Abstract
The prevalence of bacteriophages was investigated in 24 strains of four species of plant growth-promoting rhizobacteria belonging to the genus Azospirillum. Upon induction by mitomycin C, the release of phage particles was observed in 11 strains from three species. Transmission electron microscopy revealed two distinct sizes of particles, depending on the identity of the Azospirillum species, typical of the Siphoviridae family. Pulsed-field gel electrophoresis and hybridization experiments carried out on phage-encapsidated DNAs revealed that all phages isolated from A. lipoferum and A. doebereinerae strains had a size of about 10 kb whereas all phages isolated from A. brasilense strains displayed genome sizes ranging from 62 to 65 kb. Strong DNA hybridizing signals were shown for most phages hosted by the same species whereas no homology was found between phages harbored by different species. Moreover, the complete sequence of the A. brasilense Cd bacteriophage (ΦAb-Cd) genome was determined as a double-stranded DNA circular molecule of 62,337 pb that encodes 95 predicted proteins. Only 14 of the predicted proteins could be assigned functions, some of which were involved in DNA processing, phage morphogenesis, and bacterial lysis. In addition, the ΦAb-Cd complete genome was mapped as a prophage on a 570-kb replicon of strain A. brasilense Cd, and a region of 27.3 kb of ΦAb-Cd was found to be duplicated on the 130-kb pRhico plasmid previously sequenced from A. brasilense Sp7, the parental strain of A. brasilense Cd.
Bacteriophages are likely to be numerically the most prominent biological systems on earth, with an estimated population size of ≥1030 phage particles (57). Bacteriophages are ubiquitous in nature, and it has been suggested that they are environmentally important both in controlling the number of bacteria and in facilitating gene transfer. Indeed, bacteriophages represent one of the three major mobile genetic elements that contribute significantly to horizontal gene transfer in bacterial genomes by transduction. It was calculated that 1024 bacteria could be infected per second, which implies that a continuous flow of genetic material occurs between bacteria sharing the same environments (60). Seventy percent of complete bacterial genomes investigated contain prophage sequences of >10 kb (16), and it is believed that well over 50% of bacteria isolated from various environments contain prophages or are lysogenized. The integrated viral DNA, referred to as a prophage, can harbor genes that contribute to increasing the fitness of bacterial cells, such as those encoding phage-encoded virulence factors of some pathogenic bacteria (15). Consequently, the majority of the data concerning prophages come from studies of gram-positive bacteria and gammaproteobacteria of medical and industrial interest (16), but very little information is available concerning phages isolated from commensal or symbiotic bacteria.
Plant-beneficial bacteria, including some strains belonging to the genus Azospirillum, are usually referred to as plant growth-promoting rhizobacteria. Members of the genus Azospirillum represent a group of free-living alphaproteobacteria whose promotion of the growth of cereals and grasses leads to an increase of crop yield of up to 30%. The actual benefit deriving from biological nitrogen fixation has been questioned, and plant growth promotion by Azospirillum spp. seems to be due mainly to production of phytohormones (7, 20). The most abundant phytohormone produced is indole-3-acetic acid, allowing an increase in the number of lateral roots and root hairs; this results in a higher absorption of water and minerals from the soil (7). Although many studies of Azospirillum spp. are related to bacterial physiology and ecology, few genomic data are available. It has been shown that the genomes of Azospirillum strains differ in size (from 4.8 to 9.7 Mb) and in architecture, one of the main features being the presence of numerous plasmids (14, 44). Some plasmids were shown to be involved in major genomic rearrangements occurring during phenotypic switching, indicating that genomes of Azospirillum are highly dynamic (63). To date, only two temperate phages were described for this genus. The first one, termed Al-1, was isolated from Brazilian soil and is able to form plaques on A. lipoferum Br17 (24). The second one was characterized after spontaneous or mitomycin C induction in an A. brasilense Sp7 culture (28, 30).
In the frame of investigation of the role of phages in the biology of Azospirillum spp., we first studied their prevalence by examining the ability of 24 Azospirillum strains from four species to release phage particles after mitomycin C treatment. Phage particles were isolated, and morphological studies were performed by transmission electron microscopy (TEM). Phage DNA was then purified, and the molecular relatedness of phages was investigated by Southern hybridization experiments. Finally, the complete genome sequence of the lytic phage ΦAb-Cd from A. brasilense Cd strain was obtained and analyzed.
MATERIALS AND METHODS
Media and bacterial growth conditions.
The Azospirillum strains used in this study are presented in Table 1. Azospirillum strains were grown at 28°C in modified Luria-Bertani medium (LBm) (containing only 5 g liter−1 of NaCl) or in nitrogen-free basal medium supplemented with 0.025% LBm (46). Rhizobium etli CFN42 was grown in tryptone-yeast extract medium supplemented with 6 mM CaCl2 (9). For cloning purposes, Escherichia coli strains containing recombinant plasmids were grown in 2YT (yeast extract [5 g liter−1], tryptone [16 g liter−1], Nacl [5 g liter−1]) broth or agar at 37°C with chloramphenicol at 100 μg ml−1 (53).
TABLE 1.
Azospirillum strains used in this study
| Strain | Relevant property | Source or reference | Lysis with mitomycin Ca | Estimated size of the encapsidated DNA (kb) |
|---|---|---|---|---|
| A. brasilense | ||||
| Aba2 | Strain isolated from soil (Senegal) | Laboratory collection | − | |
| Cd | Strain isolated from Cynodon dactylon (United States) | 25 | + | 62 |
| L4 | Strain isolated from sorghum (Mali) | 35 | − | |
| NC16 | Strain isolated from soil (Mali) | Laboratory collection | + | NDb |
| NC20 | Strain isolated from soil (Mali) | Laboratory collection | − | |
| R5(15) | Strain isolated from rice (Cuba) | Laboratory collection | − | |
| Sp7 | Strain isolated from Digitaria decumbens (Brazil) | 59 | + | 62 |
| Sp245 | Strain isolated from wheat (Brazil) | 4 | + | 65 |
| Wb1 | Strain isolated from wheat (Pakistan) | Laboratory collection | + | 62 |
| Wb3 | Strain isolated from wheat (Pakistan) | Laboratory collection | + | ND |
| Wn1 | Strain isolated from irrigated wheat (Pakistan) | Laboratory collection | − | |
| A. doebereinerae GSF71 | Strain isolated from Miscanthus sinensis (Germany) | 22 | + | 10 |
| A. irakense KBC1 | Strain isolated from rice (Iraq) | 36 | − | |
| A. lipoferum | ||||
| 4B | Strain isolated from rice rhizosphere (France) | 5 | + | 10 |
| 4T | Strain isolated from rice rhizosphere (France) | 5 | + | 10 |
| 4VI | Phase variant of 4B | 1 | + | 10 |
| B518 | Strain isolated from rice (Japan) | 23 | + | 10 |
| B510 | Strain isolated from rice (Japan) | 23 | + | 10 |
| Br17 | Strain isolated from maize (Brazil) | 59 | − | |
| CRT1 | Strain isolated from corn (France) | 34 | − | |
| MRB16 | Strain isolated from rice (Bangladesh) | Laboratory collection | − | |
| NC4 | Strain isolated from soil (Mali) | 35 | − | |
| RSWT1 | Strain isolated from rice (Pakistan) | Laboratory collection | − | |
| TVV3 | Strain isolated from rice (Vietnam) | 61 | + | 10 |
+, presence; −, absence.
ND, no phage DNA could be purified.
Induction and purification of phage particles.
For kinetics analysis of bacterial lysis, Azospirillum strains were grown in 100 ml LBm until the optical density at 580 nm reached 0.3. The cultures were then split equally into two flasks (50 ml in each), with one receiving mitomycin C (final concentration, 0.1 μg ml−1) and the other serving as a control. Incubation of both flasks was continued, and growth was monitored by measuring the optical density at 580 nm of subsamples every hour. For subsequent studies, incubation was carried out for 16 h after the addition of mitomycin C. Purification of phage particles from all strains was carried out by clearing the induced bacterial lysate (25 ml or 50 ml) by centrifugation two times at 5,000 × g for 10 min at 20°C. The supernatant was treated with 100 μg of DNase (Roche Diagnostics, Indianapolis, IN) for 1 h at 37°C to digest DNA of the bacterial host. NaCl was next added to achieve a final concentration of 1 M, and the culture supernatant was stirred until complete dissolution of the salt occurred, followed by centrifugation at 5,000 × g for 10 min at 20°C. The supernatant was transferred to a new tube, and 10% (wt/vol) polyethylene glycol 8000 (Fluka, Buchs, Switzerland) was added. The suspension was then agitated for 2 h at 28°C, and the precipitated phages were pelleted by centrifugation at 5,000 × g for 20 min at 20°C and finally resuspended in 2 × 0.5 ml of suspension buffer (100 mM NaCl, 10 mM MgSO4, 50 mM Tris-HCl [pH 7.5]). All nucleic acids present in the suspension outside the phage particles were eliminated by performing successive incubations as follows: (i) 30 min at 37°C with a mixture of DNase I and RNase (Euromedex, Mundolsheim, France) (final concentrations, 600 μg ml−1 each); (ii) 30 min with DNase I (final concentration, 1.2 mg ml−1); and (iii) 1 h with DNase I (final concentration, 2.4 mg ml−1). To isolate phage DNA, purified phage particles were first treated for 15 min at 65°C in a lysis solution containing 10 μl of proteinase K (Fermentas, Mundolsheim, France) (20 mg ml−1), 65 μl of EDTA (0.25 M), and 20 μl of sodium dodecyl sulfate (10%). Phage DNA was finally extracted using a phenol-and-chloroform procedure (53), and the DNA pellet was dissolved in 35 μl of ultra-pure water per 25 ml of starting culture.
TEM.
The production of phage particles in Azospirillum cultures was induced as described above. Then, bacterial lysate (12 ml) was cleared by two centrifugations at 5,000 × g for 10 min at 20°C and filtration through a 0.45-μm-pore-size filter (type CA; Millipore, Billerica, MA). The phage suspension was then transferred into a polyallomer centrifuge tube (14 by 89 mm; Beckman Coulter, Roissy, France) and centrifuged at 4°C for 5 h at 75,000 × g. The phage pellet was resuspended in 30 μl of suspension buffer without pipetting. A drop of phage suspension was applied to a 300-mesh carbon/formvar-coated grid. The sample was stained with either aqueous 1% phosphotungstic acid or 2% uranyl acetate for 30 s. Excess stain was removed with a filter paper, and the grids were allowed to air dry prior to examination using a Philips CM 120 transmission electron microscope at 100 kV.
PFGE of phage DNA and hybridization experiments.
Phage DNA was extracted from purified particles as described above. A total of 100 ng of undigested DNA was deposited per well of 1% agarose gel (pulsed-field certified agarose; Bio-Rad Laboratories) in 0.5× Tris-borate-EDTA buffer (Euromedex, Mundolsheim, France). Pulsed-field gel electrophoresis (PFGE) was run using a Chef-DRIII system (Bio-Rad Laboratories) with the following parameters: 0.5× Tris-borate-EDTA running buffer at 14°C, 1 s initial switch time, 6 s final switch time, 6 V cm−1, 15 h run time, and 14°C buffer temperature. The gels were stained with ethidium bromide and visualized using Gel-doc 2000 (Bio-Rad Laboratories, Hercules, CA). For Southern blot experiments, phage DNAs were transferred to GeneScreen Plus nylon filters (PerkinElmer, Zaventem, Belgium). Phage DNA randomly labeled with [α-32P]dCTP by use of a Redi-prime DNA labeling system (Amersham plc, Little Chalfont, England) was used as a probe. Southern blotting and hybridization were carried out according to established protocols (53).
DNA sequencing and bioinformatics analyses.
The DNA template used for sequencing the A. brasilense Cd phage was purified from phage particles as described above. The template was subjected to mechanical shearing and cloned as fragments of 10 kb into low-copy-number pCNS home vector (pSU18 modified) (6). Plasmid DNAs were purified and end sequenced using dye-terminator chemistry on ABI3730xl DNA analyzer sequencers. A total of 1,500 sequences were generated from both ends of the clones. The Phred/Phrap/Consed software package (www.phrap.com) was used for sequence assembly and quality assessment (26, 27, 31). Open reading frames (ORFs) in the final genome sequences were predicted using Glimmer 2.0 software (19) and annotated with ARTEMIS 8 software (52). In our analyses, putative ORFs contained AUG (methionine), UUG (leucine), or GUG (valine) as the starting codon. The predicted proteins were compared to those listed in the NCBI protein databases by using the BLASTX algorithm (2). Searches for putative conserved domains of identified proteins were done using the INTERPROSCAN and CD-Search programs (43, 67).
Phylogenetic analysis of functional genes.
The translated amino acid sequences of putative genes encoding terminase (ORF29), a phage major capsid protein (ORF33), lysozyme (ORF53), and integrase (ORF72) were used to construct phylogenetic trees. The BLAST program was used to identify sequences homologous to the sequences of these ORFs in the draft of the A. brasilense Sp245 genome (available at http://genomics.ornl.gov/research/azo). These amino acid sequences were aligned with sequences of viruses found in the GenBank database, using the program CLUSTAL W, and phylogenetic analyses were conducted using MEGA 4.0 software (58). A Blosum 30 matrix was calculated with a gap penalty of 10.0. Distance trees were constructed with the neighbor-joining algorithm, and the bootstrap method was employed with 1,000 replicates to estimate the robustness of the tree topologies.
Plasmid content and prophage localization.
Plasmids were separated by modified Eckhardt agarose gel electrophoresis (33) as applied by Vial and coworkers (63). Plasmid sizes were estimated by comparison with those of A. brasilense Sp7 (14) and Rhizobium etli CFN42 (51). Hybridization experiments were carried out on plasmid profiles as previously described (63). To obtain a probe specific to the A. brasilense Cd phage, several ORFs were first amplified by PCR using template DNA from strain Cd as follows: ORF40 was amplified with primers F4908 (5′-AGAAGATCAACGCCAGCTTC-3′) and F4909 (5′-GACTGTGTGCCTGCGTAGAA-3′), ORF50 with primers F5561 (5′-GCAAACCACAATCACCACAG-3′) and F5562 (5′-AAGCATCTGGTAACCGTTGG-3′), and ORF53 with primers F4906 (5′-GGGCTGTATCTGACCGCATA-3′) and F4907 (5′-CATTGACGGTGGCGTAGAC-3′). The amplification cycle consisted of an initial 5 min at 95°C and 35 cycles of 30 s at 95°C, 30 s at annealing temperature, and 30 s at 72°C followed by a final 7 min extension at 72°C. Then, ORF50 was chosen, randomly prime labeled as described above, and used as a probe to hybridize plasmid profiles. For a probe specific to the pRhico plasmid, the exoC gene was amplified with primers F5559 (5′-CCTTCAGGTCGACGAGATTC-3′) and F5560 (5′-GGACTACAACGGCATCAAGA-3′).
Nucleotide sequence accession number.
The accession number for the phage DNA sequence obtained in this study is CU468217.
RESULTS
Induction of temperate phages from different Azospirillum strains.
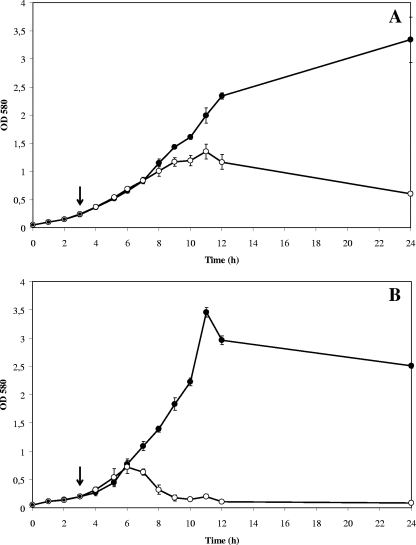
As phage-like elements had been induced previously in cultures of A. brasilense Sp7 treated with 0.1 μg ml−1 mitomycin C (28), a DNA-damaging agent able to activate the RecA-dependent induction of temperate bacteriophages in bacteria, the same procedure was applied in this study. Kinetics of bacterial lysis was monitored with several A. brasilense and A. lipoferum strains. For the A. brasilense Cd strain, the growth rate slowed down 6 h after the addition of mitomycin C and a decrease in cell biomass with the appearance of cell debris corresponding to bacterial lysis was observed 8 h after the beginning of the treatment (Fig. 1A). A similar curve was obtained with A. brasilense Sp245 (data not shown). For the A. lipoferum strains tested, the growth stopped 3 h after the addition of mitomycin C and this was immediately followed by a dramatic decline in cell biomass, pointing to the release of phage particles (see Fig. 1B for A. lipoferum B518 data; data not shown for A. lipoferum TVV3 and 4B). For these strains, the eclipse period (i.e., the time between the addition of mitomycin C and the release of phage particles) was always shorter and bacterial lysis was more striking and was nearly completed 9 h after contact with mitomycin C. For subsequent analysis of the other Azospirillum strains, induction was carried out during 16 h before cell lysis was examined. Overall, bacterial lysis was observed for 6 A. brasilense strains (out of 11), for 6 A. lipoferum strains (out of 11), and for the unique A. doebereinerae GSF71 strain tested (Table 1). As addition of mitomycin C was not accompanied by cell lysis for some strains, including the unique A. irakense strain, these findings demonstrate that cell lysis was not the result of bias due to the toxicity of mitomycin C. These data indicate that when an SOS response is triggered by the addition of mitomycin C, production of temperate phages might be induced in some Azospirillum strains.
FIG. 1.
Effect of mitomycin C treatment on growth of Azospirillum bacteria. (A) A. brasilense strain Cd; (B) A. lipoferum strain B518. Cell growth was followed by measurement of optical densities at 580 nm in a culture treated with 0.1 μg/ml mitomycin C (open circles) and in a control culture without treatment (closed circles). The addition of mitomycin is indicated by arrows.
Morphological study of Azospirillum bacteriophages.
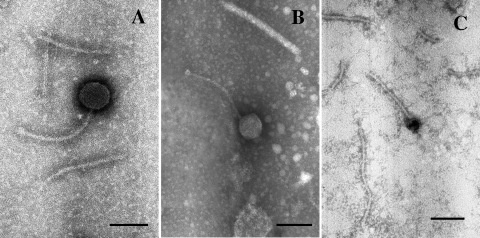
To ascertain that bacterial lysis was associated with the release of phage particles, the purification of phage-like particles from lysates of all strains was undertaken. TEM analysis was then performed on lysates from two A. brasilense and two A. lipoferum strains. Phage particles and/or phage parts (i.e., tail, head, or ghost) were detected in the lysates of the four bacterial strains. For all samples, the ultrastructures of phage particles appeared to be similar, displaying long noncontractile tails and polyhedral heads, a morphology that is typical of members of the Siphoviridae family of bacteriophages. Phage particles isolated from the two A. brasilense strains Cd and Wb1 exhibited heads of similar diameters (70 nm for both) and tails of similar sizes (with an average length of 210 nm) (Fig. 2A and B). Baseplates could sometimes be observed for those two samples. Observation of entire phage particles isolated from the two A. lipoferum strains was more difficult to achieve; phage heads had a diameter of 30 to 35 nm for both isolates, half the size of the heads of phages isolated from A. brasilense strains. The tail lengths were estimated at about 190 nm and 130 nm for phage particles isolated from A. lipoferum strains B518 (Fig. 2C) and TVV3 (data not shown), respectively. Thus, the bacterial lysis initially observed in the mitomycin C-treated cultures can be attributed to the release of temperate phages that are putative members of the Siphoviridae family. Those bacteriophages were named accordingly to the strain species and name; i.e., ΦAb-Cd designates the phage isolated from the strain A. brasilense Cd, ΦAl-B518 designates the phage isolated from the strain A. lipoferum B518, etc.
FIG. 2.
TEM micrographs of negatively stained bacteriophage particles of Azospirillum bacteria. After 0.1 μg ml−1 mitomycin C was added, phage particles were observed in the lysates of A. brasilense Cd (A), A. brasilense Wb1 (B), and A. lipoferum B518 (C). Bars, 100 nm.
Relatedness of Azospirillum bacteriophages at the genomic level.
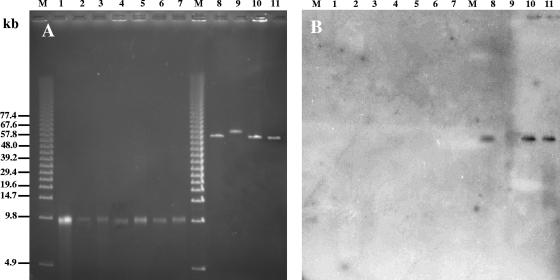
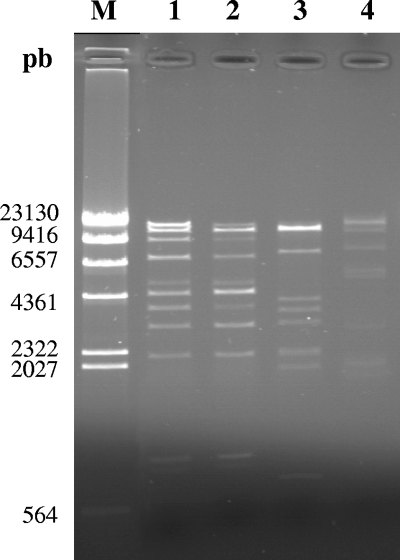
PFGE has been previously used for determining the size of phage genomes (8, 54). DNA was extracted from all Azospirillum phage particles, and PFGE analysis confirmed the presence of DNase-protected extragenomic DNA after mitomycin C induction in all but two Azospirillum strains analyzed (Fig. 3A). The absence of extragenomic DNA for those two strains indicated either that mitomycin C was able to trigger the SOS response independently of the presence of prophages or that the number of particles released was too low for purification of DNA to occur. The analysis also revealed that all phages isolated from A. lipoferum and A. doebereinerae strains showed approximately the same size (estimated at about 10 kb) (Fig. 3A, lanes 1 to 7). All phages isolated from A. brasilense strains displayed bigger genome sizes ranging from 62 to 65 kb (Fig. 3A, lanes 8 to 11). From these results, a correlation between the size of each phage genome and the identity of the bacterial species hosting the phage could be drawn. Moreover, these observations are consistent with the larger sizes of phage particles isolated from A. brasilense strains, as observed with TEM. To analyze whether the similarities observed above (with respect to the sizes of phage particles and the sizes of phage genomes) were also reflected at the genomic level, hybridization experiments were carried out on all phage DNA molecules separated by PFGE. Using the entire DNA molecule extracted from ΦAb-Cd as a probe, hybridization signals were clearly obtained exclusively with DNA of phages ΦAb-Cd, ΦAb-Sp7, and ΦAb-Wb1 (Fig. 3B, lanes 8, 10, and 11), suggesting DNA homology between those three phage genomes. When the ΦAl-B518 10-kb DNA molecule was used as a probe, hybridization signals were observed only with DNA from smaller phages isolated from A. doebereinerae and A. lipoferum strains (data not shown). In addition, identical restriction patterns were observed for ΦAb-Cd and ΦAb-Sp7 (Fig. 4), suggesting similar architectures for these two phages. This would indeed be consistent with the genetic relatedness of these two A. brasilense strains, strain Cd being isolated as a scarlet variant of strain Sp7 after inoculation on a plant (25, 59). The restriction pattern of ΦAb-Wb1 (Fig. 4, lane 4) was unrelated to those of ΦAb-Cd and ΦAb-Sp7 despite a strong hybridization signal; this result could be attributed either to the presence of only a few identical genes or to genome recombination. Comparison of restriction profiles of A. lipoferum and A. doebereinerae phage DNAs was not possible, as only smears were obtained whatever the endonuclease used (data not shown).
FIG. 3.
PFGE of DNA molecules isolated from phage particles (A) and hybridization experiments (B). DNA was isolated from the following phage particles: a phage isolated from A. doebereinerae GSF71 and named ΦAd-GSF71 (lane 1); phages isolated from A. lipoferum 4T (ΦAl-4T; lane 2), TVV3 (ΦAl-TVV3; lane 3), B510 (ΦAl-B510; lane 4), B518 (ΦAl-B518; lane 5), 4VI (ΦAl-4VI; lane 6), and 4B (ΦAl-4B; lane 7); and phages isolated from A. brasilense Wb1 (ΦAb-Wb1; lane 8), Sp245 (ΦAb-Sp245; lane 9), Cd (ΦAb-Cd; lane 10), and Sp7 (ΦAb-Sp7; lane 11). Lanes M, 5 kb ladder DNA size standard. The probe used for the hybridization experiment was the DNA molecule extracted from ΦAb-Cd (B).
FIG. 4.
Restriction analysis of phage DNAs. Lane M, λ DNA digested with HindIII. Phage DNA digested with EcoRI: ΦAb-Cd (lane 1), ΦAb-Sp7 (lane 2), ΦAb-Sp245 (lane 3), and ΦAb-Wb1 (lane 4).
Complete nucleotide sequence of the ΦAb-Cd genome.
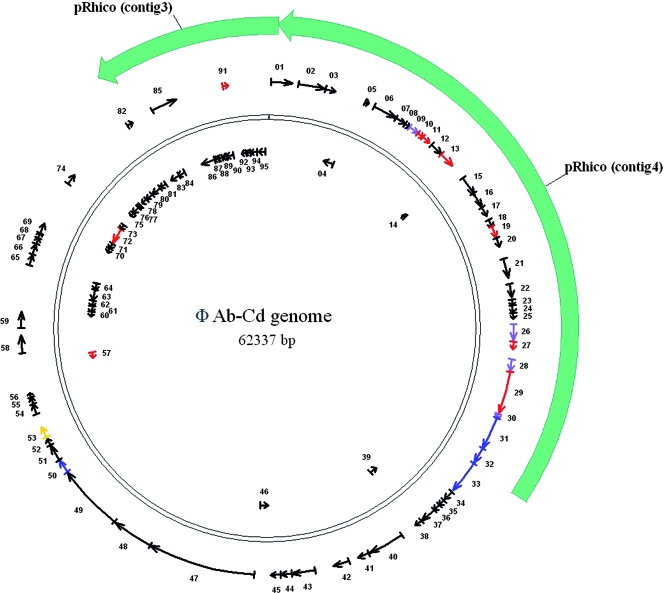
As A. brasilense Cd is one of the most studied Azospirillum strains for its abilities to colonize root-hair zones of different plants and to promote plant growth (20), the complete genome sequence of ΦAb-Cd was determined using a shotgun strategy followed by an assembly process with phrap software and annotation with ARTEMIS 8 software (52). The ΦAb-Cd genome represents a double-stranded DNA circular molecule of 62,337 pb with a G+C content of 63.93%, which is slightly lower than that described for Azospirillum spp. (69% to 71%) (59). The coding sequence represents 86.35% of the genome, with approximately 1.5 gene per kpb of nucleotide sequence, indicating a close-packed genome. A total of 95 ORFs were predicted to encode proteins larger than 40 amino acids. Searches in databases indicated that 65 (68.42%) ORFs code for hypothetical proteins, 16 (16.8%) ORFs display sequence similarity to proteins with unknown functions, and only 14 (14.73%) ORFs could be assigned functions based on protein homology (Table 2 and Fig. 5). No tRNA genes have been identified.
TABLE 2.
Genome organization of A. brasilense phages ΦAb-Cd
| ORF | Start base position | End base position | % GC | No. of aaa (kDa) | Identity with pRhicob; GenBank accession no. | Predicted functionc | INTERPRO familyd | Most significant database match; GenBank or RefSeq accession no.; % aa identity (E value)e |
|---|---|---|---|---|---|---|---|---|
| 01 | 43 | 930 | 67.57 | 295 (29.31) | pRhico093; gb AAS83081.1 | Unknown | ||
| 02 | 1165 | 2259 | 60.82 | 364 (39.27) | pRhico092; gb AAS83080.1 | Unknown | ||
| 03 | 2264 | 2689 | 64.32 | 141 (15.04) | pRhico091; gb AAS83079.1 | Unknown | ||
| 04 | 3667 | 3116 | 61.96 | 183 (20) | Unknown | Hypothetical protein PdenDRAFT_1787 (Paracoccus denitrificans PD1222); gb ABL71813.1; 35% (9E-12) | ||
| 05 | 3961 | 4110 | 61.33 | 49 (5.09) | pRhico090; gb AAS83078.1 | Unknown | ||
| 06 | 4352 | 5353 | 63.27 | 333 (36.95) | pRhico089; gb AAS83077.1 | Unknown | ||
| 07 | 5353 | 5793 | 63.72 | 146 (16.02) | pRhico088; gb AAS83076.1 | Unknown | ||
| 08 | 5790 | 6011 | 66.22 | 93 (8.08) | pRhico087; gb AAS83075.1 | Unknown | ||
| 09 | 6011 | 6430 | 67.14 | 139 (15.48) | pRhico086; gb AAS83074.1 | Phage protein4 | IPR009414 | Phage-like protein (Sinorhizobium medicae WSM419); gb EAU04543.1; 41% (2E-14) |
| 10 | 6427 | 6675 | 56.63 | 82 (9.56) | pRhico085; gb AAS83073.1 | Unknown1 | IPR010978 | Hypothetical protein FP2506_05451 (Fulvimarina pelagi HTCC2506); gb EAU42258.1; 62% (9E-23) |
| 11 | 6685 | 7041 | 68.35 | 118 (12.8) | pRhico084; gb AAS83072.1 | Epoxidase-like protein1 | IPR001387 | Helix-turn-helix domain protein, putative (Roseobacter denitrificans OCh 114); gb ABG33351.1; 37% (3E-04) |
| 12 | 7148 | 7708 | 69.16 | 186 (19.9) | pRhico083; gb AAS83071.1 | Unknown | ||
| 13 | 7705 | 8346 | 68.54 | 213 (23.31) | Unknown1 | Gp42 (bacteriophage phiKO2); gb AAR83058.1; 36% (6E-25) | ||
| 14 | 8772 | 8551 | 64.86 | 73 (7.12) | Unknown | Hypothetical protein BradDRAFT_2696; ref ZP_00862035.1; 45% (4E-07) | ||
| 15 | 8986 | 9861 | 63.01 | 291 (32.26) | Unknown | Protein of unknown function DUF1376 (Paracoccus denitrificans PD1222); gb EAN65374.1; 29% (2E-12) | ||
| 16 | 9737 | 10420 | 69.3 | 227 (24.99) | pRhico082; gb AAS83070.1 | Unknown | ||
| 17 | 10402 | 10803 | 64.68 | 133 (24.99) | pRhico081; gb AAS83069.1 | Unknown | ||
| 18 | 10981 | 11316 | 66.96 | 111 (12.38) | Unknown | Regulatory protein, LuxR:response regulator receiver (Frankia sp. strain EAN1pec); gb EAN13980.1; 37% (4E-05) | ||
| 19 | 11271 | 11774 | 69.64 | 167 (18.08) | pRhico080; gb AAS83068.1 | dCMP deaminase1 | IPR002125 | Deoxycytidylate deaminase (Magnetospirillum magneticum AMB-1); ref YP_422060.1; 50% (4E-19) |
| 20 | 11771 | 12160 | 67.18 | 129 (14.13) | Unknown | Similar to hypothetical protein ABO_0763 (Alcanivorax borkumensis SK2); ref YP_692483.1; 40% (2E-19) | ||
| 21 | 12672 | 13472 | 68.66 | 266 (29.6) | pRhico079; gb AAS83067.1 | Unknown | ||
| 22 | 13712 | 14257 | 65.2 | 181 (19.78) | pRhico078; gb AAS83066.1 | Unknown | ||
| 23 | 14353 | 14589 | 64.56 | 78 (8.6) | Unknown | |||
| 24 | 14586 | 14876 | 63.92 | 96 (10.06) | Unknown | |||
| 25 | 14873 | 15136 | 66.29 | 87 (9.22) | pRhico077; gb AAS83065.1 | Unknown | ||
| 26 | 15333 | 16004 | 61.16 | 223 (25.78) | pRhico076; gb AAS83064.1 | Putative prophage protein4 | Uncharacterized protein L246 (Acanthamoeba polyphaga mimivirus); gb AAV50518.1; 26% (3E-18) | |
| 27 | 16038 | 16376 | 64.01 | 112 (12.55) | HNH endonuclease1 | IPR003615 | HNH endonuclease (Burkholderia phymatum STM815); gb EAU97146.1; 35% (2E-17) | |
| 28 | 16756 | 17223 | 61.97 | 155 (17.42) | pRhico075; gb AAS83063.1 | Putative prophage protein4 | Hypothetical protein NB311A_02637 (Nitrobacter sp. strain Nb-311A); gb EAQ33369.1; 31% (5E-11) | |
| 29 | 17261 | 19012 | 62.44 | 583 (63.69) | pRhico074; gb AAS83062.1 | Putative terminase1 | IPR005021 | Hypothetical phage terminase large subunit (Roseobacter sp. strain SK209-2-6); gb EBA18390.1; 43% (9E-124) |
| 30 | 19027 | 19206 | 67.78 | 44 (4.46) | pRhico073; gb AAS83061.1 | Putative prophage protein4 | ||
| 31 | 19206 | 20561 | 64.38 | 451 (49.02) | pRhico072; gb AAS83060.1 | Portal protein2 | IPR006427 | Putative portal protein (Sinorhizobium medicae WSM419); gb EAU08565.1; 43% (2E-93) |
| 32 | 20558 | 21247 | 62.75 | 229 (24.7) | Phage head maturation protease2 | IPR006433 | Peptidase U35, phage prohead HK97 (Xanthobacter autotrophicus Py2); gb EAS16378.1; 48% (7E-41) | |
| 33 | 21260 | 22636 | 66.09 | 458 (48.93) | Phage major capsid protein2 | IPR006444 | Phage major capsid protein, HK97 (Xanthobacter autotrophicus Py2); gb EAS16379.1; 61% 1E-144 | |
| 34 | 22706 | 23131 | 65.96 | 141 (14.86) | Unknown | Hypothetical protein Mpe_A0539 (Methylibium petroleiphilum PM1); gb ABM93501.1; 53% (5E-32) | ||
| 35 | 23170 | 23427 | 66.28 | 85 (8.13) | Unknown | |||
| 36 | 23427 | 23732 | 66.99 | 101 (10.8) | Unknown | |||
| 37 | 23735 | 24400 | 67.27 | 221 (23.84) | Unknown | Hypothetical protein mlr8529 (Mesorhizobium loti MAFF303099); ref NP_108605.1; 28% (4E-10) | ||
| 38 | 24397 | 24720 | 62.96 | 107 (11.65) | Unknown | |||
| 39 | 24938 | 24522 | 62.35 | 138 (15.22) | Unknown | |||
| 40 | 25382 | 26854 | 63.75 | 490 (51.88) | Unknown | gp78 (bacteriophage ΦJL001); gb AAT69470.1; 27% (2E-14) | ||
| 41 | 26902 | 27393 | 65.65 | 163 (17.89) | Unknown | |||
| 42 | 27769 | 28452 | 63.16 | 227 (25.17) | Unknown | Hypothetical protein mlr8531 (Mesorhizobium loti MAFF303099); ref NP_108607.1; 37% (4E-09) | ||
| 43 | 29170 | 30147 | 62.27 | 325 (35.78) | Unknown | Hypothetical protein EcolE_01003641 (Escherichia coli E24377A); ref ZP_00703609.1; 47% (1E-22) | ||
| 44 | 30128 | 30574 | 59.73 | 148 (16.57) | Unknown | Hypothetical protein XautDRAFT_4299 (Xanthobacter autotrophicus Py2); ref ZP_01196156.1; 32% (1E-09) | ||
| 45 | 30581 | 31009 | 65.27 | 142 (15.39) | Unknown | |||
| 46 | 31526 | 31071 | 49.78 | 151 (16.61) | Unknown | |||
| 47 | 31654 | 36042 | 62.36 | 1462 (154.37) | Unknown | |||
| 48 | 36039 | 37733 | 66.02 | 564 (60.05) | Unknown | |||
| 49 | 37792 | 40539 | 63.54 | 915 (95.53) | Unknown | |||
| 50 | 40549 | 41130 | 64.43 | 193 (19.75) | Tail fiber protein2 | Tail fiber protein, putative (Polaromonas sp. strain JS666); ref YP_550528.1; 29% (5E-10) | ||
| 51 | 41140 | 41826 | 60.55 | 228 (24.33) | Unknown | Hypothetical cytosolic protein (Syntrophus aciditrophicus SB); ref YP_460612.1; 37% (4E-08) | ||
| 52 | 41830 | 42114 | 65.26 | 94 (9.55) | Unknown | |||
| 53 | 42188 | 42634 | 66.44 | 148 (15.71) | Phage-related lysozyme3 | IPR002196 | Phage-related lysozyme (Magnetospirillum magneticum AMB-1); ref YP_423349.1; 56% (1E-28) | |
| 54 | 43206 | 43628 | 56.97 | 140 (43.21) | Unknown | |||
| 55 | 43632 | 43901 | 61.11 | 89 (9.78) | Unknown | |||
| 56 | 43915 | 44091 | 63.84 | 58 (6.22) | Unknown | |||
| 57 | 45392 | 45069 | 58.33 | 107 (12.08) | Unknown1 | IPR001387 | Hypothetical protein MELB17_01400 (Marinobacter sp. strain ELB17); gb EBA01392.1; 43% (5E-15) | |
| 58 | 45736 | 46467 | 67.08 | 243 (25.27) | Unknown | |||
| 59 | 47405 | 46746 | 65.45 | 219 (23.81) | Unknown | |||
| 60 | 47662 | 47402 | 66.28 | 86 (9.36) | Unknown | |||
| 61 | 47997 | 47659 | 63.42 | 112 (11.96) | Unknown | |||
| 62 | 48293 | 47994 | 66.67 | 99 (10.85) | Unknown | |||
| 63 | 48921 | 48286 | 62.74 | 211 (23.13) | Unknown | |||
| 64 | 49291 | 48914 | 62.17 | 125 (13.37) | Unknown | Hypothetical protein NB311A_07313 (Nitrobacter sp. strain Nb-311A); ref ZP_01044934.1; 43% (5E-05) | ||
| 65 | 49306 | 49743 | 63.47 | 145 (16.75) | Unknown | |||
| 66 | 49766 | 50185 | 63.81 | 139 (15) | Unknown | |||
| 67 | 50213 | 50446 | 65.81 | 77 (8.27) | Unknown | |||
| 68 | 50476 | 50682 | 59.42 | 68 (7.49) | Unknown | |||
| 69 | 50672 | 51115 | 64.19 | 147 (16.13) | Unknown | Conserved hypothetical protein (Escherichia coli); gb ABF67882.1; 32% (2E-07) | ||
| 70 | 51396 | 51145 | 70.63 | 83 (8.56) | Unknown | |||
| 71 | 51701 | 51393 | 63.11 | 102 (11.69) | Unknown | |||
| 72 | 52642 | 51755 | 61.37 | 295 (33.47) | Integrase1 | IPR002104 | Integrase (Magnetospirillum magneticum AMB-1); ref YP_422072.1; 37% (2E-39) | |
| 73 | 52919 | 52653 | 64.04 | 88 (9.58) | Unknown | |||
| 74 | 52957 | 53424 | 64.1 | 155 (16.56) | Unknown | |||
| 75 | 53783 | 53547 | 67.51 | 78 (8.33) | Unknown | |||
| 76 | 54219 | 53776 | 64.19 | 147 (15.76) | Unknown | |||
| 77 | 54398 | 54216 | 60.11 | 60 (6.78) | Unknown | |||
| 78 | 54831 | 54514 | 70.13 | 105 (11.6) | Unknown | |||
| 79 | 55269 | 54838 | 66.2 | 143 (15.03) | Unknown | |||
| 80 | 55862 | 55293 | 56.14 | 189 (21.17) | Unknown | |||
| 81 | 56251 | 55901 | 61.54 | 116 (13.1) | Unknown | |||
| 82 | 56325 | 56579 | 63.14 | 84 (9.27) | Unknown | |||
| 83 | 57121 | 56576 | 63.92 | 181 (20.11) | pRhico071; gb AAS83033.1 | Unknown | ||
| 84 | 57507 | 57085 | 65.72 | 140 (15.82) | Unknown | |||
| 85 | 58574 | 57504 | 63.49 | 356 (39.15) | pRhico070; gb AAS83032.1 | Unknown | ||
| 86 | 59328 | 58528 | 68.04 | 266 (28.65) | pRhico069; gb AAS83031.1 | Unknown | ||
| 87 | 59528 | 59325 | 68.14 | 67 (7.23) | Unknown | |||
| 88 | 59770 | 59534 | 64.14 | 78 (8.34) | pRhico068; gb AAS83030.1 | Unknown | ||
| 89 | 60028 | 59861 | 64.29 | 55 (5.73) | pRhico067; gb AAS83029.1 | Unknown | ||
| 90 | 60408 | 60025 | 63.02 | 127 (14.36) | pRhico066; gb AAS83028.1 | Unknown | ||
| 91 | 60512 | 60745 | 61.97 | 77 (8.4) | Unknown1 | IPR002052 | ||
| 92 | 61144 | 60881 | 62.88 | 87 (9.81) | Unknown | |||
| 93 | 61377 | 61141 | 70.04 | 78 (8.34) | Unknown | |||
| 94 | 61756 | 61379 | 65.08 | 125 (13.74) | pRhico065; gb AAS83027.1 | Unknown | ||
| 95 | 62234 | 61839 | 66.92 | 131 (14.34) | Unknown |
aa, amino acid.
For ORFs displaying 100% identity with previously identified pRhico ORFs, the second most significant database match is shown.
For the ORFs with assigned functions (shown in bold) and for the ORFs possessing a conserved domain, the encoded proteins could be grouped into four classes: class 1, proteins involved in DNA processing (indicated by a superscript 1); class 2, proteins involved in phage morphogenesis (superscript 2); class 3, proteins related to bacterial lysis (superscript 3); class 4, putative phage proteins (superscript 4).
The analysis was done using INTERPROSCAN software (67).
These results were obtained using the BLASTX search.
FIG. 5.
Genome map of ΦAb-Cd and homology with pRhico. The double black circle represents the circular DNA molecule of the ΦAb-Cd genome. The predicted ORFs are depicted by thin arrows or arrowheads pointing in the direction of transcription and are numbered consecutively (see Table 2). ORFs encoding hypothetical proteins (no homologs in BLAST-P searches) are represented by black thin arrows. The ORFs encoding proteins with assigned functions are represented by colored thin arrows as follows: red arrows, DNA processing; blue, phage morphogenesis; yellow, bacterial lysis; purple, putative phage proteins. Homologies between the ΦAb-Cd sequence and contigs of pRhico are represented by thick green arrows.
At the onset of the annotation process, it was found that a region of 27,292 pb displayed 100% identity with part of the previously sequenced pRhico plasmid of strain A. brasilense Sp7, a strain highly related to A. brasilense Cd (see above) (25). The pRhico plasmid, also named p90 due to its molecular size, which is estimated at 90 MDa, was shown to be present in several A. brasilense strains, including A. brasilense Cd (45, 64). Approximately 95% (151.3 kb) of the pRhico sequence has been described as present in five contigs that were not assembled (62). Hence, the whole sequence of pRhico contig 3 (6,171 pb) and 98% of pRhico contig 4 (21,130 out of 21,577 kb) matched a part of the ΦAb-Cd sequence (Fig. 5). Two explanations could account for this finding: (i) ΦAb-Cd is integrated as a prophage in the pRhico plasmid (but this would imply that the pRhico sequence has not been entirely described), or (ii) ΦAb-Cd is integrated elsewhere in the genome as a prophage and part (18.3%) of ΦAb-Cd is duplicated within the pRhico. Subsequent experiments (see below) will allow retaining one of these hypotheses. ORF predictions performed in our study allowed the identification of ORFs similar to those already predicted for contigs 3 and 4 of pRhico, but additional ORFs were found (Table 2).
Even if the majority of the putative ORFs identified code for hypothetical proteins or for proteins without assigned functions, some genes encode putative phage proteins that have conserved functional domains and that could be grouped in four classes: proteins involved in DNA processing, proteins involved in phage morphogenesis, proteins related to bacterial lysis, and putative phage proteins. Nine ORFs (ORF10, ORF11, ORF13, ORF19, ORF27, ORF29, ORF57, ORF72, and ORF91) could be ascribed a function related to DNA processing (see Table 2). The deduced protein encoded by ORF10, although displaying 82% identity with conserved bacterial proteins with unassigned functions (COG3750.2), possesses a conserved domain matching the tRNA-binding arm, a domain found in aminoacyl tRNA synthetases. ORF11 could encode a protein showing homology with epoxidase-like protein. A search for conserved domains revealed a putative helix-turn-helix type 3 DNA binding domain (Interpro IPR 001387), identified in proteins belonging to the Lambda repressor-like family; thus, the product encoded by ORF11 could act as a transcriptional repressor like the regulators Cro and cI that control the life cycle of the Lambda bacteriophage. Site-specific DNA methylase domains involved in DNA modification have been identified in the two putative proteins encoded by ORF13 and ORF91. The product encoded by ORF19 exhibits high similarities with deoxycytidylate deaminase-like proteins that catalyze the deamination of dCMP into dUMP. The deduced product of ORF27 is homologous to endonucleases of the HNH family found in several phages (such as LambdaSa2 from Streptococcus agalactiae and bIL170 from Lactococcus spp.). ORF29 is likely to encode the phage terminase large subunit, an enzyme that is part of a large nucleoprotein complex dedicated to packaging. The integrase function allowing the DNA recombination and phage integration process was ascribed to the product encoded by ORF72, showing best identities with alphaproteobacteria like Oceanicola granulosus HTCC2516 (35% identity; RefSeq accession number ZP_01157933) and Magnetospirillum magneticum AMB-1 (37% identity; RefSeq accession number YP_422072), a bacterium taxonomically related to the Azospirillum genera.
Deduced proteins encoded by ORF31, ORF32, ORF33, and ORF50 were revealed to be structural proteins commonly found in phage particles. ORF31 encodes a putative portal protein, the best identity being with a putative portal protein of Sinorhizobium medicae WSM419 (43% identity; GenBank accession number ZP_01412930). The deduced amino acid sequence of ORF32 displays strong similarity to phage prohead proteases that are involved in the processing of the prohead protein. ORF33 may encode the major component of the phage capsid, as it belongs to the family of head proteins of many bacteriophages, such as HK97, phi-105, and P27 (Interpro IPR 006444). The deduced protein of ORF50 shares similarity with phage tail proteins, but no conserved domain could be identified.
As for functions related to bacterial lysis, only one product encoded by ORF53 could be assigned to this process; indeed, this protein revealed similarities to members of the glycoside hydrolase family, i.e., lysozyme (Interpro IPR 002196). Finally, the fourth class of ORFs encoding putative phage proteins includes ORF9, ORF26, ORF28, and ORF30.
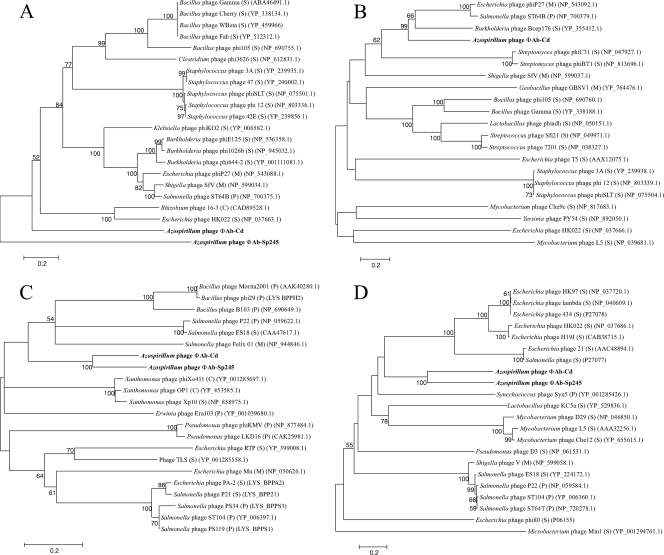
Phylogenetic analysis of functional genes.
Phage particle observations have revealed a morphology that is typical of members of the Siphoviridae family of bacteriophages. Information about the genus can be established by analyzing phylogenetic relationships of ΦAb-Cd with other functional Caudovirales phages. It has been suggested that phylogenetic analysis should be performed at the level of individual genes in order to resolve viral taxonomical issues (41). Indeed, as bacteriophages are known for horizontal exchange of functional genetic elements drawn from a large shared pool (32, 55), any phage-specific protein could be obtained by a single modular interchange during evolution. Therefore, four proteins of ΦAb-Cd, including terminase (ORF29), phage major capsid protein (ORF33), lysozyme (ORF53), and integrase (ORF72), which are commonly present in tailed phages, were chosen to build phylogenetic trees. Homologous protein sequences other than that of the phage major capsid protein have been identified from the draft of the genome of the Sp245 strain and are likely to be components of ΦAb-Sp245, as corresponding genes lie in a region containing phage ORFs whose length matches the estimated size of ΦAb-Sp245 observed here by PFGE. These sequences were compared with those of functional Caudovirales phages deposited in GenBank in order to construct the phylogenetic trees shown in Fig. 6. ΦAb-Cd and ΦAb-Sp245 lie on the same branch in the tree based on integrase, lysozyme, and terminase data but are not as closely related as other phages isolated from the same bacterial species, such as Salmonella or Staphylococcus phages, are related to each other. All trees show that ΦAb-Cd and ΦAb-Sp245 are deeply rooted, suggesting that they are quite different from previously described phages. The low bootstrap values obtained for the deep branches indicate that these phage proteins have diverged to an extent that prevents determining their evolutionary relatedness and therefore prevents classification of Azospirillum brasilense phages in known genera.
FIG. 6.
Unrooted neighbor-joining trees based on the aligned amino acid sequences encoded by four phage genes. Only genome data obtained from virions were used. (A) Terminase; (B) major capsid protein; (C) lysozyme; (D) integrase from ΦAb-Cd, from ΦAb-Sp245 (no protein homologous to a major phage protein could be identified in the A. brasilense Sp245 draft genome), and from 20 other phages. Amino acid sequences from A. brasilense phages are highlighted in bold. A Blosum 30 matrix was calculated, with a gap penalty of 10.0. The numbers at the nodes are bootstrap values based on 1,000 resamplings. Bootstrap values less than 50 are not shown. Scale bar, 0.2 amino acid substitutions per residue. M, Myoviridae; S, Siphoviridae; P, Podoviridae; C, unclassified Caudovirales.
Genome localization of the ΦAb-Cd prophage.
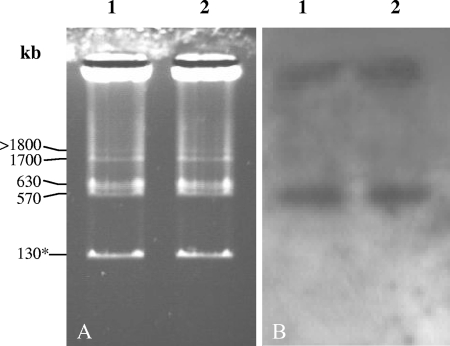
Genome comparison of ΦAb-Cd and pRhico sequences have revealed that the two entities encompassed regions that were 43.8% (27.3 kb) identical. Given the relatedness between strain A. brasilense Cd and strain A. brasilense Sp7 (strain Cd being isolated as a scarlet variant of strain Sp7 after inoculation on a plant) (25, 59) and the homology between the two phages isolated from those strains (see above), the same genetic organization could be expected in both strains. This implied either that ΦAb-Cd was integrated as a prophage in plasmid pRhico or that a region of the prophage was duplicated in the A. brasilense Cd genome. To test this hypothesis, PCR amplifications of ORF40, ORF50, and ORF53 of ΦAb-Cd were carried out with DNA isolated from ΦAb-Cd, ΦAb-Sp7, ΦAb-Wb1, and ΦAb-Sp245. PCR fragments of the expected size were obtained from DNA isolated from ΦAb-Cd and from ΦAb-Sp7, sustaining the hypothesis that the ΦAb-Cd and ΦAb-Sp7 genomes are strongly related (data not shown). No amplification products could be obtained from DNA isolated from ΦAb-Wb1 and ΦAb-Sp245, suggesting that these ORFs are absent from these phages or have divergent sequences. To determine on which replicons ΦAb-Cd and ΦAb-Sp7 were integrated, a separation of plasmids was performed (see Materials and Methods) followed by Southern hybridization experiments. First, identical plasmid patterns were observed for strains A. brasilense Cd and Sp7 that coincided with the pattern previously described, with sizes of replicons estimated at 130, 570, 630, 1,700, and >1,800 kb (Fig. 7A) (14). Second, a hybridization experiment performed with exoC, an ORF specific to pRhico (62), as a probe, enabled us to identify pRhico as being the 130-kb plasmid of both the Sp7 and Cd strains (data not shown). Third, ORF50 of ΦAb-Cd, when used as a probe, clearly hybridized with the 570-kb plasmid of both Sp7 and Cd strains, but no signal was obtained with the pRhico plasmid (Fig. 7B). These observations indicate that ΦAb-Cd and ΦAb-Sp7 are not integrated as prophages in pRhico but are present in a larger replicon of 570 kb. These results also imply that a region of 27.3 kb spanning contigs 3 and 4 of pRhico (Fig. 5) is duplicated in the genomes of the two strains and that this duplication is located on the pRhico plasmid.
FIG. 7.
Genome localization of ΦAb-Cd. Plasmids of strains A. brasilense Cd (lane 1) and Sp7 (lane 2) were separated (A) and hybridized with ORF50 of ΦAb-Cd (B). The pRhico plasmid is indicated by an asterisk.
DISCUSSION
Prevalence and morphology of Azospirillum bacteriophages.
This study clearly demonstrates the prevalence of bacteriophages among members of the Azospirillum genus. When cells were exposed to mitomycin C, bacterial lysis due to the release of phage particles was observed for 11 strains out of the 24 strains analyzed (i.e., 4 of 11 A. brasilense strains, 6 of 11 A. lipoferum, 1 of 1 A. doebereinerae, and 0 of 1 A. irakense). The induction of bacteriophages was not observed for 11 strains, indicating either that these strains do not host any prophage or that some Azospirillum-hosted prophages are not induced by the SOS response. Cell lysis could not be clearly correlated to the release of phage particles for only two strains. Cell lysis upon mitomycin C treatment was previously described for several strains of A. brasilense and A. lipoferum, but the production of phage particles was clearly evidenced only for A. brasilense Sp7 (28). The spontaneous induction of bacteriophages during growth of A. brasilense Sp7 in complex media was also reported (30).
TEM analysis revealed two types of bacteriophages displaying ultrastructural features that are typical of members of the Siphoviridae family. Identical features were previously described for the bacteriophage of A. brasilense Sp7 (30). The two types can be distinguished not only by the sizes of particles but also by the sizes of encapsidated DNA and by the durations of the eclipse period upon induction by mitomycin C. Moreover, each type of bacteriophages seems to be associated with a specific bacterial species, as only “big” phages were evidenced for A. brasilense and “small” phages for A. lipoferum strains. This finding could be related to the usually narrow host range of a given bacteriophage, limited to a bacterial species or strain (18, 30).
Relatedness among Azospirillum phage genomes.
Hybridization experiments suggested DNA homology between the phage genomes of ΦAb-Cd, ΦAb-Sp7, and ΦAb-Wb1. Restriction profiles of ΦAb-Cd and ΦAb-Sp7 are identical, and, given the relatedness between those two strains, one can speculate that these two phages are identical. The homology between ΦAb-Cd and ΦAb-Wb1 is less striking, as their restriction profiles display only a few common bands. Genomes of members of the Siphoviridae family are usually organized into functional modules (i.e., multigenic interchangeable elements whose genes are involved in the same process, such as the lysogeny module, tail gene module, etc.), and such modules appear to be conserved among prophages hosted by the same species, as demonstrated by genome analysis of six phages isolated from Streptococcus thermophilus (12, 15). However, the organization of modules can differ from one phage genome to another due to the accumulation of mutations and recombination events, leading to mosaic genomes. Such an organization might be found in the A. brasilense phages, explaining the differences in restriction profiles despite the presence of strong DNA hybridization signals.
Strain A. brasilense Sp245 seems to host a prophage that is not genetically linked to ΦAb-Cd, as shown by the absence of hybridization signal. Preliminary analysis of the draft Sp245 genome (available at http://genomics.ornl.gov/research/azo) is in accordance with this finding, as some putative phage ORFs display low or no similarity with the ΦAb-Cd sequence obtained in this study. Comparative genomic analysis of 18 Pseudomonas aeruginosa bacteriophages and 27 Staphylococcus aureus bacteriophages showed that some prophages hosted by strains of the same species are highly homologous whereas others display very little homology (38, 39). Therefore, phage diversity within a bacterial species is not restricted to the A. brasilense species. This diversity is even more obvious at the genus level, as prophages hosted by A. lipoferum strains do not share any common sequences with prophages hosted by the A. brasilense species. In contrast, the phage hosted by the A. doebereinerae strain might share common sequences with ΦAl-B518, and if so, this can probably be attributed to the genetic proximity between those two species (22).
One of the most striking features of the A. lipoferum phages induced in this study is the small size of encapsidated DNA (about 10 kb). Genome sizes of members of the Siphoviridae family usually range from 18 kb to 500 kb (17); indeed, the smallest genome of a Siphoviridae phage described in NCBI is 14,957 pb (phage bIL310 of Lactococcus lactis IL-1403; GenBank accession number AF323671). Attempts to establish the restriction profiles of A. lipoferum phage DNAs by use of various enzymes always resulted in smears; moreover, attempts to clone and sequence those DNAs failed to give data that could be assembled. All these observations seem to imply the presence of prophage-like gene transfer agents rather than of real prophages. In contrast to functional prophages, gene transfer agents typically package bacterial genome fragments and atypically package a portion of their own genome and constitute conspicuous mechanisms of generalized transduction (56). Bioinformatic analysis showed that gene transfer agents are widely distributed within alphaproteobacteria, especially in members of the Rhodobacterales family (40). However, this hypothesis needs to be experimentally validated.
Phage genome sequence.
The complete genome sequence of ΦAb-Cd obtained in this study constitutes the first description of an Azospirillum bacteriophage. The process of annotation of the ΦAb-Cd genome revealed many ORFs (81 out of 95) encoding either hypothetical proteins or conserved proteins with unknown functions; this result might be attributable to the low number of alphaproteobacteria bacteriophages actually described. Indeed, only three sequences of phages were obtained through complete genome sequencing, i.e., phage ΦJL001 of a marine alphaproteobacterium, phage SIO1 of Roseobacter SIO67, and phage PBC5 of Sinorhizobium meliloti (42, 50). Of the 14 ORFs of ΦAb-Cd to which a function could be attributed, 11 display homology with phage ORFs found in alphaproteobacterium whole genomes but none was homologous to ORFs specific to phages of other bacterial classes. This could be due to high rates of sequence divergence and to the narrow host range of bacteriophages. Interestingly, a function can be attributed to only about 25% of the ORFs even when several sequences of prophages hosted by the same species are available, as illustrated with prophages of S. aureus or Mycobacterium tuberculosis (38, 39, 48). Thus, phages can be considered to be a rich reservoir for new proteins. Some of the ORFs encoding hypothetical proteins could be genes of lysogenic conversion, contributing to increase bacterial fitness; such a case is observed for many bacterial pathogens whose virulence factors are phage encoded (13, 16). However, some of the ORFs identified here encode proteins with functions that are essential in the lytic phage cycle, such as proteins for particle formation (structural proteins, terminase), proteins for bacterial lysis (lysozyme), and proteins for integration/excision (integrase). Using such proteins to resolve the taxonomy of ΦAb-Cd proved to be unsuccessful, a result that emphasizes the problems associated with mosaicism of viral genomes when using sequence data for taxonomical purposes (41). The identification of functional modules remained difficult due to the low number of ORFs with an attributed function, but ORF26 to ORF33 could constitute a conserved genetic module encoding structural and maturation proteins.
Phages and dynamics of bacterial genomes and bacterial community.
ΦAb-Cd is found integrated on a 570-kb plasmid; whether this plasmid is conjugative or not is not known, but this could imply a putative horizontal transfer of phage DNA by conjugation. A 27.3-kb fragment of ΦAb-Cd was also recovered within another replicon, the pRhico plasmid, indicating that genomic rearrangements occurred between plasmids and phages, leading to a duplication event. Major genomic rearrangements were previously characterized for several Azospirillum strains undergoing phenotypic switching, demonstrating genome plasticity among members of the Azospirillum genus (63). Bacteriophages could thus account for the dynamics and evolution of Azospirillum genomes and could be major contributors to the genetic individuality of Azospirillum strains, as described previously for other species (15, 37, 47).
Phage particle formation was induced here in vitro by the SOS-inducing agent mitomycin C; it would be of interest to study phage-Azospirillum interactions within the rhizosphere, the natural habitat of Azospirillum spp. Indeed, environmental abiotic factors, such as nutrients and temperature, were shown to induce production of bacteriophages in A. brasilense Sp7 (30). Eukaryote-bacterium interactions may also influence the cycle of phages; for example, a lysogenic avian E. coli pathogen showed an upregulation of phage gene expression when injected into a chicken (21) and phage genes were shown to respond to molecular cues emitted by the mammalian host of the pathogen S. pyogenes (10). Moreover, upregulation of phage-derived genes has been observed during promoter trap studies of Ralstonia solanacearum grown in the presence of tomato plants and of A. brasilense induced by wheat extracts (11, 49). The fact that bacteriophages can be induced in natural conditions has a considerable impact on bacterial densities; such an impact has been demonstrated for viruses in aquatic environments (57, 65, 66) and by artificial release of a Serratia liquefaciens lysogen into soil samples (3). Despite a previous study showing no effect on the declines in the viability of strains A. brasilense Sp7 and Cd in soil inoculated with the Sp7 phage (29), the potential role of bacteriophages in population dynamics of Azospirillum spp. and of rhizospheric bacterial communities cannot be excluded. Also, the presence of prophages might provide immunity to infection by any other bacterial virus. Finally, this work might help optimize the use of bacteriophages as a tool for Azospirillum genetic studies.
Acknowledgments
We thank P. Bustos (Centro de Ciencias Genómicas, Universidad Nacional Autónoma de México, Cuernavaca, Mexico) for her contribution to the phage genome analysis and I. B. Zhulin (The University of Tennessee and U.S. Oak Ridge National Laboratory) for access to preliminary data of the A. brasilense Sp245 genome sequence project, which is supported by the U.S. National Science Foundation (grant EF-0412186 to I. B. Zhulin). We are also thankful to P. Normand (UMR5557, Ecologie Microbienne) for helpful discussions. Part of this work was made possible by an agreement between the Laboratoire d'Ecologie Microbienne and the service of Cooperation Scientifique et Technologique of the French Embassy of Mexico. Electron microscopy was performed at the Centre Technologique des Microstructures of Université Lyon 1.
This work was supported by a fellowship from the French Ministère de l'Education Nationale de la Recherche et des Nouvelles Technologies awarded to M.B.
Footnotes
Published ahead of print on 7 December 2007.
REFERENCES
- 1.Alexandre, G., and R. Bally. 1999. Emergence of a laccase-positive variant of Azospirillum lipoferum occurs via a two-step phenotypic switching process. FEMS Microbiol. Lett. 174:371-378. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ashelford, K. E., S. J. Norris, J. C. Fry, M. J. Bailey, and M. J. Day. 2000. Seasonal population dynamics and interactions of competing bacteriophages and their host in the rhizosphere. Appl. Environ. Microbiol. 66:4193-4199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baldani, V. L. D., M. A. B. Alvarez, J. I. Baldani, and J. Doëbereiner. 1986. Establishment of inoculated Azospirillum spp. in the rhizosphere and in roots of field grown wheat and sorghum. Plant Soil 90:37-40. [Google Scholar]
- 5.Bally, R., D. Thomas-Bauzon, T. Heulin, J. Balandreau, J. Richard, and J. de Ley. 1983. Determination of the most frequent N2-fixing bacteria from the rhizosphere of rice. Can. J. Microbiol. 29:881-887. [Google Scholar]
- 6.Bartolomé, B., Y. Jubete, E. Martínez, and F. de la Cruz. 1991. Construction and properties of a family of pACYC184-derived cloning vectors compatible with pBR322 and its derivatives. Gene 102:75-78. [DOI] [PubMed] [Google Scholar]
- 7.Bashan, Y., G. Holguin, and L. E. de-Bashan. 2004. Azospirillum-plant relationships: physiological, molecular, agricultural, and environmental advances (1997-2003). Can. J. Microbiol. 50:521-577. [DOI] [PubMed] [Google Scholar]
- 8.Beilstein, F., and B. Dreiseikelmann. 2006. Bacteriophages of freshwater Brevundimonas vesicularis isolates. Res. Microbiol. 157:213-219. [DOI] [PubMed] [Google Scholar]
- 9.Beringer, J. F. 1974. R factor transfer in Rhizobium leguminosarum. J. Gen. Microbiol. 84:188-198. [DOI] [PubMed] [Google Scholar]
- 10.Broudy, T. B., V. Pancholi, and V. A. Fischetti. 2001. Induction of lysogenic bacteriophage and phage-associated toxin from group a streptococci during coculture with human pharyngeal cells. Infect. Immun. 69:1440-1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brown, D. G., and C. Allen. 2004. Ralstonia solanacearum genes induced during growth in tomato: an inside view of bacterial wilt. Mol. Microbiol. 53:1641-1660. [DOI] [PubMed] [Google Scholar]
- 12.Brüssow, H., and F. Desiere. 2001. Comparative phage genomics and the evolution of Siphoviridae: insights from dairy phages. Mol. Microbiol. 39:213-223. [DOI] [PubMed] [Google Scholar]
- 13.Brüssow, H., and R. W. Hendrix. 2002. Phage genomics: small is beautiful. Cell 108:13-16. [DOI] [PubMed] [Google Scholar]
- 14.Caballero-Mellado, J., L. López-Reyes, and R. Bustillos-Cristales. 1999. Presence of 16S rRNA genes in multiple replicons in Azospirillum brasilense. FEMS Microbiol. Lett. 178:283-288. [Google Scholar]
- 15.Canchaya, C., G. Fournous, and H. Brüssow. 2004. The impact of prophages on bacterial chromosomes. Mol. Microbiol. 53:9-18. [DOI] [PubMed] [Google Scholar]
- 16.Canchaya, C., C. Proux, G. Fournous, A. Bruttin, and H. Brüssow. 2003. Prophage genomics. Microbiol. Mol. Biol. Rev. 67:238-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Casjens, S. R. 2005. Comparative genomics and evolution of the tailed-bacteriophages. Curr. Opin. Microbiol. 8:451-458. [DOI] [PubMed] [Google Scholar]
- 18.Chibani-Chennoufi, S., A. Bruttin, M.-L. Dillmann, and H. Brüssow. 2004. Phage-host interaction: an ecological perspective. J. Bacteriol. 186:3677-3686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Delcher, A. L., D. Harmon, S. Kasif, O. White, and S. L. Salzberg. 1999. Improved microbial gene identification with GLIMMER. Nucleic Acids Res. 27:4636-4641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dobbelaere, S., A. Croonenborghs, T. Amber, D. Ptacek, J. Vanderleyden, P., C. L.-G. Dutto, J. Caballero-Mellado, J. F. Aguirre, Y. Kapulnik, S. Brener, S. Burdman, D. Kadouri, S. Sarig, and Y. Okon. 2001. Responses of agronomically important crops to inoculation with Azospirillum. Aust. J. Plant Physiol. 28:1-9. [Google Scholar]
- 21.Dozois, C. M., F. Daigle, and R. Curtiss III. 2003. Identification of pathogen-specific and conserved genes expressed in vivo by an avian pathogenic Escherichia coli strain. Proc. Natl. Acad. Sci. USA 100:247-252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eckert, B., O. B. Weber, G. Kirchhof, A. Halbritter, M. Stoffels, and A. Hartmann. 2001. Azospirillum doebereinerae sp. nov., a nitrogen-fixing bacterium associated with the C4-grass Miscanthus. Int. J. Syst. Evol. Microbiol. 51:17-26. [DOI] [PubMed] [Google Scholar]
- 23.Elbeltagy, A., K. Nishioka, T. Sato, H. Suzuki, B. Ye, T. Hamada, T. Isawa, H. Mitsui, and K. Minamisawa. 2001. Endophytic colonization and in planta nitrogen fixation by a Herbaspirillum sp. isolated from wild rice species. Appl. Environ. Microbiol. 67:5285-5293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Elmerich, C., B. Quiviger, C. Rosenberg, C. Franche, P. Laurent, and J. Döbereiner. 1982. Characterization of a temperate bacteriophage for Azospirillum. Virology 122:29-37. [DOI] [PubMed] [Google Scholar]
- 25.Eskew, D. L., D. D. Focht, and I. P. Ting. 1977. Nitrogen fixation, denitrification, and pleomorphic growth in a highly pigmented Spirillum lipoferum. Appl. Environ. Microbiol. 34:582-585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ewing, B., and P. Green. 1998. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 8:186-194. [PubMed] [Google Scholar]
- 27.Ewing, B., L. Hillier, M. C. Wendl, and P. Green. 1998. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 8:175-185. [DOI] [PubMed] [Google Scholar]
- 28.Franche, C., and C. Elmerich. 1981. Physiological properties and plasmid content of several strains of Azospirillum brasilense and A. lipoferum. Ann. Microbiol. (Paris) 132:3-18. [PubMed] [Google Scholar]
- 29.Germida, J. J. 1986. Population dynamics of Azospirillum brasilense and its bacteriophage in soil. Plant Soil 90:117-128. [Google Scholar]
- 30.Germida, J. J. 1984. Spontaneous induction of bacteriophage during growth of Azospirillum brasilense in complex media. Can. J. Microbiol. 30:805-808. [Google Scholar]
- 31.Gordon, D., C. Abajian, and P. Green. 1998. Consed: a graphical tool for sequence finishing. Genome Res. 8:195-202. [DOI] [PubMed] [Google Scholar]
- 32.Hendrix, R. W. 2003. Bacteriophage genomics. Curr. Opin. Microbiol. 6:506-511. [DOI] [PubMed] [Google Scholar]
- 33.Hynes, M. F., and N. F. McGregor. 1990. Two plasmids other than the nodulation plasmid are necessary for formation of nitrogen-fixing nodules by Rhizobium leguminosarum. Mol. Microbiol. 4:567-574. [DOI] [PubMed] [Google Scholar]
- 34.Jacoud, C., D. Job, P. Wadoux, and R. Bally. 1999. Initiation of root growth stimulation by Azospirillum lipoferum CRT1 during maize seed germination. Can. J. Microbiol. 45:339-342. [Google Scholar]
- 35.Kabir, M., D. Faure, T. Heulin, W. Achouak, and R. Bally. 1995. Azospirillum populations in soils infested by a parasitic weed (Striga) under Sorghum cultivation in Mali, West Africa. Eur. J. Soil Biol. 32:157-163. [Google Scholar]
- 36.Khammas, K. M., E. Ageron, P. A. Grimont, and P. Kaiser. 1989. Azospirillum irakense sp. nov., a nitrogen-fixing bacterium associated with rice roots and rhizosphere soil. Res. Microbiol. 140:679-693. [DOI] [PubMed] [Google Scholar]
- 37.Kuroda, M., T. Ohta, I. Uchiyama, T. Baba, H. Yuzawa, I. Kobayashi, L. Cui, A. Oguchi, K. Aoki, Y. Nagai, J. Lian, T. Ito, M. Kanamori, H. Matsumaru, A. Maruyama, H. Murakami, A. Hosoyama, Y. Mizutani-Ui, N. K. Takahashi, T. Sawano, R. Inoue, C. Kaito, K. Sekimizu, H. Hirakawa, S. Kuhara, S. Goto, J. Yabuzaki, M. Kanehisa, A. Yamashita, K. Oshima, K. Furuya, C. Yoshino, T. Shiba, M. Hattori, N. Ogasawara, H. Hayashi, and K. Hiramatsu. 2001. Whole genome sequencing of methicillin-resistant Staphylococcus aureus. Lancet 357:1225-1240. [DOI] [PubMed] [Google Scholar]
- 38.Kwan, T., J. Liu, M. DuBow, P. Gros, and J. Pelletier. 2006. Comparative genomic analysis of 18 Pseudomonas aeruginosa bacteriophages. J. Bacteriol. 188:1184-1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kwan, T., J. Liu, M. DuBow, P. Gros, and J. Pelletier. 2005. The complete genomes and proteomes of 27 Staphylococcus aureus bacteriophages. Proc. Natl. Acad. Sci. USA 102:5174-5179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lang, A. S., and J. T. Beatty. 2007. Importance of widespread gene transfer agent genes in alpha-proteobacteria. Trends Microbiol. 15:54-62. [DOI] [PubMed] [Google Scholar]
- 41.Lawrence, J. G., G. F. Hatfull, and R. W. Hendrix. 2002. Imbroglios of viral taxonomy: genetic exchange and failings of phenetic approaches. J. Bacteriol. 184:4891-4905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lohr, J. E., F. Chen, and R. T. Hill. 2005. Genomic analysis of bacteriophage ΦJL001: insights into its interaction with a sponge-associated alpha-proteobacterium. Appl. Environ. Microbiol. 71:1598-1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Marchler-Bauer, A., J. B. Anderson, P. F. Cherukuri, C. DeWeese-Scott, L. Y. Geer, M. Gwadz, S. He, D. I. Hurwitz, J. D. Jackson, Z. Ke, C. J. Lanczycki, C. A. Liebert, C. Liu, F. Lu, G. H. Marchler, M. Mullokandov, B. A. Shoemaker, V. Simonyan, J. S. Song, P. A. Thiessen, R. A. Yamashita, J. J. Yin, D. Zhang, and S. H. Bryant. 2005. CDD: a conserved domain database for protein classification. Nucleic Acids Res. 33:D192-D196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Martin-Didonet, C. C., L. S. Chubatsu, E. M. Souza, M. Kleina, F. G. Rego, L. U. Rigo, M. G. Yates, and F. O. Pedrosa. 2000. Genome structure of the genus Azospirillum. J. Bacteriol. 182:4113-4116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Michiels, K., P. De Troch, I. Onyeocha, A. Van Gool, C. Elmerich, and J. Vanderleyden. 1989. Plasmid localization and mapping of two Azospirillum brasilense loci that affect exopolysaccharide synthesis. Plasmid 21:142-146. [DOI] [PubMed] [Google Scholar]
- 46.Nelson, L. M., and R. Knowles. 1978. Effect of oxygen and nitrate on nitrogen fixation and denitrification by Azospirillum brasilense grown in continuous culture. Can. J. Microbiol. 24:1395-1403. [DOI] [PubMed] [Google Scholar]
- 47.Ohnishi, M., K. Kurokawa, and T. Hayashi. 2001. Diversification of Escherichia coli genomes: are bacteriophages the major contributors? Trends Microbiol. 9:481-485. [DOI] [PubMed] [Google Scholar]
- 48.Pedulla, M. L., M. E. Ford, J. M. Houtz, T. Karthikeyan, C. Wadsworth, J. A. Lewis, D. Jacobs-Sera, J. Falbo, J. Gross, N. R. Pannunzio, W. Brucker, V. Kumar, J. Kandasamy, L. Keenan, S. Bardarov, J. Kriakov, J. G. Lawrence, W. R. Jacobs, Jr., R. W. Hendrix, and G. F. Hatfull. 2003. Origins of highly mosaic mycobacteriophage genomes. Cell 113:171-182. [DOI] [PubMed] [Google Scholar]
- 49.Pothier, J. F., F. Wisniewski-Dyé, M. Weiss-Gayet, Y. Moënne-Loccoz, and C. Prigent-Combaret. 2007. Promoter trap identification of wheat seed extract-induced genes in the plant growth-promoting rhizobacterium Azospirillum brasilense Sp245. Microbiology 153:3608-3622. [DOI] [PubMed] [Google Scholar]
- 50.Rohwer, F., A. Segall, G. Steward, V. Seguritan, M. Breitbart, F. Wolven, and F. Azam. 2000. The complete genomic sequence of the marine phage roseophage SIO1 shares homology with non-marine phages. Limnol. Oceanogr. 45:408-418. [Google Scholar]
- 51.Romero, D., S. Brom, J. Martinez-Salazar, M. L. Girard, R. Palacios, and G. Dávila. 1991. Amplification and deletion of a nod-nif region in the symbiotic plasmid of Rhizobium phaseoli. J. Bacteriol. 173:2435-2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rutherford, K., J. Parkhill, J. Crook, T. Horsnell, P. Rice, M. A. Rajandream, and B. Barrell. 2000. Artemis: sequence visualization and annotation. Bioinformatics 16:944-945. [DOI] [PubMed] [Google Scholar]
- 53.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 54.Schnabel, E. L., and A. L. Jones. 2001. Isolation and characterization of five Erwinia amylovora bacteriophages and assessment of phage resistance in strains of Erwinia amylovora. Appl. Environ. Microbiol. 67:59-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Silander, O. K., D. M. Weinreich, K. M. Wright, K. J. O'Keefe, C. U. Rang, P. E. Turner, and L. Chao. 2005. Widespread genetic exchange among terrestrial bacteriophages. Proc. Natl. Acad. Sci. USA 102:19009-19014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stanton, T. B. 2007. Prophage-like gene transfer agents—novel mechanisms of gene exchange for Methanococcus, Desulfovibrio, Brachyspira, and Rhodobacter species. Anaerobe 13:43-49. [DOI] [PubMed] [Google Scholar]
- 57.Suttle, C. A. 2005. Viruses in the sea. Nature 437:356-361. [DOI] [PubMed] [Google Scholar]
- 58.Tamura, K., J. Dudley, M. Nei, and S. Kumar. 2007. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596-1599. [DOI] [PubMed] [Google Scholar]
- 59.Tarrand, J. J., N. R. Krieg, and J. Döbereiner. 1978. A taxonomic study of the Spirillum lipoferum group, with descriptions of a new genus, Azospirillum gen. nov., and two species, Azospirillum lipoferum (Beijerinck) comb. nov. and Azospirillum brasilense sp. nov. Can. J. Microbiol. 24:967-980. [DOI] [PubMed] [Google Scholar]
- 60.Tettelin, H., V. Masignani, M. J. Cieslewicz, C. Donati, D. Medini, N. L. Ward, S. V. Angiuoli, J. Crabtree, A. L. Jones, A. S. Durkin, R. T. Deboy, T. M. Davidsen, M. Mora, M. Scarselli, I. Margarit y Ros, J. D. Peterson, C. R. Hauser, J. P. Sundaram, W. C. Nelson, R. Madupu, L. M. Brinkac, R. J. Dodson, M. J. Rosovitz, S. A. Sullivan, S. C. Daugherty, D. H. Haft, J. Selengut, M. L. Gwinn, L. Zhou, N. Zafar, H. Khouri, D. Radune, G. Dimitrov, K. Watkins, K. J. O'Connor, S. Smith, T. R. Utterback, O. White, C. E. Rubens, G. Grandi, L. C. Madoff, D. L. Kasper, J. L. Telford, M. R. Wessels, R. Rappuoli, and C. M. Fraser. 2005. Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: implications for the microbial “pan-genome.” Proc. Natl. Acad. Sci. USA 102:13950-13955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Trân Vân, V., O. Berge, D. Faure, R. Bally, P. Hebbar, and T. Heulin. 1997. A simple method for the isolation of Azospirillum strains associated with the rhizosphere of rice. Can. J. Microbiol. 43:486-490. [Google Scholar]
- 62.Vanbleu, E., K. Marchal, M. Lambrecht, J. Mathys, and J. Vanderleyden. 2004. Annotation of the pRhico plasmid of Azospirillum brasilense reveals its role in determining the outer surface composition. FEMS Microbiol. Lett. 232:165-172. [DOI] [PubMed] [Google Scholar]
- 63.Vial, L., C. Lavire, P. Mavingui, D. Blaha, J. Haurat, Y. Moënne-Loccoz, R. Bally, and F. Wisniewski-Dyé. 2006. Phase variation and genomic architecture changes in Azospirillum. J. Bacteriol. 188:5364-5373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Vieille, C., and C. Elmerich. 1990. Characterization of two Azospirillum brasilense Sp7 plasmid genes homologous to Rhizobium meliloti nodPQ. Mol. Plant-Microbe Interact. 3:389-400. [DOI] [PubMed] [Google Scholar]
- 65.Weinbauer, M. G. 2004. Ecology of prokaryotic viruses. FEMS Microbiol. Rev. 28:127-181. [DOI] [PubMed] [Google Scholar]
- 66.Wommack, K. E., and R. R. Colwell. 2000. Virioplankton: viruses in aquatic ecosystems. Microbiol. Mol. Biol. Rev. 64:69-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zdobnov, E. M., and R. Apweiler. 2001. InterProScan—an integration platform for the signature-recognition methods in InterPro. Bioinformatics 17:847-848. [DOI] [PubMed] [Google Scholar]