Abstract
Legionella pneumophila, the agent of Legionnaires' disease, is an intracellular parasite of aquatic amoebae and human macrophages. A key factor for L. pneumophila in intracellular infection is its type II protein secretion system (Lsp). In order to more completely define Lsp output, we recently performed a proteomic analysis of culture supernatants. Based upon the predictions of that analysis, we found that L. pneumophila secretes two distinct aminopeptidase activities encoded by the genes lapA and lapB. Whereas lapA conferred activity against leucine, phenylalanine, and tyrosine aminopeptides, lapB was linked to the cleavage of lysine- and arginine-containing substrates. To assess the role of secreted aminopeptidases in intracellular infection, we examined the relative abilities of lapA and lapB mutants to infect human U937 cell macrophages as well as Hartmannella vermiformis and Acanthamoeba castellanii amoebae. Although these experiments identified a dispensable role for LapA and LapB, they uncovered a previously unrecognized role for the type II-dependent ProA (MspA) metalloprotease. Whereas proA mutants were not defective for macrophage or A. castellanii infection, they (but not their complemented derivatives) were impaired for growth upon coculture with H. vermiformis. Thus, ProA represents the first type II effector implicated in an intracellular infection event. Furthermore, proA represents an L. pneumophila gene that shows differential importance among protozoan infection models, suggesting that the legionellae might have evolved some of its factors to especially target certain of their protozoan hosts.
Legionella pneumophila is the agent of Legionnaires' disease pneumonia (29). In fresh waters, this gram-negative bacterium exists in protozoan hosts and as a part of biofilms. Disease occurs when inhaled legionellae, possibly including those still associated with protozoa or protozoan vesicles (11, 12), invade alveolar macrophages. Although many factors promote the ecology and pathogenesis of L. pneumophila, protein secretion stands out for its multifaceted significance. L. pneumophila possesses type II secretion, type IVB secretion, and type IVA secretion (7, 17), and genome sequencing suggests the existence of type I and type V secretion systems (14, 40). Among these pathways, the Lsp type II system is arguably implicated in the broadest array of phenotypes (17). Indeed, this pathway is required for secretion during growth in bacteriologic media at 37°C, extracellular survival in broth/water at 12 to 25°C, colony morphology, intracellular infection of protozoa (acanthamoebae and hartmannellae), optimal intracellular infection of human macrophages and monocytes, and virulence in a murine model of pneumonia (37, 44, 55, 61, 62, 68, 69). Although type II secretion exists in many gram-negative organisms, including various animal and plant pathogens, L. pneumophila is the only intracellular pathogen known to possess a functional type II system (17). Type II secretion is a two-step process in which nascent proteins are first translocated across the inner membrane and then, after what might be a very short period, are transported from the periplasm to the exterior through a dedicated outer membrane pore (42). In Pseudomonas aeruginosa and L. pneumophila, the transport of type II substrates across the inner membrane is mediated by the Sec or Tat pathway (42, 60). Proteins secreted via the L. pneumophila type II system were first identified by enzymatic activities in wild-type but not lsp mutant culture supernatants, i.e., a metalloprotease, acid phosphatases, lipases, phospholipase A, phospholipase C, lysophospholipase A, cholesterol acyltransferase, and RNase (2-4, 8, 22, 30, 31, 37, 44, 61, 62). A two-dimensional polyacrylamide gel electrophoresis comparison of wild-type and lsp mutant supernatants then showed that the type II secretome includes at least 25 proteins (23). Among the new proteins are (i) a chitinase that promotes persistence in lungs, (ii) proteins predicted to have aminopeptidase, cellulase, nucleotidase, or decarboxylase activity, (iii) proteins that show their greatest similarity to eukaryotic proteins, and (iv) proteins with no homology to known proteins. In silico analysis of the L. pneumophila genome suggested that the type II secretome may actually encompass as many as 60 proteins (23).
For many years, it has been known that amino acids are the main carbon and energy source for growing legionellae (74), and early reports described a variety of secreted aminopeptidase and protease activities in L. pneumophila culture supernatants (10, 20, 25, 36, 53, 54, 75). However, the genes responsible for the production and secretion of these activities have remained unknown, other than the loci associated with the above-mentioned metalloprotease, which is variously known as ProA or MspA (37, 44, 50, 57, 73). By cloning two proteins identified in our proteomic screen and characterizing new mutants, we now show that L. pneumophila does secrete aminopeptidases through its type II system. While examining the role of the aminopeptidases, we also discovered that ProA promotes L. pneumophila infection of Hartmannella vermiformis amoebae, an issue that was not addressed in earlier studies on that exoprotein (51, 73).
MATERIALS AND METHODS
Strains, growth media, and chemicals.
L. pneumophila strain 130b (ATCC strain BAA-74, also known as AA100 or Wadsworth) served as our wild type (27). Mutants of 130b containing a kanamycin resistance (Kmr) cassette inserted into lspF (NU275) or proA (AA200) as well as a complemented lspF mutant were previously described (51, 62). Legionellae were routinely cultured in buffered yeast extract (BYE) broth or on buffered charcoal yeast extract (BCYE) agar (62). Growth in broth was assessed by measuring the optical density of the culture at 660 nm. Escherichia coli DH5α and DH5α λ pir (Invitrogen, Carlsbad, CA), hosts for recombinant plasmids, were grown on LB agar (6). Antibiotics were added to media at the following concentrations (in μg per ml): ampicillin, 100; chloramphenicol (Cm), 6 for L. pneumophila and 30 for E. coli; gentamicin, 2.5; kanamycin, 25 for L. pneumophila and 50 for E. coli. Chemicals were purchased from Sigma-Aldrich (St. Louis, MO).
Sequence analysis, gene cloning, and mutant constructions.
DNA and protein sequences were analyzed using Lasergene software (DNASTAR, Madison, WI). The Clustal method of Lasergene Megalign was used for protein alignments. Database searches were done using programs based on the BLAST algorithm (1). Genomic DNA was isolated from L. pneumophila as described previously (18). Based on data from the L. pneumophila strain Paris genome (14), pairs of primers were designed for amplifying genes from 130b DNA. Primers OR140lpp2866 (5′-CAGGCTTGCTCCAACAGTTA) and OR141lpp2866 (5′-CTCTAACCGACGACGCTAAT) yielded a 1,879-bp fragment containing lapA, and primers OR142lpp0031 (5′-TCCTGCGAGAACGTGATGAA) and OR143lpp0031 (5′-AGATTGCAAGTCACTCGCTG) yielded a 1,957-bp fragment for lapB. Primers were obtained from Integrated DNA Technologies (Coralville, IA). To facilitate construction of L. pneumophila mutants, the PCR fragments encoding lapA and lapB were ligated into pGem-T Easy (Promega, Madison, WI), yielding plasmids pGlapA and pGlapB, respectively. pGlapA was digested with MfeI, which cuts 91 bp after the lapA start codon, and MscI, which cuts 245 bp before the end of the gene, and then, after Klenow treatment, was ligated to a Kmr gene isolated from pMB2190 upon HincII digestion (34) to yield pGlapA::Km. Next, a SphI-XbaI fragment of pGlapA::Km containing the disrupted gene was cloned into the Cmr, sacB-containing suicide vector pRE112 (26), to give pRlapA::Km. pGlapB was digested with BstBI, which cuts 383 bp after the lapB start codon, and then, after Klenow treatment, was ligated to a gentamicin resistance (Gmr) gene isolated from pX1918GT after HincII and PvuII digestion (65) to give pGlapB::Gm. Following a SphI digest of pGlapB::Gm, disrupted lapB was cloned into pRE112, yielding pRlapB::Gm. L. pneumophila was transformed with plasmids by electroporation as previously described (19). Following electroporation of pRlapA::Km and pRlapB::Gm into strain 130b, mutants were selected based on Cm sensitivity, sucrose resistance, and Kmr (pRlapA::Km) or Gmr (pRlapB::Gm), indicative of the introduction of the mutated gene into the chromosome and loss of the pRE112 vector by allelic exchange. To construct a lapA lapB double mutant, a Gmr lapB mutant was electroporated with pRlapA::Km. To mutate proA, Kmr-disrupted proA from strain AA200 was amplified using primers OR115lipA (5′-GGCATGGTCATTCTTCACAC) and OR121proA (5′-GTGCACCAGATAATTCCGTC). The PCR fragment was then cloned into pGem-T Easy to give pGproA::Km. The latter plasmid was digested with NotI, and then, after Klenow treatment, the fragment encoding the disrupted gene was ligated into the SmaI site of plasmid pRE112, yielding pRproA::Km. To make a lapB proA double mutant, a Gmr lapB mutant was electroporated with pRproA::Km. Verification of mutant genotypes was carried out by PCR (data not shown).
Complementation analysis.
To facilitate complementation of the proA mutant, a 3,297-bp fragment specifically containing proA was amplified by PCR from strain 130b DNA using primers OR115lipA and OR121proA and then subcloned into pGem-T Easy. Following a SphI digest of the resulting plasmid, proA was cloned under the control of the tac promoter in pMMB2002 (62), yielding pMproA. To confirm expression from this construct, we observed that pMproA restored the ability of AA200 to degrade casein. For complementation of the lapB mutant, a 1,729-bp fragment specifically containing lapB was amplified by PCR from strain 130b DNA using primers JDlapBfr (5′-CGTTCAAG ATCATCGAGCTATTCCATAG) and JDlapBrv (5′-TCCCTAACAAAAAAATGGATA GCAAGAG) and then subcloned into pGem-T Easy. Following a SalI and SphI digest of the resulting plasmid, lapB was directionally cloned under the control of the tac promoter in pMMB2002, yielding pMlapB. Confirmation of this final construct was done by PCR using primers OR77pMMB2002 (5′-TCGGCTCGTATAATGTGTGG) and JDlapBrv.
Analysis of L. pneumophila-secreted enzymatic activities.
Cell-free, filter-sterilized supernatants were obtained from L. pneumophila cultures grown in BYE broth to late log phase (3). Leucine and lysine aminopeptidase activities were assayed by following the release of p-nitroanilide (pNI) from l-leucine p-NI (Leu-pNI) and l-lysine p-NI (Lys-pNI), respectively (13, 36, 56, 70). Briefly, 20 μl of sample was added to 180 μl of assay buffer (50 mM Tris-HCl [pH 8], 100 mM NaCl, 1 mM CaCl2) containing 3 mM Leu-pNI or Lys-pNI, and then the increase in absorbance at 405 nm was monitored over time of incubation at 37°C (SpectraMAX 190; Molecular Devices, Sunnyvale, CA). Alanine, arginine, cystine, phenylalanine, and tyrosine aminopeptidase activities were assayed by following the release of β-naphthylamide (βNA) from l-alanine-βNA (L-Ala-βNA), l-arginine-βNA (L-Arg-βNA), l-cystine di-βNA (L-Cys-βNA), l-phenylalanine-βNA (L-Phe-βNA), and l-tyrosine-βNA (L-Tyr-βNA), respectively (53, 54). A 10-μl aliquot of sample was added to 190 μl of 0.1 M phosphate buffer (pH 6.8) and then, to start the reaction, 3 mM of the amino acid-βNA substrate was added in a volume of 100 μl. Upon incubation at 37°C, released βNA was recorded fluorometrically (SpectraMAX GeminiXS; Molecular Devices) over time at 410 nm with an excitation wavelength of 340 nm (45, 66, 80). One unit of aminopeptidase was defined as that which yielded 1 μM of pNI or βNA in 1 min, based upon standard curves generated with pNA and pNI (66, 80). Protease activity was determined on casein agar, and acid phosphatases were monitored by the release of p-nitrophenol (p-NP) from p-NP phosphate (2, 3). RNase activity was assayed by monitoring the release of nucleotides from Baker's yeast type III RNA (62), and lipolytic activities were determined by p-NP palmitate and p-NP caprylate hydrolysis (3, 4).
Intracellular infection of protozoa and macrophages.
To examine the ability of L. pneumophila to grow in protozoa, H. vermiformis (ATCC strain 50237) and Acanthamoeba castellanii (ATCC 30234) were infected as previously described (19, 51, 77). Thus, ca. 104 CFU were added to wells containing 105 amoebae and then, at various times postinoculation, the numbers of bacteria per coculture were determined by plating dilutions on BCYE agar. To assess growth in macrophages, differentiated human U937 cells were infected as previously described (18, 62). Monolayers containing 106 macrophages were inoculated with 105 CFU, incubated for 2 h to allow bacterial entry, and then washed to remove unincorporated bacteria. At various times postinoculation, dilutions of saponin-lysed monolayers were plated on BCYE to determine the numbers of bacteria per monolayer.
Pulmonary infection of A/J mice with L. pneumophila.
As described before, female 6- to 8-week-old A/J mice (Jackson Lab, Bar Harbor, ME) were inoculated intratracheally with 25 μl of a bacterial suspension containing 106 CFU of a 1:1 ratio of wild-type and mutant strains (62). Three days later, infected lungs were homogenized, and the numbers of viable bacteria and the ratio of wild type to mutants were determined by plating dilutions on standard and antibiotic-supplemented BCYE (23, 62). Animal experiments were approved by the Animal Care and Use Committee of Northwestern University.
RESULTS
Identification and mutation of lapA and lapB in L. pneumophila.
In our previous proteomic analysis, we identified two proteins that are present in wild-type 130b but not lspF mutant supernatants and are hypothesized to be aminopeptidases (23). The first of these proteins, which we now designate as LapA, for Legionella aminopeptidase A, had been annotated as a leucine aminopeptidase. In the sequenced strains Philadelphia, Paris, and Lens, lapA is designated as lpg2814, lpp2866, and lpl2729, respectively (14, 16). In wild-type supernatants, we observed a 35-kDa LapA, a form of the protein that predicts cleavage of a signal sequence, secretion by the type II pathway, and processing to a smaller form (23). The second of these proteins, which we named LapB, also showed similarity to known aminopeptidases, although its level of similarity was not sufficient to warrant prior annotation as an aminopeptidase. In strains Philadelphia, Paris, and Lens, lapB is designated as lpg0032, lpp0031, and lpl0032, respectively (14, 16). As was the case for LapA, a ca. 35-kDa LapB was observed in wild-type supernatants (23). LapA and LapB exhibit 42% identity and 59% similarity with each other (data not shown).
In order to determine whether LapA and LapB are, in fact, aminopeptidases, we cloned each of their genes and then used allelic exchange to construct the corresponding specific mutants of strain 130b. Two independently derived mutants were obtained for each gene, i.e., Kmr NU320 and NU321 for lapA and Gmr NU322 and NU323 for lapB. We also constructed two double mutants that were defective for both lapA and lapB, i.e., Kmr Gmr NU324 and NU325. All new mutants grew normally in BYE broth and on BCYE agar at 37 and 25°C (data not shown), indicating that lapA and lapB are not required for extracellular growth in standard medium. When cultured in broth, supernatants from mutant cultures contained normal levels of acid phosphatase, lipase, RNase, and protease (data not shown), indicating that the strains do not have general defects in type II secretion.
Influence of lapA, lapB, and lspF on secreted aminopeptidase activities.
To examine secreted aminopeptidase activities, we grew legionellae in BYE broth to late log phase and then assayed culture supernatants for the ability to cleave peptide substrates. Wild-type 130b supernatants consistently cleaved arginine-, alanine-, leucine-, lysine-, phenylalanine-, and tyrosine-containing substrates (Fig. 1). As observed previously with the Corby strain of L. pneumophila (20), this activity appeared the greatest when using the leucine- and lysine-containing substrates. No activity was seen against Cys-βNA (data not shown). When the wild type was grown in BYE broth that had 25% of its usual yeast extract content, there was a ca. 97% reduction in activity in supernatants compared to the activity in supernatants from standard BYE. Hence, as might be expected, secreted activity is responsive to the amount of potential substrate in the growth medium. These data confirm that L. pneumophila strains secrete activity against a variety of aminopeptides.
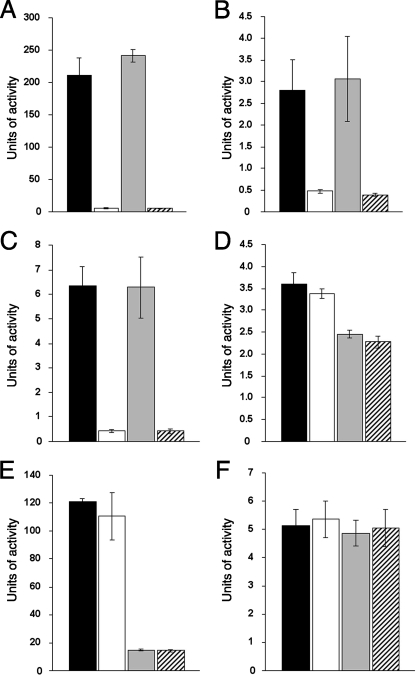
FIG. 1.
Secreted aminopeptidase activities of the wild type versus lapA and lapB mutants of L. pneumophila. Cell-free supernatants obtained from late log cultures of wild-type 130b (black bars), lapA mutant NU320 (white bars), lapB mutant NU322 (gray bars), and lapA lapB double mutant NU324 (hatched bars) were tested for their ability to cleave Leu-pNI (A), Phe-βNA (B), Tyr-βNA (C), Arg-βNA (D), Lys-pNI (E), and Ala-βNA (F). The values presented are the means and standard deviations obtained from triplicate samples and are representative of at least four independent experiments. The reduced levels of activity seen for the lapA mutants (A to C) and the lapB mutants (D and E) were statistically significant (P < 0.05; Student's t test).
As predicted from the annotation, supernatants derived from lapA mutant NU320 lacked leucine aminopeptidase activity (Fig. 1A). Since the independently derived lapA mutant NU321 also completely lacked this activity and since lapA is monocistronic, these data indicate that this defect was due to loss of LapA. We next observed that the lapA mutant also completely lacked activity against Phe-βNA and L-Tyr-βNA (Fig. 1B and C), indicating that the ability of LapA to cleave is not specific for leucine aminopeptides. In contrast to the lapA mutant, the lapB mutant did not show any loss of activity against leucine, phenylalanine, or tyrosine substrates (Fig. 1A to C). Supernatants from the independently derived lapA lapB double mutant lacked activity against leucine, phenylalanine, and tyrosine aminopeptides in the same way as did the lapA mutants (Fig. 1A to C). When testing activity against L-Arg-βNA and Lys-pNI, we observed that the LapB mutant, but not the LapA mutant, lacked activity (Fig. 1D and E). LapB mutant NU322 showed a complete loss of lysine aminopeptidase activity and a partial reduction in arginine aminopeptidase activity. Supernatants from the lapA lapB double mutant lacked activity against arginine and lysine aminopeptides in the same way as did the lapB mutant (Fig. 1D and E). When an intact copy of lapB was reintroduced into NU322 on complementing plasmid pMlapB, there was restoration of aminopeptidase activity (data not shown). Thus, the losses in arginine and lysine aminopeptidase activities in the lapB mutant were due to the absence of LapB. None of the single or double lap mutants lacked alanine aminopeptidase activity in their supernatants (Fig. 1F). Taken together, these data indicate that, under standard growth conditions, LapA is entirely responsible for secreted leucine, phenylalanine, and tyrosine aminopeptidase activities, whereas LapB is entirely responsible for the lysine aminopeptidase activity and at least partly responsible for the organism's secreted arginine aminopeptidase activity.
Supporting the fact that LapA and LapB are secreted via the type II secretion system, we observed that the leucine and lysine aminopeptidase activities were diminished in the supernatants of an lspF mutant (Fig. 2) but at wild-type levels in supernatants of a complemented lspF mutant (data not shown). Interestingly, the alanine aminopeptidase activity that is not dependent upon LapA or LapB was always fully present within the supernatants of the lspF mutant (Fig. 2), suggesting that it is secreted or released by a mechanism other than standard type II secretion.
FIG. 2.
Secreted aminopeptidase activities of the wild type versus an lspF mutant of L. pneumophila. Cell-free supernatants obtained from late log cultures of wild-type 130b (black boxes) and lspF mutant NU275 (white bars) were tested, as indicated, for their ability to cleave Leu-pNI, Lys-pNI, and Ala-βNA. The values presented are the means and standard deviations obtained from triplicate samples and are representative of at least four independent experiments. The lspF mutant's reduced levels of activity against Leu-pNI and Lys-pNI were statistically significant (P < 0.05; Student's t test).
Roles of lapA and lapB in L. pneumophila infection.
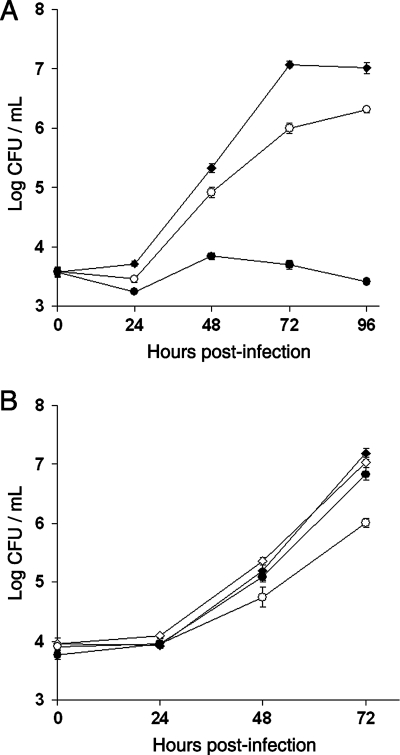
On many occasions, we and others have observed that lsp mutants of strain 130b are impaired in their ability to infect H. vermiformis amoebae and human U937 cell macrophages (23, 44, 55, 60-62). Others have shown that lsp mutants of strain Philadelphia-1 are unable to grow in A. castellanii amoebae (37). Thus, to assess the role of aminopeptidases in intracellular infection, we compared strain 130b and the LapA and LapB mutants for their abilities to infect these three disparate hosts. Mutants lacking one or both of the aminopeptidases showed no defect in H. vermiformis amoebae, A. castellanii amoebae, or U937 cell macrophages (Fig. 3). Thus, the losses of LapA and LapB do not account for the previously reduced inability of lsp mutants to grow within host cells in vitro. To ascertain the in vivo significance of the type II-secreted aminopeptidases, we assayed the ability of the lapA lapB double mutant NU324 to grow in the lungs of A/J mice. Unlike an lspF mutant (23, 62), NU324 grew and displayed a recoverability from the lungs that was similar to wild type (data not shown). These data indicate that lapA and lapB are not required for infection.
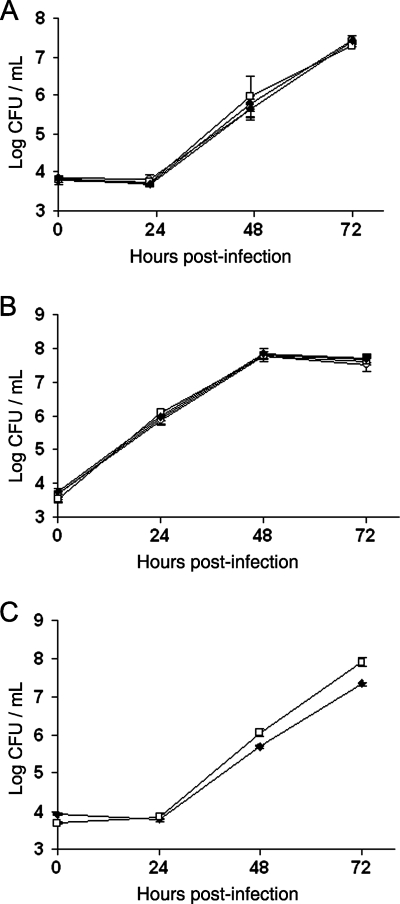
FIG. 3.
Intracellular infection of amoebae and macrophages by wild-type and aminopeptidase mutants of L. pneumophila. H. vermiformis (A), U937 cells (B), and A. castellanii (C) were infected with wild-type 130b (⧫), lapA mutant NU320 (○), lapB mutant NU322 (•), or lapA lapB double mutant NU324 (□) and then, at various times postinoculation, the numbers of bacteria per well were determined. The values presented are the means and standard deviations obtained from four (A) or three (B and C) infected wells and are representative of at least two independent experiments.
Role for the secreted zinc metalloprotease in intracellular infection of protozoa.
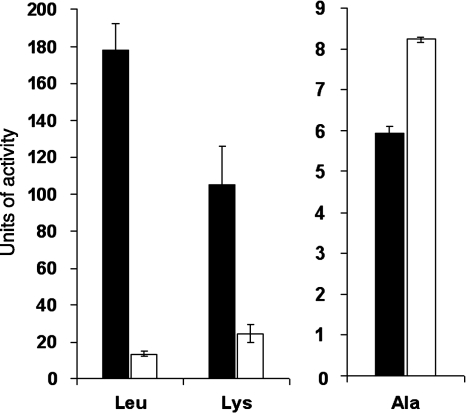
While testing the Lap mutants, we thought it would be instructive to also examine a 130b mutant lacking another type of proteolytic enzyme, i.e., the secreted ProA/MspA metalloprotease. At the outset, we viewed the protease mutant as another control. Indeed, when tested in the aminopeptidase assays, mutant AA200 did not display a reduction in activity against Leu-pNI, Phe-βNA, Tyr-βNA, or Ala-βNA (Fig. 4A). Supernatants from the proA mutant did show losses in activity against Arg-βNA and Lys-pNI that were comparable to those seen for the lapB mutant (Fig. 4B), suggesting that the protease may be needed for activation of LapB as it is for the expression of some other type-II secreted enzymes (8, 31). For intracellular infection, the protease mutant AA200 behaved like the wild type did when grown in U937 cells (data not shown) and A. castellanii strain 30234 (Fig. 5). As expected (see above), our lspF mutant of strain 130b showed a greatly reduced ability to grow in A. castellanii (Fig. 5). The U937 cell infection results obtained for the protease mutant were also in agreement with the earlier work of others, which showed that ProA is dispensable for infection of the HL-60 macrophage cell line and explanted guinea pig macrophages (51, 73). Similarly, Moffat et al. also previously observed that the proA mutant AA200 is not defective for growth in A. castellanii 30234 (51). Using the assay methods of Moffat et al., Hales and Shuman observed that a mspA mutant derived from L. pneumophila strain Philadelphia-1 does not show optimal growth in A. castellanii 30234 (37). It is not clear why we and Moffat et al. obtained a result that is different from that of Hales and Shuman. On the one hand, it is possible that proA/mspA is important for infection of acanthamoebae by some (e.g., Philadelphia-1) but not all (e.g., 130b) strains of L. pneumophila. On the other hand, since only a single mspA mutant derived from Philadelphia-1 was tested and no subsequent complementation analysis was done, it is possible that the growth defect seen was due not to the loss of mspA/proA but to a second-site mutation.
FIG. 4.
Secreted aminopeptidase activities of the wild type versus a proA mutant of L. pneumophila. Cell-free supernatants obtained from late log cultures of wild-type 130b (black boxes) and proA mutant AA200 (white bars) were tested, as indicated, for their ability to cleave Phe-βNA, Tyr-βNA, Ala-βNA, and Leu-pNI (A) or Arg-βNA and Lys-pNI (B). The values presented are the means and standard deviations obtained from triplicate samples and are representative of at least three (A) or four (B) independent experiments. Mutant AA200's reduced levels of activity against Arg-βNA and Lys-pNI (B) were statistically significant (P < 0.05; Student's t test).
FIG. 5.
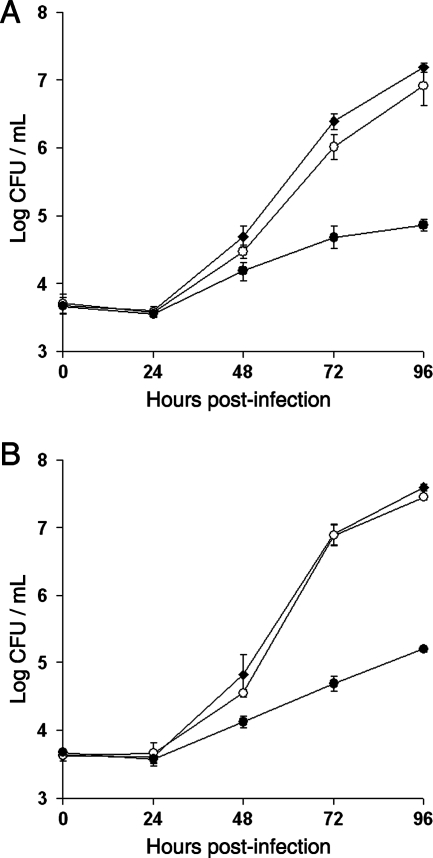
Intracellular infection of A. castellanii by the wild type, an lspF mutant, and a proA mutant of L. pneumophila. Acanthamoebae, cultured in standard proteose peptone-yeast extract-glucose medium (A) or 1034 medium (B), were infected with wild-type 130b (⧫), lspF mutant NU275 (•), and proA mutant AA200 (○) and then, at various times postinoculation, the numbers of bacteria per well were determined. The values presented are the means and standard deviations obtained from three infected wells and are representative of at least two independent experiments. At 48 to 96 h postinoculation, and under both growth conditions, the recovery of the lspF mutant was significantly less than that of the wild type and the other mutant (P < 0.05; Student's t test). In contrast, the slightly reduced recoveries of the proA mutant relative to the wild type seen at some points were not significant.
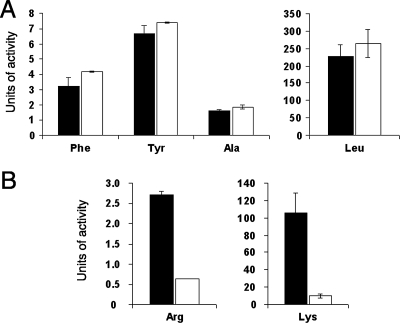
When we examined AA200 for its ability to infect H. vermiformis, a type of amoebae that had not been examined previously with any proA or mspA mutant, we observed a statistically significant defect. Indeed, the proA mutant consistently showed a ca. 10-fold-reduced recovery after 48 to 96 h of incubation with these amoebae (Fig. 6A). The mutant did not exhibit any reduced survivability when incubated in the 1034 culture medium alone (data not shown). Importantly, complementation of the mutant phenotype occurred when an intact copy of proA was introduced on a plasmid (Fig. 6B). Also, an independently derived mutant (NU327) inactivated for both lapB and proA displayed a defect in H. vermiformis that was identical to that of AA200 (data not shown). That AA200 was not defective when infecting acanthamoebae that were cultured in 1034 medium (Fig. 5B) indicates that the reduced recoverability of the proA mutant from H. vermiformis was not simply an artifact of the particular medium used to grow those amoebae. Together, these data indicate that the secreted zinc metalloprotease of L. pneumophila promotes optimal intracellular infection but that its importance is dependent upon the host system being used. Since the proA mutant was not as defective in H. vermiformis as the lspF mutant (Fig. 6A), there are likely to be more type II effectors that facilitate intracellular infection.
FIG. 6.
Intracellular infection of H. vermiformis by the wild type and metalloprotease mutants of L. pneumophila. Hartmannellae in 1034 medium were infected and then, at various times postinoculation, the numbers of bacteria per well were determined. The values presented are the means and standard deviations obtained from four infected wells. (A) Infection comparison between wild-type 130b (⧫), lspF mutant NU275 (•), and proA mutant AA200 (○). At 48 to 96 h postinfection, significant differences in bacterial recovery were obtained between the wild type and both of the mutants (P < 0.01; Student's t test). (B) Infection comparison between 130b(pMMB2002) (⧫), 130b(pMproA) (⋄), AA200(pMMB2002) (○), and AA200(pMproA) (•). pMMB2002 is the vector into which proA was cloned for the purposes of this complementation experiment. At 72 h, significant differences were obtained between AA200(pMMB2002) and all other strains (P < 0.01). The experiments presented here are representative of at least six (A) or three (B) other independent experiments. The proA mutant was similarly defective in an additional experiment that used a multiplicity of infection equal to 1.0 (versus 0.1).
DISCUSSION
Bacteria produce many types of aminopeptidases, but most of these enzymes are cell associated (32, 41, 47). Indeed, secreted aminopeptidases have only been described for Aeromonas, Bacillus, Clostridium, Pseudomonas, Streptomyces, and Vibrio (5, 13, 32, 35, 76). We have now confirmed that L. pneumophila also secretes aminopeptidases. LapA and LapB are the second and third examples of aminopeptidases being secreted via a type II secretion system, with the first example being from P. aeruginosa (13, 48). LapA and LapB are within the M28 family of peptidases, based on shared homology that encompasses amino acid residues 195 to 401 in LapA and residues 189 to 393 in LapB. The best-characterized members of the family are the secreted aminopeptidases of Streptomyces griseus and Vibrio proteolyticus (15, 33). When LapA is aligned with these aminopeptidases, all of the conserved residues of the active site (H217, D230, E265, D292, and H370) and 9 of the 10 residues that line the hydrophobic pocket adjacent to the zinc-binding site are found. When LapB is aligned with the Vibrio enzyme, residues in the active site (H213, D226, E261, D288, and H366) and residues along the hydrophobic pocket are again conserved. Based on the different types of losses in activity that we observed for the lapA and lapB mutants, LapA appears to be directed against hydrophobic aminopeptides, whereas LapB targets positively charged, hydrophilic aminopeptides. Thus, LapA and LapB perform distinct yet complementary enzymatic activities for L. pneumophila.
As a group, bacterial aminopeptidases perform a variety of functions, including the catabolism of exogenous peptides, the modulation of intracellular protein turnover, the N-terminal cleavage of nascent proteins, and the repression of transcription (32, 41, 47). Because they are secreted, LapA and LapB are most likely involved in the catabolism of exogenous peptides. Although the lapA and lapB mutants were not impaired for growth in BYE broth, U937 cells, acanthamoebae, or hartmannellae, we did not conclude that LapA and LapB are irrelevant for growth. For example, it is possible that these enzymes are critical for growth in other specialized niches or conditions. L. pneumophila might also produce other aminopeptidases or proteases that compensate for the absence of LapA and LapB. In support of the latter scenario, the L. pneumophila genome predicts the existence of a third, type II-secreted aminopeptidase that is related to LapA and LapB (23). Also, as observed in the present study, wild-type supernatants contain an alanine aminopeptidase activity that is not type II dependent. Finally, carboxypeptidase activity has been seen in L. pneumophila culture supernatants (9, 10), and other secreted (nonamino) peptidases and proteases have been predicted from the genome (14, 16, 23).
An arguably unexpected finding of our study is that L. pneumophila infection of H. vermiformis amoebae is affected by the absence of the type II-secreted metalloprotease, since earlier studies had not shown a role for ProA/MspA during infection of macrophages and A. castellanii amoebae (51, 73). Thus, our work not only uncovers a previously overlooked role for ProA/MspA but illustrates how the significance of a Legionella trait can vary depending on the host being used. Though a good number of L. pneumophila factors display differences in importance when comparing protozoan and mammalian hosts (52, 72), our observations indicate that distinctions can also be made between amoeba hosts. L. pneumophila infects at least seven genera and 17 species of protozoa (28, 67) and, therefore, it is reasonable to think that certain bacterial factors might have evolved to have greater importance in only certain protozoa. Previous studies have found that the growth and uptake of wild-type L. pneumophila can vary between Acanthamoeba, Hartmannella, and Naegleria hosts (24, 38, 55, 71). But, few past studies have examined specific mutants of L. pneumophila for their capacity to infect multiple protozoa (55, 77). One of these studies described mutants that exhibit different behaviors in different amoebae, but the insertion mutations described therein were not backcrossed or complemented (55). Thus, proA/mspA represents an L. pneumophila gene that clearly shows differential importance among protozoan models. Although a need for ProA is only evident from infections of H. vermiformis, previous studies showed that proA and its protein product are expressed during infection of A. castellanii and macrophages (51, 59). Thus, we hypothesize that in some hosts, such as acanthamoebae and macrophages, ProA/MspA is dispensable such that other proteases can, if necessary, fulfill its role, but that in other hosts, such as H. vermiformis, the need for ProA/MspA is greater and/or cannot be complemented by alternate proteases.
The metalloprotease may be promoting optimal intracellular infection by helping the bacterium to obtain amino acid nutrients. In support of this scenario, amino acid transporters in L. pneumophila and the host cell are required for optimal intracellular growth (64, 78). On the other hand, it is possible that ProA/MspA degrades host factors that are designed to control bacterial growth or cleaves other secreted Legionella proteins that directly promote intracellular infection. In infected macrophages, the protease is seen in both the Legionella phagosome and the adjacent host cytoplasm (59), supporting the possibility of it having multiple intracellular targets. Clearly, the protease's greater role in Hartmannella hosts should facilitate future investigations into the protein's intracellular role. Since ProA promotes optimal infection of H. vermiformis, an amoeba that is an environmental reservoir for legionellae (19, 28), we can now conclude that the protease plays a role in the ecology of L. pneumophila. Coupled with this environmental role, ProA contributes to disease by mediating lung damage and by inhibiting cytokine, phagocyte, NK cell, and T-cell function (21, 23, 39, 49, 58, 63, 79). Thus, ProA promotes the natural history and pathogenesis of Legionnaires' disease in multiple ways.
Finally, the results presented here increase our appreciation and understanding of type II secretion. First, with the characterization of LapA and LapB, the L. pneumophila type II secretome is now expanded to include at least 14 enzymatic activities and as such is arguably one of the best-characterized type II secretion systems (2-4, 8, 22, 30, 31, 37, 44, 61, 62). Second, with the demonstration of a role for ProA/MspA in H. vermiformis infection, we now have the first example of a type II-secreted protein promoting optimal intracellular infection. This confirms our previous hypothesis that the intracellular defects of lsp mutants are due, at least in part, to the loss of secreted effectors, as opposed to being strictly due to potential cell-associated (e.g., envelope) changes. Since a proA mutant is not as defective in H. vermiformis as an lsp mutant, we believe that there are more type II effectors that facilitate infection. Indeed, the relatively modest effect of the proA mutation is compatible with a scenario in which the importance of type II secretion derives from an additive effect of multiple secreted proteins. A similar conclusion has been drawn concerning the Dot/Icm type IV secretion system of L. pneumophila in which some effectors, analogous to LapA and LapB, are completely dispensable, while others, like ProA, contribute but in a relatively small way (43, 46).
Acknowledgments
We acknowledge members of the Cianciotto lab for helpful comments and advice. We again thank Jennifer Moffat and Lucy Tompkins for having previously sent us their proA mutant.
This work was supported by NIH grant AI43987, awarded to N.P.C.
Footnotes
Published ahead of print on 14 December 2007.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Aragon, V., S. Kurtz, and N. P. Cianciotto. 2001. Legionella pneumophila major acid phosphatase and its role in intracellular infection. Infect. Immun. 69:177-185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aragon, V., S. Kurtz, A. Flieger, B. Neumeister, and N. P. Cianciotto. 2000. Secreted enzymatic activities of wild-type and pilD-deficient Legionella pneumophila. Infect. Immun. 68:1855-1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aragon, V., O. Rossier, and N. P. Cianciotto. 2002. Legionella pneumophila genes that encode lipase and phospholipase C activities. Microbiology 148:2223-2231. [DOI] [PubMed] [Google Scholar]
- 5.Arima, J., Y. Uesugi, M. Uraji, S. Yatsushiro, S. Tsuboi, M. Iwabuchi, and T. Hatanaka. 2006. Modulation of Streptomyces leucine aminopeptidase by calcium: identification and functional analysis of key residues in activation and stabilization by calcium. J. Biol. Chem. 281:5885-5894. [DOI] [PubMed] [Google Scholar]
- 6.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.). 1989. Current protocols in molecular biology, vol. 1. Wiley, New York, NY.
- 7.Bandyopadhyay, P., S. Liu, C. B. Gabbai, Z. Venitelli, and H. M. Steinman. 2007. Environmental mimics and the Lvh type IVA secretion system contribute to virulence-related phenotypes of Legionella pneumophila. Infect. Immun. 75:723-735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Banerji, S., M. Bewersdorff, B. Hermes, N. P. Cianciotto, and A. Flieger. 2005. Characterization of the major secreted zinc metalloprotease-dependent glycerophospholipid:cholesterol acyltransferase, PlaC, of Legionella pneumophila. Infect. Immun. 73:2899-2909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Berdal, B. P., K. Bovre, O. Olsvik, and T. Omland. 1983. Patterns of extracellular proline-specific endopeptidases in Legionella and Flavobacterium spp. demonstrated by use of chromogenic peptides. J. Clin. Microbiol. 17:970-974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Berdal, B. P., O. Olsvik, S. Myhre, and T. Omland. 1982. Demonstration of extracellular chymotrypsin-like activity from various Legionella species. J. Clin. Microbiol. 16:452-457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bouyer, S., C. Imbert, M. H. Rodier, and Y. Hechard. 2007. Long-term survival of Legionella pneumophila associated with Acanthamoeba castellanii vesicles. Environ. Microbiol. 9:1341-1344. [DOI] [PubMed] [Google Scholar]
- 12.Brieland, J., M. McClain, M. LeGendre, and C. Engleberg. 1997. Intrapulmonary Hartmannella vermiformis: a potential niche for Legionella pneumophila replication in a murine model of legionellosis. Infect. Immun. 65:4892-4896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cahan, R., I. Axelrad, M. Safrin, D. E. Ohman, and E. Kessler. 2001. A secreted aminopeptidase of Pseudomonas aeruginosa. Identification, primary structure, and relationship to other aminopeptidases. J. Biol. Chem. 276:43645-43652. [DOI] [PubMed] [Google Scholar]
- 14.Cazalet, C., C. Rusniok, H. Bruggemann, N. Zidane, A. Magnier, L. Ma, M. Tichit, S. Jarraud, C. Bouchier, F. Vandenesch, F. Kunst, J. Etienne, P. Glaser, and C. Buchrieser. 2004. Evidence in the Legionella pneumophila genome for exploitation of host cell functions and high genome plasticity. Nat. Genet. 36:1165-1173. [DOI] [PubMed] [Google Scholar]
- 15.Chevrier, B., C. Schalk, H. D'Orchymont, J. M. Rondeau, D. Moras, and C. Tarnus. 1994. Crystal structure of Aeromonas proteolytica aminopeptidase: a prototypical member of the co-catalytic zinc enzyme family. Structure 2:283-291. [DOI] [PubMed] [Google Scholar]
- 16.Chien, M., I. Morozova, S. Shi, H. Sheng, J. Chen, S. M. Gomez, G. Asamani, K. Hill, J. Nuara, M. Feder, J. Rineer, J. J. Greenberg, V. Steshenko, S. H. Park, B. Zhao, E. Teplitskaya, J. R. Edwards, S. Pampou, A. Georghiou, I. C. Chou, W. Iannuccilli, M. E. Ulz, D. H. Kim, A. Geringer-Sameth, C. Goldsberry, P. Morozov, S. G. Fischer, G. Segal, X. Qu, A. Rzhetsky, P. Zhang, E. Cayanis, P. J. De Jong, J. Ju, S. Kalachikov, H. A. Shuman, and J. J. Russo. 2004. The genomic sequence of the accidental pathogen Legionella pneumophila. Science 305:1966-1968. [DOI] [PubMed] [Google Scholar]
- 17.Cianciotto, N. P. 2005. Type II secretion: a protein secretion system for all seasons. Trends Microbiol. 13:581-588. [DOI] [PubMed] [Google Scholar]
- 18.Cianciotto, N. P., B. I. Eisenstein, C. H. Mody, G. B. Toews, and N. C. Engleberg. 1989. A Legionella pneumophila gene encoding a species-specific surface protein potentiates initiation of intracellular infection. Infect. Immun. 57:1255-1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cianciotto, N. P., and B. S. Fields. 1992. Legionella pneumophila mip gene potentiates intracellular infection of protozoa and human macrophages. Proc. Natl. Acad. Sci. USA 89:5188-5191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Conlan, J. W., A. Baskerville, and L. A. E. Ashworth. 1986. Separation of Legionella pneumophila proteases and purification of a protease which produces lesions like those of Legionnaires' disease in guinea pig lung. J. Gen. Microbiol. 132:1565-1574. [DOI] [PubMed] [Google Scholar]
- 21.Conlan, J. W., A. Williams, and L. A. Ashworth. 1988. In vivo production of a tissue-destructive protease by Legionella pneumophila in the lungs of experimentally infected guinea-pigs. J. Gen. Microbiol. 134:143-149. [DOI] [PubMed] [Google Scholar]
- 22.DebRoy, S., V. Aragon, S. Kurtz, and N. P. Cianciotto. 2006. Legionella pneumophila Mip, a surface-exposed peptidylproline cis-trans-isomerase, promotes the presence of phospholipase C-like activity in culture supernatants. Infect. Immun. 74:5152-5160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.DebRoy, S., J. Dao, M. Soderberg, O. Rossier, and N. P. Cianciotto. 2006. Legionella pneumophila type II secretome reveals unique exoproteins and a chitinase that promotes bacterial persistence in the lung. Proc. Natl. Acad. Sci. USA 103:19146-19151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Declerck, P., J. Behets, Y. Delaedt, A. Margineanu, E. Lammertyn, and F. Ollevier. 2005. Impact of non-Legionella bacteria on the uptake and intracellular replication of Legionella pneumophila in Acanthamoeba castellanii and Naegleria lovaniensis. Microb. Ecol. 50:536-549. [DOI] [PubMed] [Google Scholar]
- 25.Dreyfus, L. A., and B. H. Iglewski. 1986. Purification and characterization of an extracellular protease of Legionella pneumophila. Infect. Immun. 51:736-743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Edwards, R. A., L. H. Keller, and D. M. Schifferli. 1998. Improved allelic exchange vectors and their use to analyze 987P fimbria gene expression. Gene 207:149-157. [DOI] [PubMed] [Google Scholar]
- 27.Engleberg, N. C., D. J. Drutz, and B. I. Eisenstein. 1984. Cloning and expression of Legionella pneumophila antigens in Escherichia coli. Infect. Immun. 44:222-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fields, B. S. 1996. The molecular ecology of legionellae. Trends Microbiol. 4:286-290. [DOI] [PubMed] [Google Scholar]
- 29.Fields, B. S., R. F. Benson, and R. E. Besser. 2002. Legionella and Legionnaires' disease: 25 years of investigation. Clin. Microbiol. Rev. 15:506-526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Flieger, A., S. Gong, M. Faigle, S. Stevanovic, N. P. Cianciotto, and B. Neumeister. 2001. Novel lysophospholipase A secreted by Legionella pneumophila. J. Bacteriol. 183:2121-2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Flieger, A., B. Neumeister, and N. P. Cianciotto. 2002. Characterization of the gene encoding the major secreted lysophospholipase A of Legionella pneumophila and its role in detoxification of lysophosphatidylcholine. Infect. Immun. 70:6094-6106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gonzales, T., and J. Robert-Baudouy. 1996. Bacterial aminopeptidases: properties and functions. FEMS Microbiol. Rev. 18:319-344. [DOI] [PubMed] [Google Scholar]
- 33.Greenblatt, H. M., O. Almog, B. Maras, A. Spungin-Bialik, D. Barra, S. Blumberg, and G. Shoham. 1997. Streptomyces griseus aminopeptidase: X-ray crystallographic structure at 1.75 Å resolution. J. Mol. Biol. 265:620-636. [DOI] [PubMed] [Google Scholar]
- 34.Grindley, N. D., and C. M. Joyce. 1980. Genetic and DNA sequence analysis of the kanamycin resistance transposon Tn903. Proc. Natl. Acad. Sci. USA 77:7176-7180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guenet, C., P. Lepage, and B. A. Harris. 1992. Isolation of the leucine aminopeptidase gene from Aeromonas proteolytica. Evidence for an enzyme precursor. J. Biol. Chem. 267:8390-8395. [PubMed] [Google Scholar]
- 36.Gul'nik, S. V., M. P. Yusupova, G. I. Lavrenova, I. S. Tartakovsky, S. V. Prozorovsky, and V. M. Stepanov. 1986. Proteinases of Legionella: phenylalanineaminopeptidase of L. pneumophila. J. Gen. Microbiol. 132:387-392. [DOI] [PubMed] [Google Scholar]
- 37.Hales, L. M., and H. A. Shuman. 1999. Legionella pneumophila contains a type II general secretion pathway required for growth in amoebae as well as for secretion of the Msp protease. Infect. Immun. 67:3662-3666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Harb, O. S., C. Venkataraman, B. J. Haack, L. Y. Gao, and Y. A. Kwaik. 1998. Heterogeneity in the attachment and uptake mechanisms of the Legionnaires' disease bacterium, Legionella pneumophila, by protozoan hosts. Appl. Environ. Microbiol. 64:126-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hell, W., A. Essig, S. Bohnet, S. Gatermann, and R. Marre. 1993. Cleavage of tumor necrosis factor-alpha by Legionella exoprotease. APMIS 101:120-126. [DOI] [PubMed] [Google Scholar]
- 40.Jacobi, S., and K. Heuner. 2003. Description of a putative type I secretion system in Legionella pneumophila. Int. J. Med. Microbiol. 293:349-358. [DOI] [PubMed] [Google Scholar]
- 41.Jankiewicz, U., and W. Bielawski. 2003. The properties and functions of bacterial aminopeptidases. Acta Microbiol. Pol. 52:217-231. [PubMed] [Google Scholar]
- 42.Johnson, T. L., J. Abendroth, W. G. Hol, and M. Sandkvist. 2006. Type II secretion: from structure to function. FEMS Microbiol. Lett. 255:175-186. [DOI] [PubMed] [Google Scholar]
- 43.Laguna, R. K., E. A. Creasey, Z. Li, N. Valtz, and R. R. Isberg. 2006. A Legionella pneumophila-translocated substrate that is required for growth within macrophages and protection from host cell death. Proc. Natl. Acad. Sci. USA 103:18745-18750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liles, M. R., P. H. Edelstein, and N. P. Cianciotto. 1999. The prepilin peptidase is required for protein secretion by and the virulence of the intracellular pathogen Legionella pneumophila. Mol. Microbiol. 31:959-970. [DOI] [PubMed] [Google Scholar]
- 45.Little, G. H., W. L. Starnes, and F. J. Behal. 1976. Human liver aminopeptidase. Methods Enzymol. 45:495-503. [DOI] [PubMed] [Google Scholar]
- 46.Liu, Y., and Z. Q. Luo. 2007. The Legionella pneumophila effector SidJ is required for efficient recruitment of endoplasmic reticulum proteins to the bacterial phagosome. Infect. Immun. 75:592-603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Matsui, M., J. H. Fowler, and L. L. Walling. 2006. Leucine aminopeptidases: diversity in structure and function. Biol. Chem. 387:1535-1544. [DOI] [PubMed] [Google Scholar]
- 48.Michel, G. P., E. Durand, and A. Filloux. 2007. XphA/XqhA, a novel GspCD subunit for type II secretion in Pseudomonas aeruginosa. J. Bacteriol. 189:3776-3783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mintz, C. S., R. D. Miller, N. S. Gutgsell, and T. Malek. 1993. Legionella pneumophila protease inactivates interleukin-2 and cleaves CD4 on human T cells. Infect. Immun. 61:3416-3421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Moffat, J. F., W. J. Black, and L. S. Tompkins. 1994. Further molecular characterization of the cloned Legionella pneumophila zinc metalloprotease. Infect. Immun. 62:751-753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Moffat, J. F., P. H. Edelstein, D. P. Regula, Jr., J. D. Cirillo, and L. S. Tompkins. 1994. Effects of an isogenic Zn-metalloprotease-deficient mutant of Legionella pneumophila in a guinea-pig pneumonia model. Mol. Microbiol. 12:693-705. [DOI] [PubMed] [Google Scholar]
- 52.Molmeret, M., M. Horn, M. Wagner, M. Santic, and Y. Abu Kwaik. 2005. Amoebae as training grounds for intracellular bacterial pathogens. Appl. Environ. Microbiol. 71:20-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Muller, H. E. 1981. Enzymatic profile of Legionella pneumophila. J. Clin. Microbiol. 13:423-426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nolte, F. S., G. E. Hollick, and R. G. Robertson. 1982. Enzymatic activities of Legionella pneumophila and Legionella-like organisms. J. Clin. Microbiol. 15:175-177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Polesky, A. H., J. T. Ross, S. Falkow, and L. S. Tompkins. 2001. Identification of Legionella pneumophila genes important for infection of amoebas by signature-tagged mutagenesis. Infect. Immun. 69:977-987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Prescott, J. M., and S. H. Wilkes. 1976. Aeromonas aminopeptidase. Methods Enzymol. 45:530-543. [DOI] [PubMed] [Google Scholar]
- 57.Quinn, F. D., and L. S. Tompkins. 1989. Analysis of a cloned sequence of Legionella pneumophila encoding a 38 kD metalloprotease possessing haemolytic and cytotoxic activities. Mol. Microbiol. 3:797-805. [DOI] [PubMed] [Google Scholar]
- 58.Rechnitzer, C., M. Diamant, and B. K. Pedersen. 1989. Inhibition of human natural killer cell activity by Legionella pneumophila protease. Eur. J. Clin. Microbiol. Infect. Dis. 8:989-992. [DOI] [PubMed] [Google Scholar]
- 59.Rechnitzer, C., A. Williams, J. B. Wright, A. B. Dowsett, N. Milman, and R. B. Fitzgeorge. 1992. Demonstration of the intracellular production of tissue-destructive protease by Legionella pneumophila multiplying within guinea-pig and human alveolar macrophages. J. Gen. Microbiol. 138:1671-1677. [DOI] [PubMed] [Google Scholar]
- 60.Rossier, O., and N. P. Cianciotto. 2005. The Legionella pneumophila tatB gene facilitates secretion of phospholipase C, growth under iron-limiting conditions, and intracellular infection. Infect. Immun. 73:2020-2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rossier, O., and N. P. Cianciotto. 2001. Type II protein secretion is a subset of the PilD-dependent processes that facilitate intracellular infection by Legionella pneumophila. Infect. Immun. 69:2092-2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rossier, O., S. Starkenburg, and N. P. Cianciotto. 2004. Legionella pneumophila type II protein secretion promotes virulence in the A/J mouse model of Legionnaires' disease pneumonia. Infect. Immun. 72:310-321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sahney, N. N., J. T. Summersgill, J. A. Ramirez, and R. D. Miller. 2001. Inhibition of oxidative burst and chemotaxis in human phagocytes by Legionella pneumophila zinc metalloprotease. J. Med. Microbiol. 50:517-525. [DOI] [PubMed] [Google Scholar]
- 64.Sauer, J. D., M. A. Bachman, and M. S. Swanson. 2005. The phagosomal transporter A couples threonine acquisition to differentiation and replication of Legionella pneumophila in macrophages. Proc. Natl. Acad. Sci. USA 102:9924-9929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schweizer, H. P., and T. T. Hoang. 1995. An improved system for gene replacement and xylE fusion analysis in Pseudomonas aeruginosa. Gene 158:15-22. [DOI] [PubMed] [Google Scholar]
- 66.Segarra, A. B., R. Wangensteen, M. Ramirez, I. Banegas, F. Hermoso, F. Vargas, F. Vives, R. Duran, F. Alba, M. de Gasparo, and I. Prieto. 2006. Atrial angiotensinase activity in hypothyroid, euthyroid, and hyperthyroid rats. J. Cardiovasc. Pharmacol. 48:117-120. [DOI] [PubMed] [Google Scholar]
- 67.Shadrach, W. S., K. Rydzewski, U. Laube, G. Holland, M. Ozel, A. F. Kiderlen, and A. Flieger. 2005. Balamuthia mandrillaris, free-living ameba and opportunistic agent of encephalitis, is a potential host for Legionella pneumophila bacteria. Appl. Environ. Microbiol. 71:2244-2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Söderberg, M. A., and N. P. Cianciotto. 2006. The type II protein secretion system of L. pneumophila is important for growth in iron-rich media and survival in tap water at low temperatures, p. 214-216. In N. P. Cianciotto, Y. Abu Kwaik, P. H. Edelstein, B. S. Fields, D. F. Geary, T. G. Harrison, C. A. Joseph, R. M. Ratcliff, J. E. Stout, and M. S. Swanson (ed.), Legionella: state of the art 30 years after its recognition. ASM Press, Washington, DC.
- 69.Söderberg, M. A., O. Rossier, and N. P. Cianciotto. 2004. The type II protein secretion system of Legionella pneumophila promotes growth at low temperatures. J. Bacteriol. 186:3712-3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Spungin, A., and S. Blumberg. 1989. Streptomyces griseus aminopeptidase is a calcium-activated zinc metalloprotein. Purification and properties of the enzyme. Eur. J. Biochem. 183:471-477. [DOI] [PubMed] [Google Scholar]
- 71.Steinert, M., M. Ott, P. C. Lück, E. Tannich, and J. Hacker. 1994. Studies on the uptake and intracellular replication of Legionella pneumophila in protozoa and in macrophage-like cells. FEMS Microbiol. Ecol. 15:299-308. [Google Scholar]
- 72.Swanson, M. S., and B. K. Hammer. 2000. Legionella pneumophila pathogenesis: a fateful journey from amoebae to macrophages. Annu. Rev. Microbiol. 54:567-613. [DOI] [PubMed] [Google Scholar]
- 73.Szeto, L., and H. A. Shuman. 1990. The Legionella pneumophila major secretory protein, a protease, is not required for intracellular growth or cell killing. Infect. Immun. 58:2585-2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tesh, M. J., S. A. Morse, and R. D. Miller. 1983. Intermediary metabolism in Legionella pneumophila: utilization of amino acids and other compounds as energy sources. J. Bacteriol. 154:1104-1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Thompson, M. R., R. D. Miller, and B. H. Iglewski. 1981. In vitro production of an extracellular protease by Legionella pneumophila. Infect. Immun. 34:299-302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Toma, C., and Y. Honma. 1996. Cloning and genetic analysis of the Vibrio cholerae aminopeptidase gene. Infect. Immun. 64:4495-5000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Viswanathan, V. K., S. Kurtz, L. L. Pedersen, Y. Abu-Kwaik, K. Krcmarik, S. Mody, and N. P. Cianciotto. 2002. The cytochrome c maturation locus of Legionella pneumophila promotes iron assimilation and intracellular infection and contains a strain-specific insertion sequence element. Infect. Immun. 70:1842-1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wieland, H., S. Ullrich, F. Lang, and B. Neumeister. 2005. Intracellular multiplication of Legionella pneumophila depends on host cell amino acid transporter SLC1A5. Mol. Microbiol. 55:1528-1537. [DOI] [PubMed] [Google Scholar]
- 79.Williams, A., A. Baskerville, A. B. Dowsett, and J. W. Conlan. 1987. Immunocytochemical demonstration of the association between Legionella pneumophila, its tissue-destructive protease, and pulmonary lesions in experimental Legionnaires' disease. J. Pathol. 153:257-264. [DOI] [PubMed] [Google Scholar]
- 80.Ye, S., S. Y. Chai, R. A. Lew, and A. L. Albiston. 2007. Insulin-regulated aminopeptidase: analysis of peptide substrate and inhibitor binding to the catalytic domain. Biol. Chem. 388:399-403. [DOI] [PubMed] [Google Scholar]