ. Microbiol. 72:5610-5614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed., vol. 1-3. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 27.SAS. 1990. SAS/STAT user's guide, version 6, 4th ed. SAS Institute Inc., Cary, NC.
- 28.Seifring, S. C., M. Varma, E. Atikovic, L. J. Wymer, and R. A. Haugland. Improved real-time PCR assays for the detection of fecal indicator bacteria in surface waters with different instrument and reagent systems. J. Water Health, in press. [DOI] [PubMed]
- 29.Shanks, O. C., C. Nietch, M. T. Simonich, M. Younger, D. Reynolds, and K. G. Field. 2006. Basin-wide analysis of the dynamics of fecal contamination and fecal source identification in Tillamook Bay, Oregon. Appl. Environ. Microbiol. 72:5537-5546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shanks, O. C., J. W. Santo Domingo, and J. E. Graham. 2006. Use of competitive DNA hybridization to identify differences in the genomes of bacteria. J. Microbiol. Methods 66:321-330. [DOI] [PubMed] [Google Scholar]
- 31.Shanks, O. C., J. W. Santo Domingo, R. Lamendella, C. A. Kelty, and J. E. Graham. 2006. Competitive metagenomic DNA hybridization identifies host-specific microbial genetic markers in cow fecal samples. Appl. Environ. Microbiol. 72:4054-4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shanks, O. C., J. W. Santo Domingo, J. Lu, C. A. Kelty, and J. E. Graham. 2007. Identification of bacterial DNA markers for the detection of human fecal pollution in water. Appl. Environ. Microbiol. 73:2416-2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shepley, D. P., and D. M. Wolk. 2004. Quantitative molecular methods: result standardization, interpretation, and laboratory quality control, p. 101. In D. H. Persing (ed.), Molecular microbiology: diagnostic principles and practice. ASM Press, Washington, DC.
- 34.Siebert, P. D., and J. W. Larrick. 1992. Competitive PCR. Nature 359:557-558. [DOI] [PubMed] [Google Scholar]
- 35.Spiegelhalter, D. J., K. R. Abrams, and J. P. Myles. 2003. Bayesian approaches to clinical trials and health-care evaluation. John Wiley & Sons Inc., New York, NY.
- 36.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.U.S. Environmental Protection Agency. 1986. Ambient water quality criteria for bacteria—1986. EPA 440/5-84/002. Criteria and Standards Division, U.S. Environmental Protection Agency, Washington, DC.
- 38.U.S. Environmental Protection Agency. 2000. Improved enumeration methods for recreational water quality indicators: enterococci and Escherichia coli. EPA-821-R-97-004. U.S. Environmental Protection Agency, Washington, DC.
- 39.Walters, S. P., V. P. G. Gannon, and K. G. Field. 2007. Detection of Bacteroidales fecal indicators and the zoonotic pathogens E. coli O157:H7, Salmonella, and Campylobacter in river water. Environ. Sci. Technol. 41:1856-1862. [DOI] [PubMed] [Google Scholar]
- 40.Wheeler Alm, E., J. Burke, and A. Spain. 2003. Fecal indicator bacteria are abundant in wet sand at freshwater beaches. Water Res. 37:3978-3982. [DOI] [PubMed] [Google Scholar]
- 41.Whitman, R. L., D. A. Shively, H. Pawlik, M. B. Nevers, and M. N. Byappanahalli. 2003. Occurrence of Escherichia coli and enterococci in Cladophora (Chlorophyta) in nearshore water and beach sand of Lake Michigan. Appl. Environ. Microbiol. 69:4714-4719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang, Y., D. Zhang, L. Wenquan, J. Chen, Y. Peng, and W. Cao. 2003. A novel real-time quantitative PCR method using attached universal template probe. Nucleic Acids Res. 31:e123. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.