Abstract
Using two forest soils, we previously constructed two fosmid libraries containing 113,700 members in total. The libraries were screened to select active antifungal clones using Saccharomyces cerevisiae as a target fungus. One clone from the Yuseong pine tree rhizosphere soil library, pEAF66, showed S. cerevisiae growth inhibition. Despite an intensive effort, active chemicals were not isolated. DNA sequence analysis and transposon mutagenesis of pEAF66 revealed 39 open reading frames (ORFs) and indicated that eight ORFs, probably in one transcriptional unit, might be directly involved in the expression of antifungal activity in Escherichia coli. The deduced amino acid sequences of eight ORFs were similar to those of the core genes encoding type II family polyketide synthases, such as the acyl carrier protein (ACP), ACP synthases, aminotransferase, and ACP reductase. The gene cluster involved in antifungal activity was similar in organization to the putative antibiotic production locus of Pseudomonas putida KT2440, although we could not select a similar active clone from the KT2440 genomic DNA library in E. coli. ORFs encoding ATP binding cassette transporters and membrane proteins were located at both ends of the antifungal gene cluster. Upstream ORFs encoding an IclR family response regulator and a LysR family response regulator were involved in the positive regulation of antifungal gene expression. Our results suggested the metagenomic approach as an alternative to search for novel antifungal antibiotics from unculturable soil bacteria. This is the first report of an antifungal gene cluster obtained from a soil metagenome using S. cerevisiae as a target fungus.
Microbial secondary metabolites are good sources for the discovery of novel antimicrobial compounds, including antifungal compounds (17). Microbial metabolites exhibit versatile chemical structures with diverse biological activities that exceed the scope of synthetic organic chemicals (43). As a result of increasing environmental concerns and the development of resistance in fungal pathogens to synthetic fungicides (2, 12, 44), exploitation of antifungal agents from microbial metabolites is being considered as an approach to the identification of novel fungicides which meet environmental requirements (43, 47). Natural substances with antifungal activity could be developed as fungicides from novel microbial metabolites and could serve as potential lead compounds with a new mode of action to control plant diseases and to cure human fungal diseases (8, 10, 14, 20). The conventional strategy for discovering novel antibiotics from microorganisms depends heavily on the screening of biological activity from pure cultured microorganisms. Many antifungal bacteria and compounds have been isolated by the classical biological activity-based screening approach (22). However, this approach is not entirely effective in searches for novel antifungal compounds based on real microbial diversity. In fact, there is an ongoing controversy around the fact that the rediscovery rate of antibiotics from actinomycetes is as high as 99%, indicating that the chance for finding novel antimicrobial compounds from culturable bacteria is rare (13, 22).
The recognition of microbial diversity and the importance of unculturable bacteria in microbial ecosystems have initiated the screening of unculturable bacterial resources. This requires a genetic approach to exploring microbial resources from both aquatic and terrestrial ecosystems (4, 45). The metagenome approach includes cloning the total microbial genome isolated directly from microbial ecosystems into a culturable surrogate host and attempting to screen for valuable microbial resources (23, 39, 49). The metagenome consists primarily of the unculturable bacterial genome and a minor fraction of the culturable bacterial genome. This is the case because, in most microbial habitats, unculturable bacteria are the dominant members of the total microbial community (57). It has frequently been estimated that 0.001 to 1% of bacteria in soils and aquatic habitats are culturable and characterized as a pure culture (1). Similarly, previous studies with 16S rRNA clone analysis showed that the most abundant species in a forest was from the phylum Acidobacteria, which has few cultured representatives (19, 30, 32).
Compared to the successful discovery of novel enzymes from metagenomes (11, 18, 27, 28, 31, 45, 53), the discovery of antimicrobial compounds is a difficult task, since multiple genes and entire gene clusters involved in the production of the target compounds should be cloned together with resistance and regulatory genes in one contiguous segment (52). This implies that relatively large metagenomic DNA fragments should be cloned and expressed in a surrogate host. The genes encoding the modular enzyme type I polyketide synthase (PKS) frequently encompass DNA fragments of more than 50 kb (23). In many cases, expression of large DNA fragments would be a barrier in a host bacterium such as Escherichia coli. Nevertheless, several papers have reported the discovery of antimicrobial compounds and their respective gene clusters by a metagenomic approach from terrestrial and symbiotic microbial habitats (5, 41, 42). Alternatively, the selection of metagenomic clones based on DNA sequence homology can be conducted either by direct PCR amplification or by screening metagenome libraries with DNA probes. Soil DNA and metagenome clones encoding type I and type II PKSs have been detected from soils by sequence-based screening (16, 50, 55).
In this study, Saccharomyces cerevisiae was used as a target organism to search for antifungal activity from a forest soil metagenome. This fungus is fast growing and is suitable for screening the metagenome library using the double-agar-layer method. Corran et al. (9) reported that 67% of tested fungicides exhibited S. cerevisiae growth inhibition at 5- to 100-μg/ml concentrations and proposed that S. cerevisiae may be an excellent indicator organism to search for candidate fungicidal compounds. Therefore, it may be feasible to use S. cerevisiae as a model fungus to screen for antifungal activity from metagenomes. Here, we describe the isolation of a metagenome clone with antifungal activity using S. cerevisiae as a target fungus. The selected clone was characterized to define the gene cluster involved in antifungal activity expression.
MATERIALS AND METHODS
Microbial strains and growth conditions.
E. coli strains were grown at 37°C on Luria-Bertani (LB) agar or in LB broth supplemented with the appropriate antibiotics (35). S. cerevisiae Y-139, a wild type with natural chloramphenicol resistance, was routinely grown at 30°C on YPD agar (1% yeast extract, 2% peptone, 2% dextrose, 1.5% agar) or in LB broth containing chloramphenicol. Pseudomonas putida KT2440 (29) was purchased from the American Type Culture Collection and routinely grown at 30°C on LB agar. Bacillus subtilis strain JH642 carrying pBSK185 (a chloramphenicol-resistant plasmid) was routinely grown at 30°C on LB agar or in LB broth containing chloramphenicol (32). The following antibiotic concentrations were used for E. coli strains, B. subtilis JH642, and S. cerevisiae Y-139: chloramphenicol, 25 μg/ml; ampicillin, 100 μg/ml; and kanamycin, 25 μg/ml.
Recombinant DNA technology, plasmids, and sequencing clone pEAF66.
Plasmid preparation, restriction endonuclease digestion, DNA ligation, plasmid DNA transformation, agarose gel electrophoresis, and other standard recombinant DNA techniques were carried out following standard methods (46). DNA sequencing and primer synthesis were performed commercially at the DNA sequencing facility of GenoTech Corp. (Daejeon, South Korea). Complete DNA sequencing of the pEAF66 clone was performed by the shotgun sequencing method using plasmid DNA purified from a 500-ml culture of E. coli carrying pEAF66. A shotgun library was constructed by subcloning mechanically sheared DNA (1.5 to 4 kb) into pUC118 digested with SmaI. Insert DNAs were sequenced with a ABI 3730xl DNA analyzer (Applied Biosystems, Foster City, CA). Chromatograms were processed by the phred/phrap/consed software package (http://www.phrap.org). The sequencing gaps were closed by primer walking. The sequence data provided approximately sixfold coverage of pEAF66 DNA. DNA sequences were analyzed with the BLAST program provided by the National Center for Biotechnology Information. Open reading frames (ORFs) were analyzed with the ORF Finder software at the National Center for Biotechnology Information. The following criteria were used to identify the potential ORFs among the potential ORFs detected. Nonoverlapping sequences longer than 50 amino acids were retained. For overlapping ORFs in different reading frames, we selected those with BLAST homologues. The start codon was determined by the presence of conserved Shine-Dalgarno sequences 4 to 10 bp ahead of ATG codon for the individual ORF.
Two genes encoding response regulators were subcloned in high-copy-number plasmids such as pUC119 (59) and pUC129 (26). A 2.58-kb SphI fragment of pEAF66 was subcloned into pUC119 to generate pEJ101. A 1.25-kb XhoI fragment of pEAF66 was subcloned into pUC129 to generate pEJ102. Plasmids pEJ101 and pEJ102 carried genes encoding LysR and IclR family response regulators under the control of the lac promoter, respectively (Fig. 1).
FIG. 1.
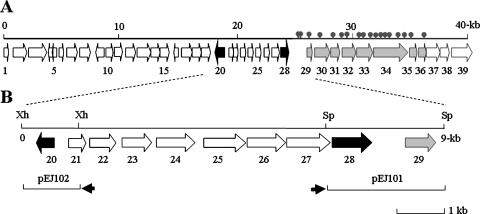
Map of a 40-kb fragment of pEAF66. (A) Map of pEAF66 carrying 39 ORFs encoding bacterial type II PKS components (gray arrows) and putative response regulators (dark arrows). Circles at the top of the map indicate the 17 transposon insertion sites causing loss of antifungal activity of the original pEAF66. (B) Enlarged map of orf20 to orf29. orf20 and orf28 (dark arrows) represent potential response regulators in the IclR family and the LysR family, respectively. Two subclones, pEJ101 and pEJ102, from among a high copy number of plasmids are indicated, and the small dark arrow in the subclones represents the lac-operon promoter. Sp and Xh, SphI and XhoI sites for subcloning, respectively.
Construction of a genomic DNA library.
The genomic DNA of P. putida KT2440 was obtained with a genomic-DNA isolation kit (Promega, Madison, WI) and size-fractionated in a 0.5% low-melting-point agarose gel, and a DNA fragment (>30 kb) was collected for library construction. The purified genomic DNA was ligated into a fosmid vector, pEpiFOS-5 (Epicentre, Madison, WI), as previously described (32). The ligation mixture was then packaged into lambda phages using MaxPlax lambda packaging extracts (Epicentre). The packaged library was transduced into E. coli EPI-100, and E. coli transformants were selected on LB agar supplemented with chloramphenicol. The presence of recombinant plasmids and the polymorphism of the insert DNA were examined by agarose gel electrophoresis of a BamHI digestion of the purified plasmids from randomly selected E. coli transformants. The library clones were stored in cryotubes as clone pools, with approximately 500 clones per pool.
Selection of antifungal metagenome clones.
To select active antifungal clones from the previously constructed and stored forest soil metagenome library (32), the library pool stocks were diluted in buffer (per liter: NaCl, 8.5 g; KH2PO4, 0.3 g; Na2HPO4, 0.6 g; MgSO4, 0.2 g; gelatin, 0.1 g), and metagenomic clones of E. coli were cultured on LB agar supplemented with chloramphenicol for 3 days at 37°C. The number of clones per plate was adjusted to approximately 500 by dilution of the library stock, and at least fivefold the number per initial stock was bioassayed using the procedure described by Rondon et al. (45). Briefly, the S. cerevisiae Y-139 culture suspension was adjusted to an optical density at 600 nm of 0.2 to 0.3 and mixed with LB soft agar at 42°C just before the medium was poured in order to maintain the viability of the yeast. The precultured E. coli clones were overlaid with 5 ml of LB soft agar containing 0.5 ml of S. cerevisiae Y-139 culture. The plates were incubated overnight at 28°C and scored for activity by looking for zones of growth inhibition in the yeast lawn. One antifungal active clone was selected and designated pEAF66. We further analyzed this clone by sequencing the whole insert DNA and transposon insertion mutagenesis. G+C content (%) was determined by using software available at http://tim.saraogtim.com/molbio/gccontent.php.
To investigate the effect of transcriptional regulators for antifungal activity expression of pEAF66, various plasmid constructs were introduced into E. coli EPI-100 carrying pEAF66. E. coli EPI-100 cells carrying pEAF66 and various subclones were grown for 2 days at 37°C on LB medium supplemented with 0.15 mM of isopropylthio-β-d-galactoside, and LB soft agar containing S. cerevisiae was overlaid as described above. Yeast growth inhibition by E. coli EPI-100 carrying various constructs was compared 2 days after incubation.
Transposon mutagenesis of the selected clone.
To define the genes involved in the antifungal activity production from pEAF66, in vitro transposon mutagenesis was carried out following the mutagenesis protocol provided with the GPS-mutagenesis system (New England BioLabs Inc., Beverly, MA). Transposon insertion mutants were selected on LB supplemented with chloramphenicol and kanamycin. The mutants were subjected to antifungal activity analysis with the S. cerevisiae Y-139 strain by the double-agar-layer method. Mutants deficient in antifungal activity were selected, and a transposon insertion site with flanking DNA fragments was subcloned into pUC119. Transposon insertion sites were determined by DNA sequencing and restriction analysis of the mutated subclones.
Antifungal activity analysis.
E. coli cells carrying pEAF66 were grown in 500 ml LB broth supplemented with chloramphenicol in a shaking incubator at 37°C for 2 days. Although the cell-free culture supernatant did not exhibit clear inhibition of S. cerevisiae by double-agar-layer assay, the culture supernatant was extracted twice serially with an equal volume of n-hexane, ethyl acetate, and butanol. The residual aqueous fraction was concentrated to dryness. The organic solvent extracts were also concentrated to dryness. All four residues were dissolved in an appropriate solvent and tested for antifungal activity against S. cerevisiae Y-139. The E. coli cells carrying pEAF66 were also grown in LB agar medium at 37°C for 2 days. The solid cultures were chopped into small pieces and submerged overnight in twice the volume of acetone. Acetone extracts were evaporated to remove the acetone. The residual aqueous fraction was serially extracted with solvents such as n-hexane, ethyl acetate, and butanol. The residual aqueous fraction was concentrated to dryness. The organic solvent extracts were also concentrated to dryness. Residues were dissolved in an appropriate solvent and tested for antifungal activity against S. cerevisiae.
Nucleotide sequence accession number.
The nucleotide sequence of the DNA insert of pEAF66 was deposited in the GenBank database under accession number EU099626.
RESULTS
Selection of the antifungal active clone.
An antifungal active clone was selected by screening the two previously constructed metagenomic libraries of 113,700 clones using the double-agar-layer method (32). The active antifungal clone pEAF66 was selected from the metagenomic library of pine tree rhizosphere soil in Yuseong forest. The E. coli colony containing pEAF66 was indistinguishable in colony morphology and growth rate from the E. coli colony with a fosmid vector. E. coli carrying pEAF66 also exhibited weak growth inhibition of B. subtilis JH642 by the double-agar-layer method. However, E. coli carrying pEAF66 did not exhibit any growth inhibition of Agrobacterium tumefaciens SK563 carrying chloramphenicol-resistant plasmid pKHt1 (37) by the double-agar-layer method. This result suggested that our clone pEAF66 expresses a selective antifungal activity. When the purified plasmid pEAF66 was reintroduced into E. coli EPI-100, the E. coli cells exhibited the same antifungal activity against S. cerevisiae. The clone (pEAF66) was further analyzed to identify the active antifungal compound. Crude culture and cell extracts of E. coli carrying pEAF66 from solid culture and broth culture were partitioned through organic extraction. Each fraction was then concentrated and tested for antifungal activity. However, neither fraction exhibited antifungal activity. To date, various extractions and partitioning to isolate the active component have been unsuccessful.
Characterization of the antifungal clone.
Since efforts to isolate and identify active antifungal components produced from clone pEAF66 were not successful, we analyzed the genes responsible for antifungal activity from pEAF66 by transposon insertional mutagenesis and subsequent bioassays of the insertion mutant clones. Among 120 mutant clones tested, 17 were deficient in activity against S. cerevisiae. All 17 mutants also lacked activity against B. subtilis, whereas the original pEAF66 clone inhibited B. subtilis growth. This result indicated that the same locus might be responsible for both antifungal and antibacterial activity. Partial DNA sequences of the transposon insertion sites were determined as described in Materials and Methods, and they were similar to the genes of bacterial type II PKSs or bacterial fatty acid synthases. Therefore, DNA sequences of the full-length insert of pEAF66 were determined and analyzed. The overall G+C content of pEAF66 was 63.7%. We defined the potential ORFs and their organization by a BLAST search (Fig. 1A). The size of the insert DNA is 40,225 bp, and 39 ORFs, including two partial ORFs at both ends of the insert, were identified. The 17 transposon insertion sites were clustered in a potential single transcriptional unit from orf29 to orf35 (Fig. 1A). Two transposon insertions occurred in the promoter area of the antifungal gene cluster. In fact, in silico promoter sequence analysis revealed that the two transposon insertions were exactly in the middle of promoter consensus sequences of the transcriptional unit (data not shown). Therefore, the gene cluster from orf29 to orf35 appeared to be directly responsible for producing the antifungal component. The close relatives of the pEAF66 genes and the identities of deduced amino acid sequences are summarized in Table 1. Since there are no phylogenetic marker genes in the insert DNA, the pEAF66 organism of origin was not known. However, most of the ORFs were highly similar to genes of Proteobacteria.
TABLE 1.
List of ORFs from antifungal clone pEAF66, gene function, and similar genes in GenBank
| ORF | Gene function | Close relative (protein, identity, organism) |
|---|---|---|
| 1 | Membrane protein, partial | Small-conductance mechanosensitive channel, 54%, Burkholderia dolosa |
| 2 | Fatty acid desaturase | Stearoyl coenzyme A desaturase oxidoreductase, 63%, R. solanacearum GMI-1000 genome |
| 3 | Putative beta-lactamase | Hypothetical beta-lactamase, 46%, Robiginitalea biformata genome |
| 4 | 50S ribosomal protein L33 | 50S ribosomal protein L33, 87%, R. solanacearum GMI-1000 genome |
| 5 | 50S ribosomal protein L28 | 50S ribosomal protein L28, 90%, R. solanacearum GMI-1000 genome |
| 6 | Transporter | Putative Mg2+ transporter, 57%, Bordetella parapertussis genome |
| 7 | Unknown, hypothetical | Unknown |
| 8 | Hypothetical protein | Hypothetical protein, 24%, Methanosarcina barkeri |
| 9 | RadC, DNA repair | DNA repair protein RadC, 64%, Azoarcus sp. |
| 10 | Peptidylprolyl isomerase | Peptidylprolyl isomerase, FKBP type, 53%, Burkholderia phytofirmans genome |
| 11 | Drug tolerant protein | Hydroxymethylbutenyl pyrophosphate reductase, 82%, Herbaspirillum seropedicae genome |
| 12 | ABC transporter | Branched amino acid transporter, 69%, R. solanacearum GMI-1000 genome |
| 13 | ABC transporter | Amino acid permease component, 62%, R. solanacearum GMI-1000 genome |
| 14 | ABC transporter | Amino acid-transporting ATPase, 73%, R. solanacearum GMI-1000 genome |
| 15 | ABC transporter | Amino acid ATP binding transporter, 70%, R. solanacearum GMI-1000 genome |
| 16 | Phasin, polyalkanate binding | Polyhydroxyalkanoate granule-associated protein, 49%, R. eutropha genome |
| 17 | Transmembrane protein | Transmembrane protein, 56%, R. solanacearum GMI-1000 genome |
| 18 | Oxidoreductase | Probable oxidoreductase, 66%, R. solanacearum GMI-1000 genome |
| 19 | Endopeptidase | d-Alanyl-d-alanine carboxypeptidase, 56%, R. solanacearum GMI-1000 genome |
| 20 | IclR regulator | Transcriptional regulator, 77%, R. solanacearum GMI-1000 genome |
| 21 | Diacylglycerolkinase | Diacylglycerol kinase. 52%, P. putida KT2440 genome |
| 22 | Hypothetical | Predicted esterase, 57%, R. solanacearum GMI-1000 genome |
| 23 | Antioxidant | Peroxiredoxin (oxidoreductase, peroxidase), 76%, R. solanacearum GMI-1000 genome |
| 24 | Permease | Transmembrane protein, 60%, R. solanacearum GMI-1000 genome |
| 25 | ABC transporter | Sulfate transporter, 76%, R. solanacearum GMI-1000 genome |
| 26 | ABC transporter | Permease component, 79%, R. solanacearum GMI-1000 genome |
| 27 | ABC transporter | Sulfate transporting ATPase, 71%, R. solanacearum GMI-1000 genome |
| 28 | LysR family regulator | CysB, activator, 71%, R. solanacearum GMI-1000 genome |
| 29 | Acyl carrier protein | Acyl carrier protein, 39%, Pseudomonas putida KT2440 genome |
| 30 | β-Ketoacyl ACP synthase II | 3-Oxoacyl-(acyl-carrier-protein) synthase II, 44%, P. putida KT2440 genome |
| 31 | β-Ketoacyl ACP synthase | β-Ketoacyl synthase, 29%, P. putida KT2440 genome |
| 32 | β-Ketoacyl ACP synthase II | 3-Oxoacyl-(acyl-carrier-protein) synthase II, 53%, P. putida KT2440 genome |
| 33 | β-Ketoacyl ACP synthase | β-Ketoacyl synthase, 31%, P. putida KT2440 genome |
| 34 | Aminotransferase | Pyridoxalphosphate dependent aminotransferase class III, 44%, P. putida KT2440 genome |
| 35 | ACP reductase | 3-Oxoacyl-(acyl-carrier-protein) reductase, 52%, P. putida KT2440 genome |
| 36 | Ketoreductase | Oxidoreductase, 52%, P. putida KT2440 genome |
| 37 | Hypothetical protein | Hypothetical protein, 32%, P. putida KT2440 genome |
| 38 | Membrane protein | Outer membrane lipoprotein-sorting protein, 32%, P. putida KT2440 genome |
| 39 | Membrane transporter, partial | Transporter, 49%, P. putida KT2440 genome |
Antifungal gene cluster and transporters.
The antifungal gene cluster as a potential single transcriptional unit contained ORFs similar to the genes of bacterial type II PKSs. Bacterial type II PKSs contain a set of three genes referred to as the minimal PKS. The three genes encode two ketoacyl synthases, KSα and KSβ (chain length factor) and the acyl carrier protein (ACP) (34). Homologs of all three core enzyme genes were identified in our gene cluster (Table 1 and Fig. 1A). Our antifungal gene cluster comprised a potential operon with 11 ORFs, since most start codons either overlapped a preceding stop codon or were several bases from the preceding stop codon. The cluster consisted of a gene for ACP (orf29) and four similar genes encoding ketoacyl synthases (from orf30 to orf33) as core enzymes of type II PKS. In addition, genes similar to those encoding aminotransferase, ACP reductase, and ketoreductase (from orf34 to orf36) were identified in the downstream region. Although four genes encoding ketoacyl synthase were detected, they were not highly similar to each other. The highest identity of deduced amino acid sequences among the four genes was 34% between orf30 and orf32 (data not shown). Ketoreductases are probably involved in the hydroxylation of the poly-keto-acyl chain before cyclization, although transposon insertion at orf36 abolishing antifungal activity was not identified. No transposon insertion mutant defective in antifungal activity was identified from orf36 to orf39. We could not find any cyclase or dehydratase homologs in our gene cluster.
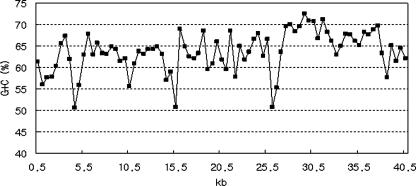
An identical organization from orf29 to orf39 was found in the operon (PP2777 to 2788) of the P. putida KT2440 genome, which is probably related to the biosynthesis of secondary metabolites such as antibiotics (38). The identity of deduced amino acid sequences between genes of pEAF66 and those of P. putida KT2440 ranged from 31% to 53% (Table 1). A fosmid library of P. putida KT2440 was constructed and subsequently screened for antifungal activity. Screening of more than 20,000 clones did not select any active antifungal clone. It is interesting that a similar gene organization of the region upstream of the antifungal gene cluster was not found in the genome of Gammaproteobacteria such as P. putida KT2440. Instead, the upstream part of the gene organization (from orf1 to orf28) was found in the genomes of Betaproteobacteria, such as Ralstonia solanacearum and Ralstonia metallidurans (Table 1). Whereas the average G+C content of pEAF66 was 63.7%, there were some regions with a relatively AT-rich (50 to 55% G+C) satellite. These were located mostly in the intergenic area, which may be the promoter region (Fig. 2). A long intergenic space (804 bp) found in front of orf29 with a G+C content of 50.6% may be the promoter area for the antifungal gene cluster. The relatively high G+C content (67.8%) in the antifungal gene cluster (orf29 to orf35) was recognized in the area from 27 kb to 36.5 kb (Fig. 2).
FIG. 2.
G+C percentage of a 40-kb insert DNA of pEAF66. Note that the G+C content in the antifungal gene cluster (from 27 to 36.5 kb) is relatively high and the potential promoter region is relatively AT rich.
The results indicate that orf38 and orf39 were similar to the transporters of orf24 to orf27, and they encode the putative permease and ATP binding cassette (ABC) transporters, respectively. Since orf38 and orf39 are physically linked to the antifungal gene cluster, they may be involved in the secretion of an antifungal component or the uptake of a primary metabolite required for biosynthesis of the antifungal component. However, transposon insertion mutants deficient in antifungal activity were not obtained from either locus.
Positive regulation for antifungal activity.
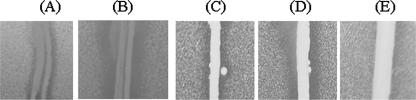
DNA sequence analysis revealed that two genes encoding transcriptional regulators were located upstream of the antifungal gene cluster. orf20 and orf28 encode IclR and LysR family response regulators, respectively. Deduced amino acid sequences of both genes exhibited a conserved DNA binding domain such as the helix-turn-helix motif (data not shown). We subcloned each gene in the high-copy-number plasmid, where the gene expression is driven by the lac promoter of the pUC plasmid. The subclone pEJ101 carried orf28, encoding the LysR family response regulator in pUC119, and pEJ102 carried orf20, encoding the IclR family response regulator in pUC129. Introduction of either pEJ101 or pEJ102 into E. coli carrying pEAF66 apparently increased the yeast growth inhibition zone by the double-agar-layer method compared to E. coli carrying pEAF66 and pUC119 or pUC129 (Fig. 3). The negative control with E. coli carrying pEPI-FOS5 and pEJ102 did not show any yeast growth inhibition (Fig. 3). Similarly, E. coli carrying pEPI-FOS5 and pEJ101 did not exhibit any yeast growth inhibition (data not shown). Repeated experiments with E. coli carrying the various subclones exhibited the consistent results.
FIG. 3.
Enhanced antifungal activity of E. coli EPI-100 carrying pEAF66 and genes for transcriptional regulators. S. cerevisiae growth inhibition by E. coli carrying pEAF66 plus pUC129 (A), pEJ102 (B), pUC119 (C), pEJ101 (D), or pEPI-FOS5 and pEJ102 (E) was assessed.
The deduced amino acid sequence of orf28 revealed over 60% identity to many LysR family response regulators from bacterial genome analysis data (data not shown). orf28 was tightly linked to the upstream region of three ORFs from orf25 to orf27. From orf25 to orf28, the stop codons of the preceding ORF overlapped with the start codon of the following ORF, indicating that at least those four ORFs constituted one transcriptional unit. orf25, orf26, and orf27 were highly similar to ABC transporter genes. The deduced amino acid sequence of orf20 revealed over 70% identity to many putative IclR family response regulators from cultured bacterial genomic sequences. Alignment of IclR from pEAF66 with other functionally characterized response regulators in the IclR family (36) showed an amino acid sequence identity of 24 to 30% (data not shown). It is likely that the orf20, encoding the IclR family response regulator, constitutes a single ORF transcriptional unit, and it is divergently transcribed from other putative operons from orf21 to orf28. Thus, it is likely that the antifungal gene cluster of pEAF66 encompasses a DNA fragment greater than 21 kb when we include the regulatory genes and potential transporters.
DISCUSSION
The metagenome constitutes a valuable microbial resource derived from many unculturable bacteria that can be used to search for antimicrobial compounds and their genes (21). Here we report the first case of selection of an antifungal clone from a metagenome using S. cerevisiae as a target fungus. The probability of obtaining desirable antifungal activity from the fosmid-based metagenome library in our selection process was very low. We obtained only one clone out of 113,700 clone libraries. Obtaining bioactive clones from metagenomes is more frequently performed by function-driven analysis based on heterologous expression of the cloned DNA (5, 6, 15, 32). Since heterologous expression of the cloned DNA still has a limited ability to obtain bioactive clones from the metagenome, the screening of a library with a PCR-amplified probe could be applied to various metagenome libraries, as shown in the case of a clone from uncultivated symbiotic bacteria of a marine sponge (41, 42). The direct PCR amplification of PKS gene fragments from soil and their phylogenetic analysis indicate that screening metagenomic libraries with a PCR-amplified probe is promising for selecting novel bioactive clones (16). Although we selected an antifungal clone from a pine tree rhizosphere metagenome by screening 113,700 clones, library construction from natural suppressive soil samples (56) may be necessary to increase the probability of obtaining the desired antifungal activity. In addition, vector modification and screening improvement would increase the chance of selecting biologically active clones (33, 51, 58).
The active antifungal clone pEAF66 exhibited clear S. cerevisiae growth inhibition. However, the intensive effort to isolate the active component failed when E. coli carrying pEAF66 or E. coli carrying pEAF66 and pEJ101 or pEJ102, which gave increased antifungal activity, was used. We speculated that the antifungal component of pEAF66 might be unstable during our extraction process. Instead of characterizing the component conferring antifungal activity, the DNA sequence analysis of pEAF66 indicated that a gene cluster encoding bacterial type II PKS homologs is involved in the antifungal activity expression in E. coli. Microbial PKSs are involved in the biosynthesis of many natural products of industrial interest and are classified as one of two structurally different types of enzymes (25). Type I PKSs are modular multifunctional enzymes, while the bacterial type II PKSs are multienzyme complexes composed of three or more separate mono- or bifunctional enzymes, which act iteratively during the synthesis of antibiotics. In this study, we identified genes encoding type II PKS homologs, suggesting that the active antifungal component from pEAF66 is probably related to the polyketides. So far, we have been unable to find an extraction procedure that can separate the active antifungal component. Although we do not know the chemical nature of the antifungal component from pEAF66, the gene cluster could serve as a genetic resource for polyketide engineering from unculturable microorganisms (7, 24, 40, 54). Regardless of the difficulty in purifying the antifungal component, it is also possible to speculate that the antifungal component could be the result of a hybrid synthesis, in which some of the enzymes originated from E. coli chromosome, while others were from our clone. Due to the presence of genes encoding three core enzymes for iteratively acting type II PKS, it is likely that E. coli carrying pEAF66 may produce polyketide antifungal compounds. However, without the chemical identity of the active substance, it is difficult to predict the structure of the potential polyketide compounds. Because of four ketoacyl synthases (from orf30 to orf33) and aminotransferase (orf34), the final product will be the result of repeated addition of two carbons to condense malonyl-ACP to a certain chain length of polyketide and subsequent addition of an amine group to the growing acyl chain of the polyketide (3, 24, 25, 40). However, the identification of the final active antifungal components will certainly provide the answer regarding the detailed process of biosynthesis.
Gene organization similar to that of the antifungal gene cluster of pEAF66 was identified from several genome sequences of Gammaproteobacteria such as P. putida KT2440. Thus, it is logical to expect similar antifungal activity against S. cerevisiae from the gene cluster of P. putida KT2440. The P. putida KT2440 operon (PP2777 to PP2788) may be related to the production of antibiotics, although the active component was not isolated nor predicted from strain KT2440 (38). Our screening of more than 20,000 fosmid clones of P. putida KT2440 did not yield any active antifungal clone, suggesting that either the operon (PP2777 to PP2788) of P. putida KT2440 was not expressed in E. coli or the expressed product of the operon does not have antifungal activity. In fact, when precultured P. putida KT2440 was overlaid with LB soft agar containing S. cerevisiae Y-139 culture, clear growth inhibition of the yeast was not detected. This result suggested that even though our antifungal gene cluster showed gene organization similar to that of the P. putida operon, it is likely that the P. putida operon encodes a different activity.
Interestingly, while the downstream part of pEAF66 involved in antifungal activity has a gene organization similar to that of the genome of Gammaproteobacteria, such as P. putida KT2440, the upstream part of the gene organization was similar to genes from the genome of Betaproteobacteria, such as R. solanacearum. This finding raised the question of whether pEAF66 is a chimeric metagenome clone that resulted from the simultaneous ligation of two random DNA fragments originating from Betaproteobacteria and Gammaproteobacteria. Through G+C content analysis, we identified a relatively AT-rich promoter region in front of the antifungal gene cluster of pEAF66. However, G+C content analysis did not clearly indicate whether two different DNA fragments were coligated to generate pEAF66. Interestingly, the IclR family response regulator (orf20) and the LysR family response regulator (orf28) appeared to play roles as positive regulators in the expression of the antifungal gene cluster. Since orf20 is somewhat distal from and not linked to the antifungal gene cluster but is found in betaproteobacterial gene organization, the role of the IclR regulator in positive regulation of antifungal gene cluster expression was surprising. The similar IclR family response regulator and the LysR family response regulator were not found in the upstream part of the operon (PP2777 to PP2788) of P. putida KT2440. The regulators of antifungal gene expression are located in the upstream part, which is similar to the betaproteobacterial genome, while structural genes for antifungal activity are located in the downstream part. Thus, it is likely that the insert DNA of pEAF66 comes from one bacterium.
The IclR family response regulators are widespread among prokaryotes and act as repressors, activators, and proteins with a dual function (36). The IclR family response regulators always have an N-terminal DNA binding domain and a C-terminal effector binding site. Since a clear consensus sequence for the IclR family response regulator has yet to be defined, we could not determine if there is a consensus sequence for IclR protein in the promoter area of the antifungal gene cluster. The LysR family response regulator is one of the most frequently found positive transcriptional regulators in prokaryotes (48). The LysR regulators contained a helix-turn-helix motif and a coinducer recognition domain. Binding of small molecules to the LysR activates transcription of target genes. At this stage, it is not clear whether IclR and LysR directly regulate the expression of the antifungal gene cluster in a cooperative manner or a hierarchical manner. A transposon insertion mutation in either orf20 or orf28 abolishing antifungal activity of pEAF66 was not identified. It is likely that the LysR and IclR families of transcriptional regulators interact with certain chemicals either by an antifungal compound precursor or by foreign signal molecules and subsequently regulate expression of the downstream antifungal gene cluster.
The flanking genes of antifungal clusters of pEAF66 were similar to those for membrane proteins and ABC transporters probably involved in the secretion of antifungal compounds or uptake of primary metabolites for the biosynthesis of the antifungal component. Since transposon insertion mutants deficient in antifungal activity were not obtained from either locus, it is not clear whether they are directly involved in the secretion of antifungal compounds by pEAF66. Although they are involved in the transport process regarding antifungal activity, if E. coli transporters can restore the transport of antifungal activity despite the mutation of orf37 to orf39, the mutation of transporters of pEAF66 would not disrupt the expression of antifungal activity.
S. cerevisiae is a fast-growing fungus that is sensitive to several fungicides. This yeast could be used as an indicator strain to search for candidate fungicides (9). In this study, we assessed the utility of S. cerevisiae in selecting clones with antifungal activity from a metagenome, and we succeeded in screening clones for such activity. On the other hand, the active antifungal component was not isolated and identified from E. coli carrying the antifungal clone. However, molecular characterization of the gene cluster indicated the presence of a novel antifungal gene cluster which may produce a polyketide family component. Further effort is necessary to isolate and identify the antifungal component encoded by the pEAF66 gene cluster. If the antifungal substance is identified as a novel compound, the substance could be used as an antifungal lead compound for novel fungicide development. To our knowledge, this is the first report of an antifungal gene cluster obtained from a soil metagenome.
Acknowledgments
This research was supported by a grant (MG 05-0103-1-0) to S.-W. Lee from the Microbial Genomics and Application Center of the 21st Century Frontier Research Program funded by the Ministry of Science and Technology of the Republic of Korea.
Footnotes
Published ahead of print on 7 December 2007.
REFERENCES
- 1.Amann, R. I., W. Ludwig, and K.-H. Schleifer. 1995. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 59:143-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson, J. B. 2005. Evolution of antifungal-drug resistance: mechanisms and pathogen fitness. Nat. Rev. Microbiol. 3:547-556. [DOI] [PubMed] [Google Scholar]
- 3.Aron, Z. D., P. C. Dorrestein, J. R. Blackhall, N. L. Kelleher, and C. T. Walsh. 2005. Characterization of a new tailoring domain in polyketide biogenesis: the amine transferase domain of MycA in the mycosubtilin gene cluster. J. Am. Chem. Soc. 127:14986-14987. [DOI] [PubMed] [Google Scholar]
- 4.Béjà, O., M. T. Suzuki, E. V. Koonin, L. Aravind, A. Hadd, L. P. Nguyen, R. Villacorta, M. Amjadi, C. Garrigues, S. B. Javanovich, R. A. Feldman, and E. F. DeLong. 2000. Construction and analysis of bacterial artificial chromosome libraries from a marine microbial assemblage. Environ. Microbiol. 2:516-529. [DOI] [PubMed] [Google Scholar]
- 5.Brady, S. F., C. J. Chao, and J. Clardy. 2002. New natural product families from an environmental DNA (eDNA) gene cluster. J. Am. Chem. Soc. 124:9968-9969. [DOI] [PubMed] [Google Scholar]
- 6.Brady, S. F., and J. Clardy. 2000. Long-chain N-acyl amino acid antibiotics isolated from heterologously expressed environmental DNA. J. Am. Chem. Soc. 122:12903-12904. [Google Scholar]
- 7.Bull, A. T., A. C. Ward, and M. Goodfellow. 2000. Search and discovery strategies for biotechnology: the paradigm shift. Microbiol. Mol. Biol. Rev. 64:573-606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Copping, L. G., and S. O. Duke. 2007. Natural products that have been used commercially as crop protection agents. Pest Manag. Sci. 63:524-554. [DOI] [PubMed] [Google Scholar]
- 9.Corran, A. J., A, Renwick, and S. J. Dunbar. 1998. Approaches to in-vitro lead generation for fungicide invention. Pestic. Sci. 54:338-344. [Google Scholar]
- 10.Daoubi, M., R. Durán-Patrón, M. Hmamouchi, R. Hernández-Galán, A. Benharref, and I. G. Collado. 2004. Screening study for potential lead compounds for natural product-based fungicides. I. Synthesis and in vitro evaluation of coumarins against Botrytis cinerea. Pest Manag. Sci. 60:927-932. [DOI] [PubMed] [Google Scholar]
- 11.DeSantis, G., Z. Zhu, W. A. Greenberg, K. Wong, J. Chaplin, S. R. Hanson, B. Farwell, L. W. Nicholson, C. L. Randi, D. P. Weiner, D. E. Robertson, and M. J. Burk. 2002. An enzyme library approach to biocatalysis: development of nitrilases for enantioselective production of carboxylic acid derivatives. J. Am. Chem. Soc. 124:9024-9025. [DOI] [PubMed] [Google Scholar]
- 12.De Waard, M. A., A. C. Andrade, K. Hayashi, H. Schoonbeek, I. Stergiopoulos, and L. Zwiers. 2006. Impact of fungal drug transporters on fungicide sensitivity, multidrug resistance and virulence. Pest Manag. Sci. 62:195-207. [DOI] [PubMed] [Google Scholar]
- 13.Firn, R. D., and C. G. Jones. 2000. The evolution of secondary metabolism—a unifying model. Mol. Microbiol. 37:989-994. [DOI] [PubMed] [Google Scholar]
- 14.Früh, T., P. Chemla, J. Ehrler, and S. Farooq. 1996. Natural products as pesticides: two examples of stereoselective synthesis. Pestic. Sci. 46:37-47. [Google Scholar]
- 15.Gillespie, D. E., S. F. Brady, A. D. Bettermann, N. P. Cianciotto, M. R. Liles, M. R., Rondon, J. Clardy, R. M. Goodman, and J. Handelsman. 2002. Isolation of antibiotics turbomycin A and B from a metagenomic library of soil microbial DNA. Appl. Environ. Microbiol. 68:4301-4306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ginolhac, A., C. Jarrin, B. Gillet, P. Robe, P. Pujic, K. Tuphile, H. Bertrand, T. M. Voget, G. Perrière, P. Simonet, and R. Nalin. 2004. Phylogenetic analysis of polyketide synthase I domains from soil metagenomic libraries allows selection of promising clones. Appl. Environ. Microbiol. 70:5522-5527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Glasby, J. S. 1992. Dictionary of antibiotic-producing organisms. Prentice Hall, New York, NY.
- 18.Gupta, R., Q. K. Beg, and P. Lorenz. 2002. Bacterial alkaline proteases: molecular approaches and industrial applications. Appl. Microbiol. Biotechnol. 59:15-32. [DOI] [PubMed] [Google Scholar]
- 19.Hackl, E., S. Zechmeister-Boltenstern, L. Bodrossy, and A. Sessitsch. 2004. Comparison of diversities and compositions of bacterial populations inhabiting natural forest soils. Appl. Environ. Microbiol. 70:5057-5065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hadacek, F., and H. Greger. 2000. Testing of antifungal natural products: methodologies, comparability of results and assay choice. Phytochem. Anal. 11:137-147. [Google Scholar]
- 21.Handelsman, J. 2004. Metagenomics: application of genomics to uncultured microorganisms. Microbiol. Mol. Biol. Rev. 68:669-685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Handelsman, J., M. R. Rondon, S. P. Brady, J. Clady, and R. M. Goodman. 1998. Molecular biological access to the chemistry of unknown soil microbes: a new frontier for natural products. Chem. Biol. 5:R245-R249. [DOI] [PubMed] [Google Scholar]
- 23.Hopwood, D. A. 1999. Forty years of genetics with Streptomyces: from in vivo through in vitro to in silico. Microbiology 145:2183-2202. [DOI] [PubMed] [Google Scholar]
- 24.Hutchinson, C. R., and I. Fujii. 1995. Polyketide synthase gene manipulation: a structure-function approach in engineering novel antibiotics. Annu. Rev. Microbiol. 49:201-238. [DOI] [PubMed] [Google Scholar]
- 25.Katz, L., and S. Donaldio. 1993. Polyketide synthesis: prospects for hybrid antibiotics. Annu. Rev. Microbiol. 47:875-912. [DOI] [PubMed] [Google Scholar]
- 26.Keen, N. T., S. Tamaki, D. Kobayashi, and D. Trollinger. 1988. Improved broad-host-range plasmids for DNA cloning in Gram-negative bacteria. Gene 70:191-197. [DOI] [PubMed] [Google Scholar]
- 27.Knietsch, A., S. Bowien, G. Whited, G. Gottschalk, and R. Daniel. 2003. Identification and characterization of coenzyme B12-dependent glycerol dehydratase- and diol dehydratase-encoding genes from metagenomic DNA libraries derived from enrichment cultures. Appl. Environ. Microbiol. 69:3048-3060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Knietsch, A., T. Waschkowitz, S. Bowien, A. Henne, and R. Daniel. 2003. Construction and screening of metagenomic libraries derived from enrichment cultures: generation of a gene bank for genes conferring alcohol oxidoreductase activity on Escherichia coli. Appl. Environ. Microbiol. 69:1408-1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kojima, Y., H. Fujisawa, A. Nakazawa, T. Nakazawa, F. Kanetsuna, H. Taniuchi, M. Nozaki, and O. Hayaishi. 1967. Studies on pyrocatechase. I. Purification and spectral properties. J. Biol. Chem. 242:3270-3278. [PubMed] [Google Scholar]
- 30.Kuske, C. R., S. M. Barns, and J. D. Busch. 1997. Diverse uncultivated bacterial groups from soils of the arid southwestern United States that are present in many geographic regions. Appl. Environ. Microbiol. 63:3614-3621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee, S.-W., K. Won, H. K. Lim, J.-C. Kim, G. J. Choi, and K. Y. Cho. 2004. Screening for novel lipolytic enzymes from uncultured soil microorganisms. Appl. Microbiol. Biotechnol. 65:720-726. [DOI] [PubMed] [Google Scholar]
- 32.Lim, H. K., E. J. Chung, J.-C. Kim, G. J. Choi, K. S. Jang, Y. R. Chung, K. Y. Cho, and S.-W. Lee. 2005. Characterization of a forest soil metagenome clone that confers indirubin and indigo production on Escherichia coli. Appl. Environ. Microbiol. 71:7768-7777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Martinez, A., S. J. Kolvek, C. L. T. Yip, J. Hopke, K. A. Brown, I. A. MacNeil, and M. S. Osburne. 2004. Genetically modified bacterial strains and novel bacterial artificial chromosome shuttle vectors for constructing environmental libraries and detecting heterologous natural products in multiple expression hosts. Appl. Environ. Microbiol. 70:2452-2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McDaniel, R., S. Ebert-Khosla, H. Fu, D. A. Hopwood, and C. Khosla. 1993. Engineering biosynthesis of novel polyketides. Science 262:1546-1550. [DOI] [PubMed] [Google Scholar]
- 35.Miller, J. H. 1972. Experiments in molecular genetics, p. 352-355. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 36.Molina-Henares, A. J., T. Krell, M. E. Guazzaroni, A. Segura, and J. L. Ramos. 2006. Members of the IclR family of bacterial transcriptional regulators function as activators and/or repressor. FEMS Microbiol. Rev. 30:157-186. [DOI] [PubMed] [Google Scholar]
- 37.Mullins, E. D., X. Chen, P. Romaine, R. Raina, D. M. Geiser, and S. Kang. 2001. Agrobacterium-mediated transformation of Fusarium oxysporum: an efficient tool for insertional mutagenesis and gene transfer. Phytopathology 91:173-180. [DOI] [PubMed] [Google Scholar]
- 38.Nelson, K. E., C. Weinel, I. T. Paulsen, R. J. Dodson, H. Hilbert, V. A. P. Martins dos Santos, D. E. Fouts, S. R. Gill, M. Pop, M. Holmes, L. Brinkac, M. Beanan, R. T. DeBoy, S. Daugherty, J. Kolonay, R. Madupu, W. Nelson, O. White, J. Peterson, H. Khouri, I. Hance, P. C. Lee, E. Holtzapple, D. Scanlan, K. Tran, A. Moazzez, T. Utterback, M. Rizzo, K. Lee, D. Kosack, D. Moestl, H. Wedler, J. Lauber, D. Stjepandic, J. Hoheisel, M. Straetz, S. Heim, C. Kiewitz, J. A. Eisen, K. N. Timmis, A. Düsterhöft, B. Tümmler, and C. M. Fraser. 2002. Complete genome sequence and comparative analysis of the metabolically versatile Pseudomonas putida KT2440. Environ. Microbiol. 4:799-808. [DOI] [PubMed] [Google Scholar]
- 39.Pettit, R. K. 2004. Soil DNA libraries for anticancer drug discovery. Cancer Chemother. Pharmacol. 54:1-6. [DOI] [PubMed] [Google Scholar]
- 40.Pfeifer, B. A., and C. Khosla. 2001. Biosynthesis of polyketides in heterologous hosts. Microbiol. Mol. Biol. Rev. 65:106-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Piel, J. 2002. A polyketide synthase-peptide synthase gene cluster from an uncultured bacterial symbiont of Paederus beetles. Proc. Natl. Acad. Sci. USA 99:14002-14007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Piel, J., D. Hui, G. Wen, D. Butzke, M. Platzer, N. Fusetani, and S. Matsunaga. 2004. Antitumor polyketide biosynthesis by an uncultivated bacterial symbiont of the marine sponge Theonella swinhoei. Proc. Natl. Acad. Sci. USA 101:16222-16227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Porter, N. 1985. Physicochemical and biophysical panel symposium on biologically active secondary metabolites. Pestic. Sci. 16:422-427. [Google Scholar]
- 44.Richardson, M. D. 2005. Changing patterns and trends in systemic fungal infections. J. Antimicrob. Chemother. 56(Suppl. 1):i5-i11. [DOI] [PubMed] [Google Scholar]
- 45.Rondon, M. R., P. R. August, A. D. Bettermann, S. F. Brady, T. H. Grossman, M. R. Liles, K. A. Loiacono, B. A. Lynch, I. A. MacNeil, C. Minor, C. L. Tiong, M. Gilman, M. S. Osburne, J. Clardy, J. Handelsman, and R. M. Goodman. 2000. Cloning the soil metagenome: a strategy for accessing the genetic and functional diversity of uncultured microorganisms. Appl. Environ. Microbiol. 66:2541-2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 47.Saxena, S., and A. K. Pandey. 2001. Microbial metabolites as eco-friendly agrochemicals for the next millennium. Appl. Microbiol. Biotechnol. 55:395-403. [DOI] [PubMed] [Google Scholar]
- 48.Schell, M. A. 1993. Molecular biology of the LysR family of transcriptional regulators. Annu. Rev. Microbiol. 47:597-626. [DOI] [PubMed] [Google Scholar]
- 49.Schloss, P. D., and J. Handelsman. 2003. Biotechnological prospects from metagenomics. Curr. Opin. Biotechnol. 14:303-310. [DOI] [PubMed] [Google Scholar]
- 50.Seow, K.-T., G. Meurer, M. Gerlitz, E. Wendt-Pienkowski, C. R. Hutchinson, and J. Davies. 1997. A study of iterative type II polyketide synthases, using bacterial genes cloned from soil DNA: a means to access and use genes from uncultured microorganisms. J. Bacteriol. 179:7360-7368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Uchiyama, T., T. Abe, T. Ikemura, and K. Watanabe. 2005. Substrate-induced gene-expression screening of environmental metagenome libraries for isolation of catabolic genes. Nat. Biotechnol. 23:88-93. [DOI] [PubMed] [Google Scholar]
- 52.Vining, L. C. 1992. Secondary metabolism, inventive evolution and biochemical diversity—a review. Gene 115:135-140. [DOI] [PubMed] [Google Scholar]
- 53.Voget, S., C. Leggewie, A. Uesbeck, C. Raasch, K.-E. Jaeger, and W. R. Streit. 2003. Prospecting for novel biocatalysts in a soil metagenome. Appl. Environ. Microbiol. 69:6235-6242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Watanabe, K., M. A. Rude, C. T. Walsh, and C. Khosla. 2003. Engineered biosynthesis of an ansamycin polyketide precursor in Escherichia coli. Proc. Natl. Acad. Sci. USA 100:9774-9778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wawrik, B., L. Kerhof, G. J. Zylstra, and J. J. Kukor. 2005. Identification of unique type II polyketide synthase genes in soil. Appl. Environ. Microbiol. 71:2232-2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Weller, D. M., J. M. Raaijmakers, B. B. McSpadden Gardener, and L. S. Thomashow. 2002. Microbial populations responsible for specific soil suppressiveness to plant pathogens. Annu. Rev. Phytopathol. 40:309-348. [DOI] [PubMed] [Google Scholar]
- 57.Whitman, W. B., D. C. Coleman, and W. J. Wiebe. 1998. Prokaryotes: the unseen majority. Proc. Natl. Acad. Sci. USA 95:6578-6583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Williamson, L. L., B. R. Borlee, P. D. Schloss, C. Guan, H. K. Allen, and J. Handelsman. 2005. Intracellular screen to identify metagenomic clones that induce or inhibit a quorum-sensing biosensor. Appl. Environ. Microbiol. 71:6335-6344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]