Abstract
The linked H19 and Igf2 genes on mouse distal chromosome 7 are subject to genomic imprinting. Competition between the promoters of the genes for transcription from shared enhancers has been proposed as an explanation for the coordinate expression and reciprocal imprinting of these two genes. To test this model, we have used Cre-loxP technology to generate in mice a conditional deletion of the H19 promoter and structural gene that leaves no transcription unit in the locus. Contrary to the prediction of enhancer competition we find that transcriptional activity from the H19 promoter is not required for the imprinted silencing of the Igf2 gene.
A small number of genes in the mammalian genome are subject to genomic imprinting, the parent-of-origin-specific expression of the two alleles of a gene. To date, nearly 30 imprinted genes have been identified in human and mouse, the majority of which are located in close proximity to at least one other imprinted gene (1, 2). This clustering has raised the possibility that imprinting may be regulated by regional signals that act over a chromosomal domain. One imprinted region that has been studied intensively lies at the distal end of mouse chromosome 7 and the corresponding region of human chromosome 11p15.5. This region spans approximately 1 Mb and includes the paternally expressed Igf2 gene and the maternally expressed H19 gene (1).
A functional link between the imprinting of H19, which encodes an untranslated RNA, and Igf2, which encodes a fetal growth factor, was suggested by the observation that the genes are coexpressed throughout development in tissues of endodermal and mesodermal origin (3, 4). We have proposed previously that the coexpression and reciprocal imprinting of Igf2 and H19 could result from the promoters of the H19 and Igf2 genes competing for enhancers located downstream of the H19 gene (5). On the maternal chromosome the H19 gene would monopolize the enhancers by virtue of its proximity and/or greater promoter strength, whereas on the paternal chromosome, allele-specific methylation of H19 would silence the gene and allow Igf2 access to the enhancers. One prediction of this model was verified when two endodermal enhancers that lie 3′ of H19 were shown to be required for transcription of both genes (6). A second prediction, that silencing of H19 by DNA methylation is required for Igf2 expression, was tested in homozygous mutant mice lacking the maintenance DNA methyltransferase (7). The activation of paternal H19 transcription that results from the loss of DNA methylation was accompanied by the silencing of Igf2 in cis, a finding consistent with enhancer competition.
The third prediction of this model is a requirement for H19 transcription on the maternal chromosome to maintain the silence of the maternal Igf2 allele. This prediction has been tested with two mutations that deleted the H19 gene and its promoter. The first mutation (H19Δ13) replaced 13 kb of the locus, encompassing the H19 transcription unit and its flank, with a neomycin resistance gene (Neo) (8). The resulting mice exhibited extensive loss of imprinting at Igf2, expressing the maternal Igf2 allele at between 30% and 100% of the paternal levels in a variety of tissues. A smaller deletion that replaced just 3 kb of the H19 gene body and promoter with a different Neo gene cassette (H19Δ3) reported a modest relaxation of the Igf2 imprint in skeletal muscle, with maternal Igf2 mRNA levels that were 25% of the paternal level (9). Although both mutations eliminated H19 transcription, they left another transcription unit at the locus, the Neo cassette, a potential competitor for Igf2 transcription. Thus, the differences in their phenotypes could have resulted from the transcriptional properties of the selectable markers, rather than the importance of the deleted DNA sequences.
The definitive test of enhancer competition requires a deletion of the H19 promoter that leaves no transcription unit, i.e., no competitor, at the locus. We have generated this deletion by using Cre-loxP gene targeting and find that, contrary to the prediction of the enhancer competition model, the absence of transcription from the H19 promoter has no effect on the imprinting of Igf2 in liver and only a small effect in skeletal muscle. Our data lead us to propose a methylation-regulated chromatin boundary as an alternative mechanism for Igf2 imprinting.
MATERIALS AND METHODS
Construction of the H19 Targeting Vector.
Two 129Sv/J genomic clones covering the H19 gene and its flanks that have been described previously were used to generate the H19loxP targeting vector (10). The 5′ homologous fragment is 5.5 kb and extends from a BglII site at −2 kb relative to the H19 transcriptional start site to a SalI site at +2.6 kb. The 3′ homologous fragment is 7.5 kb and extends from the SalI site to a BglII site at +10 kb. The 5′ arm was modified by the insertion of a loxP site into the EcoRV site at −235 bp, and the 3′ arm was modified by the insertion of the Pgk1-Neo cassette followed by a second loxP site into the SalI site. The two modified clones were ligated into pBKSII to generate the targeting construct H19loxP, which was linearized for electroporation at a unique vector NotI site.
Generation of Conditionally Targeted and Null Alleles at the H19 Locus.
The H19loxP targeting vector was electroporated into E14 embryonic stem (ES) cells followed by selection in 250 μg/ml G418. Resistant colonies were screened by Southern blotting with a 500-bp, PCR-generated H19 fragment from −2.2 kb to −2.7 kb (5′ probe) and verified at the 3′ end with a 1-kb BglII-EcoRI fragment (3′ probe). Correctly targeted clones were obtained at a frequency of 1 in 40 G418-selected clones. H19loxP ES cell clones were transiently transfected with the Cre expression vector pBS185 (11), replica-plated, and screened for survival in G418. Clones that died in G418 medium were analyzed by Southern blotting with the 5′ probe to verify excision of H19-Neo.
RNA and DNA Analysis.
All animals were analyzed between 3 and 5 days after birth. Liver and tongue (skeletal muscle) RNA was extracted with Trizol (GIBCO/BRL) for RNase protection analysis or with LiCl for reverse transcription–PCR. RNase protection analysis was performed with the RPAII kit (Ambion) by using 5 μg of total RNA for H19 assays and 10 μg of total RNA for Igf2 assays. RNase protection probes have been described previously and include a 120-bp SmaI–BamHI fragment of the first exon of the mouse H19 gene (12), a 350-bp, PCR-generated fragment of the mouse Igf2 gene 3′ untranslated region [Igf2 allele-specific (8)] and a 270-bp XhoII–DraI fragment of the mouse rpL32–4A gene (13). RNase protection gels were exposed to x-ray film and quantitated by use of a Molecular Dynamics PhosphorImager.
Southern blotting was performed with 5 μg of digested mouse genomic DNA and analyzed by electrophoresis in 0.8% agarose gels. DNA was transferred to Hybond N+ membranes and hybridized in 5× SSPE (0.18 M NaCl/10 mM phosphate, pH 7.4/1 mM EDTA) with random-primed probes. The blots were washed twice at 2× SSPE/0.1% SDS, once at 1× SSPE/0.1% SDS and once at 0.1× SSPE/0.1% SDS. Radiolabeled probes for methylation analysis were a 3.8-kb EcoRI fragment comprising from −40 to −3,800 bp of the H19 5′ flanking region, and a 717-bp fragment covering the Igf2 DMR1 region.
RESULTS AND DISCUSSION
Generation of a Floxed Allele at the H19 Locus.
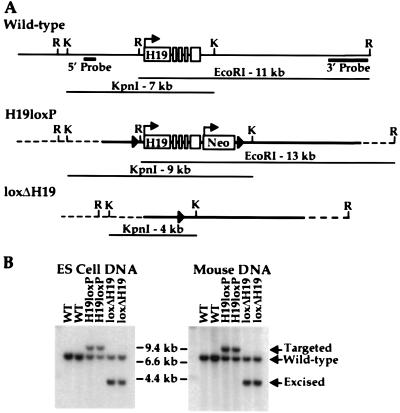
The RNA product of the H19 gene has been shown to be dispensable for its own imprinting and that of Igf2 (10). Transcription from the H19 promoter, on the other hand, could be required in the germ line during the erasure and/or resetting of the epigenetic mark, as well as in the soma to regulate Igf2 silencing. To specifically test the latter, we adopted a strategy that would allow us to delete the H19 gene and promoter after the maternal chromosome had received its appropriate epigenetic modification during female gametogenesis. A gene-targeting vector was generated in which the H19 gene was flanked with loxP sites (floxed) without altering the structural gene or its transcriptional control elements (H19loxP allele) (Fig. 1A). The lox site at the 5′ end of the gene was integrated at −235 bp, a region that is not conserved between the human and mouse genes (14). The 3′ lox site was integrated at a SalI site 320 bp downstream of the polyadenylation signal of the gene. After electroporation into ES cells, G418-resistant clones were screened for the targeted allele by Southern blotting with a 5′ probe that detects a 7-kb KpnI fragment in wild-type ES cells and a 9-kb fragment in correctly targeted ES cells (Fig. 1). Targeted clones also were checked at the 3′ end, where an 11-kb EcoRI fragment in wild-type ES cells is converted to a 13-kb fragment in targeted clones (Fig. 1A and data not shown). Correctly targeted H19loxP clones were microinjected into C57BL/6 blastocysts, and chimeras were bred to C57BL/6 mice to assay for germ-line transmission. Two independent clones that gave germ-line transmission were studied further.
Figure 1.
Conditional targeting of the H19 locus. (A) The structures of the wild-type, targeted, and excised H19 loci are shown along with diagnostic restriction fragments. The solid arrowheads represent the loxP sites, the solid lines represent DNA contained within the targeting construct, and the dashed lines represent DNA outside the targeting construct. The two probes used for Southern analysis of ES cell clones and mouse DNA are shown and are located outside of the region contained within the targeting construct. Restriction sites are abbreviated as EcoRI (R) and KpnI (K). (B) KpnI-digested ES cell and mouse genomic DNA was hybridized to the 5′ probe, which detects a 7-kb fragment in wild-type (WT) H19 alleles, a 9-kb fragment in the floxed (H19loxP) allele, and a 4-kb fragment in the deletion allele (loxΔH19).
Analysis of H19loxP Mice.
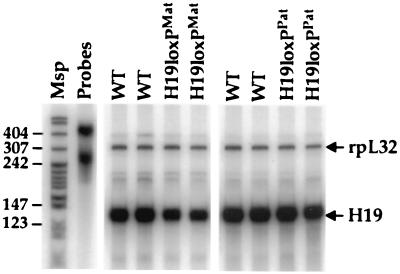
To verify that the insertion of the loxP sites and Neo cassette did not disrupt expression of the H19 gene, H19 RNA in neonatal liver was quantitated by RNase protection analysis with rpL32 as an internal standard (Fig. 2). We use the designations H19loxPMat and H19loxPPat to indicate animals inheriting the H19loxP allele maternally and paternally, respectively. H19loxPMat animals exhibited a reduction in total H19 mRNA levels in both liver and skeletal muscle to approximately 60% of that seen in wild-type littermates, whereas the expression of wild-type maternal H19 RNA in H19loxPPat mice was unaffected (Fig. 2).
Figure 2.
H19 RNA expression in H19loxP heterozygotes. A quantitative RNase protection assay was used to determine the levels of H19 RNA in livers of wild-type (WT) mice and mice carrying the H19loxPMat and H19loxPPat alleles. The upper band indicates the level of expression of an internal control RNA (rpL32). The Msp lane contains MspI-digested pBR322 as a size standard, and the Probes lane contains undigested probes.
The decrease in H19 RNA likely is due to a reduction in transcription rather than to a change in RNA stability because the loxP sites lie outside the transcription unit. It is possible that one or both of the loxP sites disrupt a hitherto unmapped regulatory element. More likely, the reduction in H19 expression is caused by the presence of the Neo gene that is integrated just downstream of H19 (Fig. 1A). Interference by the Pgk1 promoter, either directly or through competition for enhancers, could result in a decrease in total H19 RNA levels. We have observed a similar effect of the Neo cassette in two targeted alleles at the Ednrb locus (M. K. Shin and S.M.T., unpublished results), suggesting that the effect is unrelated to imprinting.
Interestingly, the H19loxPMat animals showed no relaxation of Igf2 imprinting in liver and skeletal muscle despite the decrease in H19 transcription (data not shown). Taken alone this observation does not argue for or against enhancer competition, because the combined transcription of H19 and Neo still could be sufficient to restrict Igf2 transcription.
Conditional Deletion of the H19 Gene in the Maternal Germ Line.
To examine the consequences of deleting transcription at the H19 locus in the soma, we took advantage of a transgene that expresses Cre recombinase in growing oocytes under the direction of the oocyte-specific Zp3 promoter (15). By this stage of oogenesis it is thought that the female H19 gametic mark has been established based on the erasure of H19 methylation and its biallelic expression (16, 17). Animals carrying the H19loxP allele were crossed to Zp3Cre transgenics, and compound heterozygous females were bred to Mus castaneus males to provide polymorphisms for allelic RNA analysis. When the progeny of this cross were analyzed, ≈50% of the animals carried the H19 deletion, referred to as loxΔH19Oocyte. Southern blotting demonstrated the loss of the 9-kb targeted KpnI fragment and the appearance of a new 4-kb band, diagnostic of Cre-mediated excision of H19 and Neo (Fig. 1). The loxΔH19Oocyte animals showed a complete absence of the targeted H19 DNA in both liver and skeletal muscle, indicating that excision occurred before fertilization (data not shown). This is consistent with the previous report demonstrating the efficacy of Zp3Cre in the female germ line (15).
H19 and Igf2 Expression in loxΔH19Oocyte Heterozygotes.
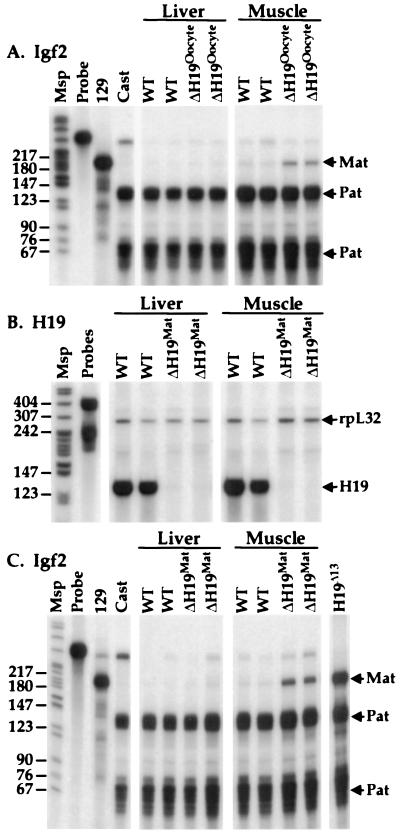
Animals in which the H19 transcription unit was deleted during oogenesis displayed no H19 RNA in liver and skeletal muscle, indicating that the deletion had no effect on the imprinting of the paternal H19 gene (data not shown). An allele-specific RNase protection assay for Igf2 mRNA was used to investigate its imprinting status after excision of the H19 gene (Fig. 3A). In contrast to the prediction of the enhancer competition model, loxΔH19Oocyte animals showed no loss of Igf2 imprinting in liver. This finding was confirmed by quantitative RNase protection assays, which revealed no difference between Igf2 mRNA levels in loxΔH19Oocyte and wild-type littermates (data not shown). The maintenance of Igf2 imprinting in the livers of loxΔH19Oocyte animals provides convincing evidence that enhancer competition between the H19 and Igf2 promoters does not underlie the silencing of the maternal Igf2 allele. In contrast to the finding in liver, loxΔH19Oocyte pups exhibited a small amount of relaxation of Igf2 imprinting in skeletal muscle (Fig. 3A). The degree of relaxation is significantly less than the 100% relaxation of Igf2 imprinting seen in the H19Δ13 maternal heterozygotes and somewhat less than the 25% relaxation reported for H19Δ3 heterozygotes (8, 9).
Figure 3.
H19 and Igf2 expression in loxΔH19Oocyte and loxΔH19Mat mice. (A) The imprinting status of the Igf2 gene in liver and skeletal muscle of wild-type (WT) mice and mice carrying the loxΔH19Oocyte deletion were examined by an allele-specific RNase protection assay. The Msp lane contains MspI-digested pBR322 as a size standard, and the Probe lane contains undigested probes. The 129 and Cast lanes contain liver RNA samples from the parental 129 and M. Castaneus mice. Mat and Pat refer to the parental origin of the RNase protection bands. (B) H19 RNA was analyzed by an RNase protection assay in wild-type (WT) mice and mice carrying the loxΔH19Mat allele. (C) The imprinting status of the Igf2 gene in liver and skeletal muscle of wild-type (WT) mice and mice carrying the loxΔH19Mat deletion was examined by an allele-specific RNase protection assay as in A. The loxΔH19Δ13 lane contains liver RNA from a mouse with maternal inheritance of the H19Δ13 deletion, where the maternal expression of Igf2 mRNA has been determined previously to be approximately 30% of paternal expression (8).
The tissue-specific relaxation of Igf2 imprinting in skeletal muscle is difficult to explain. Of the four targeted mutations that have been generated at the H19 locus, the only one that displays no relaxation of Igf2 imprinting in skeletal muscle is the H19Luc mutation (10). This mutation alone preserves the endogenous H19 promoter, and, consequently, we cannot rule out a minor role for the H19 promoter in the maintenance of the Igf2 imprint in skeletal muscle. Alternatively, the sensitivity of Igf2 imprinting in skeletal muscle to any perturbation in the H19 locus could be related to the nature or location of the tissue-specific transcriptional regulatory elements that act in mesoderm to regulate Igf2 and H19, about which very little is known.
Germ-Line Inheritance of loxΔH19Mat.
In light of the modest consequence of excising the H19 transcription unit during oogenesis, we went on to ask whether transcription from the H19 promoter is required during gametogenesis to set the sex-specific epigenetic states of the parental chromosomes. H19loxP-targeted ES cells were transiently transfected with the plasmid pBS185, which expresses the Cre recombinase (11), and clones were replica-plated into G418. G418-sensitive clones were analyzed by Southern blotting by using the 5′ probe to detect the loss of the 9-kb targeted KpnI fragment and the appearance of a new 4-kb band (Fig. 1). Two independent clones that gave germ-line transmission for the deletion, referred to as loxΔH19, were studied.
The heterozygous female offspring of male chimeras were crossed to M. castaneus males, and the allelic expression of H19 and Igf2 was assessed in their progeny. This cross tests whether the loxΔH19 allele is capable of switching from a paternal epigenotype to a maternal one during oogenesis in the absence of the H19 gene. As expected, no H19 RNA was detected in loxΔH19Mat offspring (Fig. 3B). Furthermore, as was noted with loxΔH19Oocyte progeny, Igf2 mRNA imprinting was maintained in liver, and minimal relaxation of imprinting was detected in skeletal muscle (Fig. 3C). Quantitative RNase protection demonstrated a 17% increase in total skeletal muscle Igf2 mRNA, and this increase likely accounts for the fact that the loxΔH19Mat animals are approximately 5% larger than their wild-type littermates (data not shown). The similar behaviors of loxΔH19Oocyte and loxΔH19Mat animals argue that transcription from the H19 promoter is not required for setting the maternal epigenetic mark.
Germ-Line Inheritance of loxΔH19Pat.
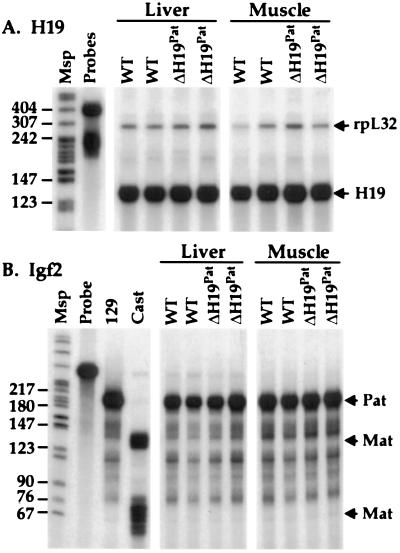
Offspring of male loxΔH19 heterozygotes were analyzed for their ability to maintain the paternal epigenetic mark in the absence of the H19 gene. No differences between wild-type and loxΔH19Pat mutant littermates were observed in the levels of H19 or Igf2 expression or imprinting (Fig. 4). Additionally, heterozygous male offspring of loxΔH19 female heterozygotes were crossed to Mus spretus H19-Igf2 congenic female mice. This cross tests the ability of the loxΔH19 allele to switch from a maternal to a paternal epigenotype. No alterations were found in the expression or imprinting of either H19 or Igf2 in the progeny of this cross. Animals homozygous for the loxΔH19 allele also were bred to Mus spretus H19-Igf2 congenic mice, and their offspring were found to display phenotypes identical to those seen in the heterozygous crosses (data not shown).
Figure 4.
H19 and Igf2 expression in loxΔH19Pat mice. (A) H19 RNA was analyzed by an RNase protection assay in wild-type (WT) mice and mice carrying the loxΔH19Pat allele. Lane designations are as in Fig. 2. (B) The imprinting status of the Igf2 gene in liver and skeletal muscle of WT mice and mice carrying the loxΔH19Pat deletion were examined by an allele-specific RNase protection assay. Lane designations are as in Fig. 3A.
Methylation Status of the H19 and Igf2 Genes in cis to loxΔH19.
To assess whether the absence of transcription from the H19 promoter had altered the ability of the chromosome to acquire the appropriate patterns of DNA methylation, we examined methylation of the H19 and Igf2 genes in loxΔH19 heterozygotes. The H19 gene is heavily methylated on the paternal chromosome and largely unmethylated on the maternal chromosome. In particular, a 2-kb region between −2 and −4 kb is methylated exclusively on the paternal allele at almost all CpG residues (17, 18). This methylation is inherited from sperm, and maintained throughout embryogenesis, and thus is believed to represent the gametic mark for H19.
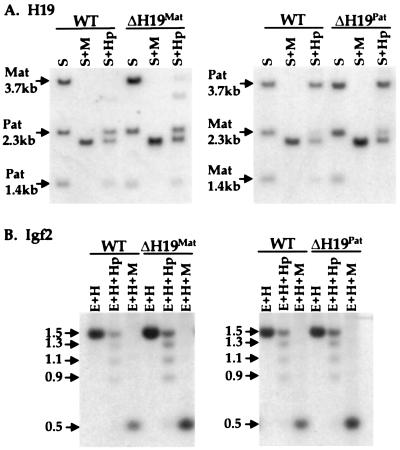
To assess the methylation status of this region in loxΔH19Mat and loxΔH19Pat animals, liver DNA was digested with SacI, an enzyme that detects an H19 polymorphism between 129 and M. castaneus mice, in the absence or presence of the methylation-sensitive restriction enzyme HpaII or its methylation-insensitive isoschizomer MspI. The maternally inherited H19 gene was unmethylated in both wild-type and the loxΔH19Mat mice, as demonstrated by the nearly complete digestion of the 3.7-kb SacI fragment (Fig. 5A). In loxΔH19Pat mice the 5′ flanking region of the mutant gene was completely methylated, as shown by the maintenance of the 3.7-kb fragment in both wild-type and loxΔH19Pat animals. These results demonstrate that transcriptional activity from the H19 promoter is not required to establish or maintain parent-of-origin-specific methylation patterns.
Figure 5.
Methylation of H19 and Igf2 in loxΔH19 heterozygotes. (A) Methylation analysis of the H19 5′ flanking region. DNA from wild-type (WT) mice and mice carrying loxΔH19Mat (Left) or loxΔH19Pat (Right) alleles was digested with SacI (S) and SacI plus HpaII (Hp) or MspI (M). The blots were hybridized to a 3.8-kb EcoRI probe from the 5′ flank of the H19 gene. The partially methylated maternal H19 DNA in loxΔH19Mat animals is more apparent than in wild-type animals because of loading differences. (B) Methylation analysis of the Igf2 DMR1 region. Liver genomic DNAs from the same animals as in A were digested with EcoRI (E) and HindIII (H) or with EcoRI–HindIII plus HpaII or MspI. The blots were hybridized to a 717-bp probe from the DMR1 region of the Igf2 5′ flank. In these animals, the 1.5-kb band represents the fully methylated paternal allele whereas the reduced methylation on the maternal allele gives a range of smaller digestion products.
The Igf2 gene displays two regions of differential methylation, the DMR1 region located upstream of promoter P1 and the DMR2 region in the 3′ part of the gene (19, 20). The allele-specific methylation of these regions is not established until after fertilization, and, unlike H19, it is the transcriptionally active allele of Igf2 that is more highly methylated. Although this methylation does not qualify as a gametic mark, nor is it required for Igf2 transcription (10), it does appear to correlate with expression of Igf2 (21).
To examine DMR1 methylation in loxΔH19 mice, liver DNA was digested with EcoRI and HindIII in the presence or absence of HpaII or MspI and analyzed by Southern blotting with a DMR1 probe (Fig. 5B). In wild-type mice, the methylated paternal allele appears as a 1.5-kb fragment whereas partial methylation of several maternal HpaII sites leads to multiple digestion products ranging from 500 bp to l.3 kb. DNA samples from loxΔH19Mat and loxΔH19Pat animals show a pattern identical to those of their wild-type littermates.
A Boundary Model for the Imprinting of H19 and Igf2.
Our data have demonstrated conclusively that transcription from the H19 promoter is not required for the silencing of the maternal Igf2 gene in cis. This rules out strict enhancer competition between the H19 and Igf2 gene promoters to explain the coexpression and reciprocal imprinting of these two genes. At best, the H19 promoter may play a minor role in silencing the maternal Igf2 allele in skeletal muscle. When considered together with the recent findings of Thorvaldson et al. (22) our results argue that the sequences that regulate Igf2 silencing must lie exclusively upstream of the H19 transcription unit. Thorvaldson et al. (22) generated a 1.6-kb deletion within the candidate gametic mark upstream of the H19 gene. Mice carrying this deletion exhibit a complete loss of imprinting of both H19 and Igf2, with both genes coexpressed at reduced levels on both parental chromosomes. Those authors argued that their findings could be reconciled with enhancer competition if the epigenetic region was required for high-level H19 transcription on the maternal chromosome. This interpretation cannot be sustained in light of our results.
What is the mechanism by which the flank of the H19 gene silences the Igf2 and Ins2 genes 90 kb away? We currently favor the idea that the region functions as a chromatin boundary or insulator element on the unmethylated maternal chromosome (23). Boundary elements are defined as cis-acting DNA sequences that delineate specialized gene expression domains in chromatin (24, 25). When placed on either side of a gene and its regulatory elements, they insulate the gene from regulatory elements of neighboring genes. In other circumstances, boundary elements are used to segregate regulatory elements that act on the same gene in different cells into separate functional domains (26). At the H19 gene, we suggest that the epigenetic control region forms a boundary on the maternal chromosome that blocks the access of Igf2 to the downstream enhancers, resulting in the exclusive expression of the H19 gene. On the paternal chromosome the boundary cannot form in the presence of DNA methylation, and, therefore, Igf2 has access to the enhancers. Transcription of H19 is inhibited by spreading of the methylation from the epigenetic region into the promoter.
A boundary can rationalize much of the experimental data that have been generated to understand the mechanism of Igf2 imprinting. The model explains the extensive relaxation of Igf2 imprinting observed with the H19Δ13 deletion (8) and the smaller gametic mark deletion (22), both of which excised the putative boundary. In contrast, the H19Δ3 deletion and our loxΔH19 deletion that left the boundary intact had no effect on Igf2 imprinting in liver and only a modest effect in muscle [Ripoche et al. (9) and L. Dandolo, personal communication]. In an experiment in which the enhancers that normally lie downstream of the boundary were moved to a site upstream of the putative boundary, the direction of the maternal imprint was reversed, with Igf2 expressed and H19 silent (27). This finding is precisely what would be expected if the epigenetic region functions as a boundary. Finally, the hypersensitivity of the 2-kb epigenetic region to nucleases on the maternal chromosome but not on the paternal chromosome is consistent with the nuclease sensitivity of well characterized boundaries in a variety of organisms (23, 28).
One way to reconcile a boundary model with a modified version of enhancer competition has been suggested by Geyer (24). She proposed that insulators could function as “decoy” or pseudo-promoters, blocking the interaction between downstream genes and their enhancers, in a directional manner, without producing functional transcripts. This explanation for insulator function has been suggested recently to explain the mechanism of action of the scs and scs’ elements in Drosophila, the latter of which coincides with the promoter for a downstream gene (29). Because previous attempts to locate transcripts emanating from the H19 5′ nuclease hypersensitive region between −2 and −4 kb using Northern analysis and RNase protection had been unsuccessful, we used the more sensitive reverse transcription–PCR to search for transcripts in the region. Using several PCR primer pairs spanning the region from −2 to −5.5 kb we detected very low level transcription across the H19 5′ differentially methylated region and extending further upstream (data not shown). Semiquantitative reverse transcription–PCR analysis indicated that these transcripts are present at <0.1% of the level of H19 RNA. These transcripts are difficult to characterize in detail because of their low abundance, but they raise the possibility that the upstream region, rather than the H19 promoter, could be a target of enhancer competition with Igf2. On the other hand, transcription that does not yield stable transcripts has been detected across the human β-globin locus control region and may be a general property of cis-acting elements (30). The presence of these additional transcripts does not affect our conclusion that transcription from the H19 promoter, as originally proposed in the enhancer competition model, is not required for Igf2 imprinting.
The chromatin boundary model may be applicable to other imprinted loci. The intragenic epigenetic mark of the Igf2r gene, which is methylated on the transcriptionally active maternal chromosome (31), could function on the paternal chromosome as a boundary for distal regulatory elements, resulting in the paternal-specific silencing of the gene. Likewise in the Prader Willi/Angelman syndromes gene cluster, the extensive maternal methylation at the 5′ end of the SNRPN gene (32, 33) could function, in part, to inactivate a boundary for a neuronal enhancer required for expression of the distal Angelman candidate gene, UBE3A, which is imprinted only in brain (34, 35). It will be important in the future to consider the relative positions of epigenetic marks and regulatory elements to imprinted genes when proposing mechanisms of imprinting regulation.
Acknowledgments
We thank Dr. G. Martin for the ZP3Cre mice and members of the Tilghman laboratory for comments on the manuscript. This work was supported by a Jane Coffin Childs postdoctoral fellowship (J.V.S.) and a grant from National Institute of General Medical Sciences (S.M.T.). S.M.T. is an investigator of the Howard Hughes Medical Institute.
ABBREVIATION
- ES
embryonic stem
References
- 1.Bartolomei M S, Tilghman S M. Annu Rev Genet. 1997;31:493–525. doi: 10.1146/annurev.genet.31.1.493. [DOI] [PubMed] [Google Scholar]
- 2.Reik W, Walter J. Curr Opin Genet Dev. 1998;8:154–164. doi: 10.1016/s0959-437x(98)80136-6. [DOI] [PubMed] [Google Scholar]
- 3.Poirier F, Chan C-T J, Timmons P M, Robertson E J, Evans M J, Rigby P W J. Development (Cambridge, UK) 1991;113:1105–1114. doi: 10.1242/dev.113.4.1105. [DOI] [PubMed] [Google Scholar]
- 4.Lee J E, Pintar J, Efstratiadis A. Development (Cambridge, U.K.) 1990;110:151–159. doi: 10.1242/dev.110.1.151. [DOI] [PubMed] [Google Scholar]
- 5.Bartolomei M S, Tilghman S M. Semin Dev Biol. 1992;3:107–117. [Google Scholar]
- 6.Leighton P A, Saam J R, Ingram R S, Stewart C L, Tilghman S M. Genes Dev. 1995;9:2079–2089. doi: 10.1101/gad.9.17.2079. [DOI] [PubMed] [Google Scholar]
- 7.Li E, Beard C, Jaenisch R. Nature (London) 1993;366:362–365. doi: 10.1038/366362a0. [DOI] [PubMed] [Google Scholar]
- 8.Leighton P A, Ingram R S, Eggenschwiler J, Efstratiadis A, Tilghman S M. Nature (London) 1995;375:34–39. doi: 10.1038/375034a0. [DOI] [PubMed] [Google Scholar]
- 9.Ripoche M-A, Kress C, Poirier F, Dandolo L. Genes Dev. 1997;11:1596–1604. doi: 10.1101/gad.11.12.1596. [DOI] [PubMed] [Google Scholar]
- 10.Jones B K, Levorse J, Tilghman S M. Genes Dev. 1998;12:2200–2207. doi: 10.1101/gad.12.14.2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sauer B, Henderson N. New Biol. 1990;2:441–449. [PubMed] [Google Scholar]
- 12.Brunkow M E, Tilghman S M. Genes Dev. 1991;5:1092–1101. doi: 10.1101/gad.5.6.1092. [DOI] [PubMed] [Google Scholar]
- 13.Dudov K P, Perry R P. Cell. 1984;37:457–468. doi: 10.1016/0092-8674(84)90376-3. [DOI] [PubMed] [Google Scholar]
- 14.Brannan C I, Dees E C, Ingram R S, Tilghman S M. Mol Cell Biol. 1990;10:28–36. doi: 10.1128/mcb.10.1.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lewandoski M, Wassarman K M, Martin G R. Curr Biol. 1997;7:148–151. doi: 10.1016/s0960-9822(06)00059-5. [DOI] [PubMed] [Google Scholar]
- 16.Szabo P E, Mann J R. Genes Dev. 1995;9:1857–1868. doi: 10.1101/gad.9.15.1857. [DOI] [PubMed] [Google Scholar]
- 17.Olek A, Walter J. Nat Genet. 1997;17:275–276. doi: 10.1038/ng1197-275. [DOI] [PubMed] [Google Scholar]
- 18.Tremblay K D, Duran K L, Bartolomei M S. Mol Cell Biol. 1997;17:4322–4329. doi: 10.1128/mcb.17.8.4322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brandeis M, Kafri T, Ariel M, Chaillet J R, McCarrey J, Razin A, Cedar H. EMBO J. 1993;12:3669–3677. doi: 10.1002/j.1460-2075.1993.tb06041.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Feil R, Walter J, Allen N D, Reik W. Development (Cambridge, UK) 1994;120:2933–2943. doi: 10.1242/dev.120.10.2933. [DOI] [PubMed] [Google Scholar]
- 21.Forne T, Oswald J, Dean W, Saam J R, Bailleul B, Dandolo L, Tilghman S M, Walter J, Reik W. Proc Natl Acad Sci USA. 1997;94:10243–10248. doi: 10.1073/pnas.94.19.10243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thorvaldson J L, Duran K L, Bartolomei M S. Genes Dev. 1998;12:3693–3702. doi: 10.1101/gad.12.23.3693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hark A T, Tilghman S M. Hum Mol Genet. 1998;7:1979–1985. doi: 10.1093/hmg/7.12.1979. [DOI] [PubMed] [Google Scholar]
- 24.Geyer P K. Curr Opin Genet Dev. 1997;7:242–248. doi: 10.1016/s0959-437x(97)80134-7. [DOI] [PubMed] [Google Scholar]
- 25.Kellum R, Elgin S C R. Curr Biol. 1998;8:R521–R524. doi: 10.1016/s0960-9822(07)00337-5. [DOI] [PubMed] [Google Scholar]
- 26.Hagstrom K, Muller M, Schedl P. Genes Dev. 1996;10:3202–3215. doi: 10.1101/gad.10.24.3202. [DOI] [PubMed] [Google Scholar]
- 27.Webber A, Ingram R I, Levorse J, Tilghman S M. Nature (London) 1998;391:711–715. doi: 10.1038/35655. [DOI] [PubMed] [Google Scholar]
- 28.Szabo P E, Pfeifer G P, Mann J R. Mol Cell Biol. 1998;18:6767–6776. doi: 10.1128/mcb.18.11.6767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Avramova Z, Tikhonov A. Trends Genet. 1999;15:138–139. doi: 10.1016/s0168-9525(99)01712-6. [DOI] [PubMed] [Google Scholar]
- 30.Ashe H L, Minks J, Wijgerde M, Fraser P, Proudfoot N J. Genes Dev. 1997;11:2494–2509. doi: 10.1101/gad.11.19.2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stoger R, Kubicka P, Liu C-G, Kafri T, Razin A, Cedar H, Barlow D P. Cell. 1993;73:61–71. doi: 10.1016/0092-8674(93)90160-r. [DOI] [PubMed] [Google Scholar]
- 32.Sutcliffe J S, Nakao M, Christian S, Orstavik K H, Tommerup N, Ledbetter D H, Beaudet A L. Nat Genet. 1994;8:52–58. doi: 10.1038/ng0994-52. [DOI] [PubMed] [Google Scholar]
- 33.Glenn C C, Saitoh S, Jong M T C, Filbrandt M M, Surti U, Driscoll D J, Nicholls R D. Am J Hum Genet. 1996;58:335–346. [PMC free article] [PubMed] [Google Scholar]
- 34.Rougeulle C, Glatt H, Lalande M. Nat Genet. 1997;17:14–15. doi: 10.1038/ng0997-14. [DOI] [PubMed] [Google Scholar]
- 35.Vu T H, Hoffmann A R. Nat Genet. 1997;17:12–13. doi: 10.1038/ng0997-12. [DOI] [PubMed] [Google Scholar]