Abstract
RT-RiboSyn measures the specific rate of ribosome synthesis in distinct microbial populations by measuring the generation rate of precursor 16S rRNA relative to that of mature 16S rRNA when precursor 16S rRNA processing is inhibited. Good agreement was demonstrated between specific rate of ribosome synthesis and specific growth rate of Acinetobacter calcoaceticus.
A new molecular-biology-based method was developed to determine the specific growth rate of a distinct microbial population in an environmental sample by measurement of the specific rate of ribosome synthesis. This new method has been named RT-RiboSyn, based upon the use of reverse transcription and primer extension (RT&PE) to measure the specific rate of ribosome synthesis as shown in equation 1:
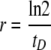 |
(1) |
where tD is the ribosome doubling time.
RT-RiboSyn utilizes the antibiotic chloramphenicol, which disrupts the processing of precursor 16S (pre-16S) rRNA to mature 16S rRNA over time (6). When exposed to chloramphenicol, the bacterial cells continue to generate pre-16S rRNA while the mature 16S rRNA remains constant (9), excluding low rates of degradation. Monitoring pre-16S rRNA synthesis has been used in previous work (2, 5, 8, 9) as an indicator of growth response, and pre-16S rRNA synthesis has been shown (5) to dramatically increase compared to the sum of pre-16S and mature 16S rRNA levels. RT-RiboSyn expands on this concept to determine a specific rate of ribosome synthesis by using a primer that specifically targets a population of interest.
Acinetobacter calcoaceticus (ATCC 23055) was cultured in nutrient broth and shaken at 250 rpm to generate four distinct growth conditions, including mid-log growth phase cultures incubated at 25, 30, and 35°C and a stationary-phase culture incubated at 30°C. Chloramphenicol (final concentration, 20 mg/liter) was added to a 50-ml sample from each culture to inhibit the secondary processing of pre-16S rRNA (6). Subsamples (4 ml) were collected from each 50-ml sample at 0, 10, 20, and 30 min of exposure to chloramphenicol, centrifuged (10,000 × g for 5 min), decanted, and stored promptly at −80°C.
The optical densities of the four cultures were measured periodically at 684 nm by using a spectrophotometer, and the specific growth rates were determined for the time of sample collection.
RNA was extracted from the subsamples by using the phenol-chloroform method (7), followed by purification with an RNAqueous kit (Ambion, Inc.). Residual DNA was removed using DNase I treatment (DNAfree kit by Ambion, Inc.). Finally, RT&PE using the ImProm-II reverse transcription system (Promega Corporation) was performed according to the manufacturer's instructions, with an MgCl2 concentration of 2.5 mM. The WellRed-labeled primers (Sigma-Genosys) used in the RT&PE reaction were Eub338 (5′ GCTGCCTCCCGTAGGAGT 3′) and Acin0659 (5′ CTGGAATTCTACCATCCTCTCCCA 3′), which target conserved sites of the pre-16S and 16S rRNA for all Eubacteria and Acinetobacter species, respectively (3, 4). The primer extension step was 1 h at 42°C and 47°C for the Eub338 and Acin0659 primers, respectively. Samples were then incubated at 85°C for 15 min to inactivate any RNase inhibitor present, followed by quenching in an ice slurry for 5 min. Samples were then mixed with RNase A (Sigma) cocktail (40 μl) and incubated at 37°C for 30 min. The RT&PE samples were analyzed by capillary electrophoresis with the CEQ 8000 genetic analysis system (Beckman-Coulter), with resulting electropherograms used for analysis (see the supplemental material). The size standards for the WellRed-labeled primers were the GenomeLab DNA size standard kit (600 nucleotides) for the Eub338 RT&PE samples and the MapMarker 1000 (Bioventures, Inc.) sizing standard for the Acin0659 RT&PE samples.
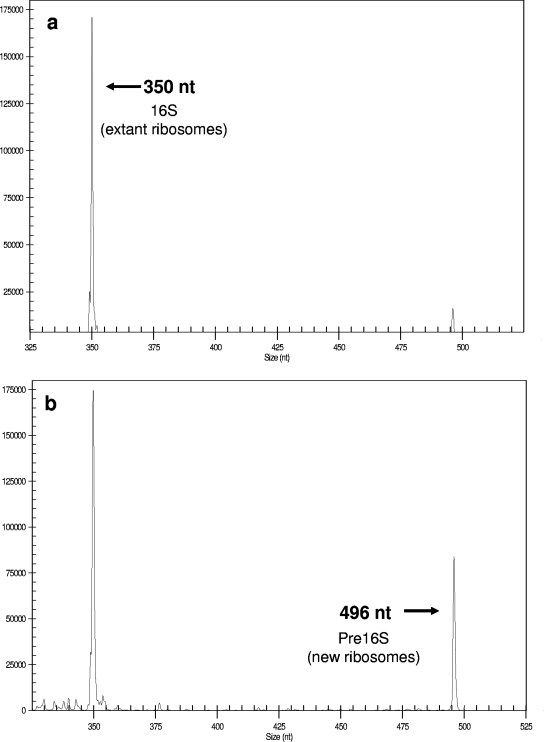
Capillary electrophoresis separates the two RT&PE products by length, and using the same primer for both products allows for the comparison of the two peaks by area (Fig. 1; also see the supplemental material). The fragment lengths correspond to the predicted lengths of the pre-16S and 16S RT&PE products (10). The ratio of the pre-16S and 16S RT&PE products (pre-16S:16S) was determined for each subsample and plotted versus time of chloramphenicol exposure. A trend line was fitted to these data, and the equation was determined. Using the slope of the equation, the ribosome doubling time was determined. Specific rates of ribosome synthesis were then determined using equation 1.
FIG. 1.
Electropherograms of RT-RiboSyn products derived from A. calcoaceticus incubated in nutrient broth at 25°C after exposure to chloramphenicol for 0 min (a) and 20 min (b). The WellRed-labeled Eub338 primer was used. nt, nucleotides.
Table 1 shows the mean pre-16S:16S values for each of the subsamples collected from the four cultures. For all subsamples, the low coefficient of variance indicates a strong reproducibility with the RT-RiboSyn method. For growing cultures, the mean pre-16S:16S values increased with longer exposure to chloramphenicol, which is consistent with earlier work (10).
TABLE 1.
General and statistical data for calculated ratios of precursor 16S rRNA to mature 16S rRNA
| Primer used | Culture temp (°C) | Timeb (min) | Mean pre-16S:16S ratio | SD | No. of samples | COVc (%) |
|---|---|---|---|---|---|---|
| Eub338 | 25 | 0 | 0.073 | 0.001 | 4 | 2.0 |
| 10 | 0.145 | 0.002 | 3 | 1.0 | ||
| 20 | 0.270 | 0.021 | 4 | 7.9 | ||
| 30 | 0 | 0.126 | 0.014 | 3 | 11.0 | |
| 10 | 0.265 | 0.013 | 4 | 4.7 | ||
| 30 | 0.571 | 0.011 | 3 | 1.9 | ||
| 35 | 0 | 0.266 | 0.006 | 3 | 2.2 | |
| 20 | 0.513 | 0.010 | 5 | 2.0 | ||
| 30 | 0.669 | 0.036 | 3 | 5.4 | ||
| 30a | 0 | 0.104 | 0.014 | 3 | 13.4 | |
| 10 | 0.104 | 0.019 | 3 | 17.8 | ||
| 20 | 0.093 | 0.012 | 3 | 12.6 | ||
| Acin0659 | 25 | 0 | 0.136 | 0.005 | 4 | 3.6 |
| 10 | 0.272 | 0.027 | 3 | 9.8 | ||
| 20 | 0.390 | 0.01 | 3 | 2.6 | ||
| 30 | 0 | 0.105 | 0.008 | 5 | 8.0 | |
| 10 | 0.195 | 0.006 | 4 | 10.6 | ||
| 20 | 0.265 | 0.002 | 3 | 2.3 | ||
| 35 | 0 | 0.473 | 0.025 | 5 | 5.3 | |
| 10 | 0.606 | 0.037 | 5 | 6.1 | ||
| 30 | 0.957 | 0.008 | 3 | 0.9 | ||
| 30a | 0 | 0.091 | 0.001 | 3 | 0.9 | |
| 10 | 0.094 | 0.003 | 3 | 2.8 | ||
| 30 | 0.088 | 0.007 | 4 | 8.2 |
Stationary-phase culture.
Time of exposure to chloramphenicol.
COV, coefficient of variance.
Table 2 provides a comparison of the specific rate of ribosome synthesis as measured by RT-RiboSyn and the specific growth rate of each culture as determined by spectrophotometry. When RT-RiboSyn was used with the Eub338 primer for the mid-log growth phase samples, the specific rates of ribosome synthesis were in good agreement (within 10.0%) with the specific growth rate measurements, while the Acin0659 primer resulted in slightly higher variation (within 21.0%).
TABLE 2.
Specific rates of ribosome synthesis and specific growth rates as calculated by RT-RiboSyn and spectrophotometry as well as percent differences between the measurements
| Culture temp (°C) | Result for indicated methoda
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Spectro photometry
|
RT-RiboSyn
|
|||||||
| Eub335
|
Acin0659
|
|||||||
| μ (h−1) | R2 | r (h−1) | R2 | % Diff | r (h−1) | R2 | % Diff | |
| 25 | 0.381 | 0.811 | 0.387 | 0.971 | 1.6 | 0.482 | 0.998 | 21.0 |
| 30 | 0.550 | 0.988 | 0.611 | 0.999 | 10.0 | 0.532 | 0.926 | −3.3 |
| 35 | 0.562 | 0.925 | 0.545 | 0.996 | −3.1 | 0.665 | 0.994 | 15.5 |
| 30b | 0.007 | 0.476 | −0.017 | 0.706 | NA | −0.004 | 0.358 | NA |
r, specific rate of ribosome synthesis; μ, specific growth rate; % diff, percent difference between the measurements; NA, not applicable. The R2 values of the linear regression are included.
Stationary-phase culture.
It is unclear why the specific rates of ribosome synthesis measured with the Acin0659 primer were different from those for the Eub338 primer. It has been shown that chloramphenicol completely prevents pre-16S rRNA degradation under all conditions in Escherichia coli (2). Assuming that this behavior holds true for A. calcoaceticus, degradation of mature 16S rRNA during accumulation of pre-16S rRNA may be the cause of the specific rate of ribosome synthesis being higher than the specific growth rate. For the 30°C stationary-phase samples, the calculated specific rates of ribosome synthesis and specific growth rates were very low for both methods. However, it is important to note that the RT-RiboSyn method clearly distinguishes between an actively growing culture and no growth.
While the RT-RiboSyn method shows promise as a useful new molecular biology tool, there are possible limitations that require further investigation. Chloramphenicol-resistant bacteria may present another limitation. Although it has been noted that the growth rates of many bacterial genera are inhibited by chloramphenicol concentrations of 1 to 10 mg/liter (1), many papers since have indicated resistance to chloramphenicol in bacteria through a variety of modes. Our experiments were performed with chloramphenicol concentrations of 20 mg/liter, which may render this resistance ineffective.
The analysis of the batch growth cultures in this study has yielded differences between the specific rate of ribosome synthesis and the specific growth rate. Ideally, RT-RiboSyn should be tested with cells collected from a chemostat operated over a broad range of specific growth rates to determine the limits of the method. However, these results from batch growth cultures are very promising. We have demonstrated good agreement between the measurement of specific ribosome synthesis by RT-RiboSyn and the measurement of specific growth rate with the conventional spectrophotometric method for A. calcoaceticus under different growth conditions using standard culture media.
Supplementary Material
Acknowledgments
We acknowledge the support from the new Florida Center of Excellence for Biomolecular Identification and Targeted Therapeutics.
Further thanks go to Stefi Depovic and the laboratory of James Garey (University of South Florida), Mary Beth Colter of the Molecular Biology Core Facility at the H. Lee Moffitt Cancer Center and Research Institute, and Samuel J. DuPont.
Footnotes
Published ahead of print on 14 December 2007.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Brock, T. D. 1961. Chloramphenicol. Bacteriol. Rev. 25:32-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cangelosi, G. A., and W. H. Brabant. 1997. Depletion of pre-16S rRNA in starved Escherichia coli cells. J. Bacteriol. 179:4457-4463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Loy, A., M. Horn, and M. Wagner. 2003. probeBase—an online resource for rRNA-targeted oligonucleotide probes. Nucleic Acids Res. 31:514-516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Oerther, D. B., J. Pernthaler, A. Schramm, R. Amann, and L. Raskin. 2000. Monitoring precursor 16S rRNAs of Acinetobacter spp. in activated sludge wastewater treatment systems. Appl. Environ. Microbiol. 66:2154-2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Oerther, D. B, M. C. M. van Loosdrecht, and L. Raskin. 2002. Quantifying the impact of wastewater micronutrient composition on in situ growth activity of Acinetobacter spp. Water Sci. Technol. 46(1-2):443-447. [PubMed] [Google Scholar]
- 6.Pace, N. R. 1973. Structure and synthesis of the ribosomal ribonucleic acid of prokaryotes. Bacteriol. Rev. 37:562-603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schmid, M., S. Schmitz-Esser, M. Jetten, and M. Wagner. 2001. 16S-23S rDNA intergenic spacer and 23S rDNA of anaerobic ammonium-oxidizing bacteria: implications for phylogeny and in situ detection. Environ. Microbiol. 3:450-459. [DOI] [PubMed] [Google Scholar]
- 8.Stahl, D. A., B. Flesher, H. R. Mansfield, and L. Montgomery. 1988. Use of phylogenetically based hybridization probes for studies of ruminal microbial ecology. Appl. Environ. Microbiol. 54:1079-1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stroot, P. G., and D. B. Oerther. 2003. Elevated precursor 16S rRNA levels suggest the presence of growth inhibitors in wastewater. Water Sci. Technol. 47(11):241-250. [PubMed] [Google Scholar]
- 10.Stroot, P. G. 2004. Novel transcription method confirms growth inhibition of bacteria exposed to domestic wastewater. Ph.D. dissertation. University of Cincinnati, Cincinnati, OH.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.