Abstract
A single missense mutation was identified in a novel, highly conserved zinc-finger gene, ZCD2, in three consanguineous families of Jordanian descent with Wolfram syndrome (WFS). It had been shown that these families did not have mutations in the WFS1 gene (WFS1) but were mapped to the WFS2 locus at 4q22-25. A G→C transversion at nucleotide 109 predicts an amino acid change from glutamic acid to glutamine (E37Q). Although the amino acid is conserved and the mutation is nonsynonymous, the pathogenesis for the disorder is because the mutation also causes aberrant splicing. The mutation was found to disrupt messenger RNA splicing by eliminating exon 2, and it results in the introduction of a premature stop codon. Mutations in WFS1 have also been found to cause low-frequency nonsyndromic hearing loss, progressive hearing loss, and isolated optic atrophy associated with hearing loss. Screening of 377 probands with hearing loss did not identify mutations in the WFS2 gene. The WFS1-encoded protein, Wolframin, is known to localize to the endoplasmic reticulum and plays a role in calcium homeostasis. The ZCD2-encoded protein, ERIS (endoplasmic reticulum intermembrane small protein), is also shown to localize to the endoplasmic reticulum but does not interact directly with Wolframin. Lymphoblastoid cells from affected individuals show a significantly greater rise in intracellular calcium when stimulated with thapsigargin, compared with controls, although no difference was observed in resting concentrations of intracellular calcium.
Wolfram syndrome (WFS [MIM 222300]) is an autosomal recessive disorder with severe neurodegeneration. Affected individuals present with juvenile-onset insulin-dependent diabetes mellitus and optic atrophy.1,2 Other neurological and endocrine manifestations include diabetes insipidus, sensorineural deafness, dementia, psychiatric illnesses, renal-tract abnormalities, and bladder atony.3,4 One gene for the disorder, WFS1, on chromosome 4p16.3, has been cloned and encodes a novel transmembrane protein located in the endoplasmic reticulum (ER) and may play a role in calcium (Ca2+) homeostasis.5–8 In addition, dominant mutations in the WFS1 gene have been shown to play a role in low-frequency sensorineural hearing loss, progressive hearing loss, and deafness with optic atrophy.9–11
The locus for WFS2 (MIM 604928) was mapped to chromosome 4q22-24 between markers D4S1591 and D4S3240 in three large, consanguineous Jordanian families.12 Although the families are not related, haplotype analysis shows a common affected haplotype among the families. An extensive phenotypic analysis showed additional symptoms in individuals with WFS2, such as significant bleeding tendency, as well as a defective platelet aggregation with collagen,13 which has not been previously described in families with WFS. In addition, a considerable number of the patients had peptic ulcer disease that was compounded by the bleeding tendency, causing gastrointestinal-tract bleeding.14
In this study, we identified a mutation causative for WFS2 in a novel, highly conserved zinc-finger gene, ZCD2, that is located within the critical gene region. In addition, evidence of the pathogenetic nature of the mutation was revealed. Additional studies of this novel gene include the characterization of its protein domains, expression, and cellular localization and studies to determine whether there is a role for this gene in Ca2+ homeostasis. To determine whether the WFS2 gene is also associated with nonsyndromic hearing loss, mutation analysis with a large cohort of individuals with hearing loss was also performed.
Material and Methods
Genetic Markers
New genetic markers in the 4q24-q25 region were identified by searching for microsatellite regions in genomic DNA with use of the program Sputnik. Four new microsatellite clusters were identified within the critical gene region. Primers to amplify the region were developed and were tested in 10 unrelated CEPH individuals (all primers here and below are shown 5′→3′) (Wpoly2F [tgactagttgatgggtgcag], Wpoly2R [ccaaacgctagtgagatgtg], Wpoly5F [caatatcccatactgagagtc], Wpoly5R [tgcatgttctgaacacgtac], Wpoly6F [actgttggtagatgtcagtc], Wpoly6R [gaccacatcttctgtgtgcc], Wpoly7F [atgctgtacgttctagccag], and Wpoly7R [gtgatcttgtattctgcaacc]). All microsatellites were found to be polymorphic, with heterozygosities of 0.79 for Wpoly2, 0.81 for Wpoly5, 0.78 for Wpoly6, and 0.82 for Wpoly7. These markers and other known markers in the region (D4S2961, D4S3043, D4S1572, D4S2913, D4S411, D4S1531, and D4S2917) were typed in the WFS2-affected families. Primers were end labeled with γ-P33, and annealing temperatures were 60°C for Wpoly2, Wpoly5, and Wpoly6 and 65°C for Wpoly7. Samples were run on sequencing gels and were exposed to x-ray film.
SSCP Analysis
Primer sets were individually optimized for single-band amplification. Samples were prepared by mixing 7 μl of PCR products with 3 μl SSCP loading dye (95% formamide, 20 mM EDTA, 0.05% xylene cyanol, and 0.05% bromophenol blue), were denatured at 95°C for 5 min, and were quickly cooled on ice. Three samples were run for each primer set: the patient PCR product, the control PCR product, and the patient plus control PCR products. Denatured samples were run through a 10% (49:1) acrylamide:bis-acrylamide gel (National Diagnostics) with 1× Tris-borate EDTA (TBE) and 10% glycerol either at 8 W for 5 h at 4°C or at 3 W for 16 h at 4°C after a 30-min prerun. Gels were stained in SYBR Gold (Molecular Probes) diluted 1:10,000 in 1× TBE, in accordance with the manufacturer’s protocol.
ZCD2 PCR and Restriction-Enzyme Digest
Four sets of primers were synthesized to amplify exons 1–3 of the ZCD2 gene from genomic DNA: Exon1F (gctcgggagaggagttgac) and Exon1R (ggatttttacgcctccttcc), Exon2aF (agcactgcagattctgacaca) and Exon2aR (cgttttagaacgccaacacc), Exon2bF (tcgcacttcttggctacctt) and Exon2bR (ggggatttaagaggcgaaac), and Exon3F (gctttctttctgagagcatttc) and Exon3R (ccagtagtaataattaagaccaccatt). PCR was performed using an annealing temperature of 65°C. The mutation was screened by digesting the 249-bp Exon2a PCR product with XmnI by use of the manufacturer’s conditions (NEB). ZCD2 exon 2 from an affected individual from each family was sequenced, to confirm the change. The use of these samples was approved by the Virginia Commonwealth University and Jordan University of Science and Technology institutional review boards (IRBs), and informed consent or assent, when appropriate, was obtained from all subjects.
TaqMan Genotyping
To calculate the allele frequency of the mutation, a Custom TaqMan SNP Genotyping Assay (Applied Biosystems) was designed specific to the detection of this single base change. This methodology uses 5′ fluorogenic chemistry and TaqMan probes. An ABI PRISM 7900 sequence detection system was used for genotyping the control cohorts. An affected individual (W2) from family WS-2 and two obligate carriers were used as homozygous and heterozygous controls, respectively, after the genotype was confirmed by sequencing. The genotype was confirmed by sequencing in 88 randomly chosen DNA samples (20%) of the samples from 440 Jordanian controls.
Control Cohorts
The three control cohorts include an ethnically matched cohort of 440 unrelated Jordanian controls. In addition, 1,064 controls obtained from the Human Genome Diversity Project (HGDP)–CEPH Human Genome Diversity Cell Line Panel15 and 86 unrelated CEPH controls were used. The HGDP-CEPH panel contains 148 individuals of the same ethnic background as the three families. These cohorts represent a total of 3,180 chromosomes, of which 1,176 are ethnically matched.
Hearing-Loss Cohorts
Of the 377 probands with hearing loss who were screened, 288 were from multiplex families indicating a recessive mode of inheritance and had severe-to-profound hearing loss. We also screened 84 probands with moderate-to-severe autosomal dominant hearing loss. The probands were classified as autosomal dominant if there is a family history of hearing loss with vertical transmission and if the hearing loss is postlingual and progressive. Although the majority had severe-to-profound hearing loss, 14 individuals had high-frequency hearing loss, but only 1 individual had low-frequency hearing loss. The use of these samples was approved by the Virginia Commonwealth University IRB, and informed consent or assent, when appropriate, was obtained from all subjects.
Cell Culture
Two control (W1 and JA) and one affected (W2) lymphoblastoid cell lines from the Jordanian population were maintained in RPMI 1640 with l-glutamate (Invitrogen) supplemented with 10% fetal bovine serum (FBS). P19 mouse embryonic carcinoma cells were grown in α–minimum essential medium (Invitrogen) supplemented with 7.5% bovine calf serum, 2.5% FBS, and 0.5% gentamicin. HEK293 human embryonic kidney cells were maintained in Dulbecco’s modified Eagle’s medium (Invitrogen) supplemented with 10% FBS and 1% penicillin and streptomycin. All cell lines were maintained in a 5% CO2 incubator that was kept at 37°C. P19 and HEK293 cells that were going to be transfected were maintained in the described media without antibiotic supplementation.
Protein Analysis
The protein sequence was analyzed through Prosite,16 and PhosphoMotif Finder of the Human Protein Reference Database to identify putative protein motifs.17 To identify homologues of the gene, the protein was searched through use of the basic local alignment search tool nucleotide database (BLAST: tBLASTn).18
RT-PCR
Total RNA was extracted from W1, W2, and JA human lymphoblastoid cells by use of Trizol (Invitrogen). Total RNA was treated with 20 U of RQ1 DNase (Promega). First-strand cDNAs were generated from 500 ng of total RNA with 5 ng/μl of both oligo dT and random primers and 0.5 mM deoxynucleotide triphosphates (dNTPs), then they were heated to 65°C for 5 min and were cooled immediately afterwards on ice. First-strand buffer (1×), 0.01 M dithiothreitol, and 40 U RNase inhibitor (Promega) were then added, and the mixture was incubated at 37°C for 2 min. An aliquot of 200 U of Moloney murine leukemia virus–reverse transcriptase (Invitrogen) was added, and the reaction was allowed to take place at 37°C for 50 min, followed by a final incubation at 70°C for 15 min.
Transcript-specific primers used for amplification of ZCD2 cDNA were pogo.aRTF (gctcgggagaggagtgga) and pogo.aRTR (tgcaggaagaaaataaattggaa). GADPH primers used were GADPHF (acttcaacagcgacacccactc) and GADPHR (ccctgttgctgtagccaaattc). PCR was performed in 12.5-μl reactions containing 1.25 μl of cDNA, 1× PCR buffer (GeneChoice), 0.2 mM dNTPs, 0.26 pmol/μl of each primer, and 0.25 U of Taq polymerase (GeneChoice). The primers were annealed at 63°C.
Expression-Vector Construct and Transfection
Two mammalian expression vectors containing a FLAG-tagged ZCD2 cDNA were constructed, one of which had the FLAG on the N-terminus, and the other on the C-terminus. The FLAG tags with restriction-enzyme ends were generated using oligonucleotides and were inserted into the destination vector pDEST26 (Invitrogen). For the N-terminus FLAG tag, the primers were ggaccatggcgtactacgactacaaagacgatgacgacaagt and gcggaccatggcgtactacgactacaaagacgatgacgacaagtctag, which create a SacII site at the 5′ end and an XbaI site at the 3′ end. For the C-terminus FLAG tag, the primers were gtacaaagtggttgactacaaagacgatgacgacaagtgagacgt and aaagtggttgactacaaagacgatgacgacaagtgag, which create a BsrGI site at the 5′ end, then an attB2 site upstream of the FLAG sequence, followed by a stop codon, and an AatII site at the 3′ end.
For the N-terminus FLAG construct, the FLAG-tag sequence was cloned into pDEST26 by use of SacII and XbaI sites. For the C-terminus FLAG-tag construct, a fragment from pDEST26 was excised using SalI and HindIII and was cloned into pBluescript KS (Stratagene); then, the C-terminus FLAG tag was cloned into the pDEST26 fragment by use of the BsrGI and AatII sites; and, finally, the pDEST26 fragment containing the FLAG tag was cloned back into the SalI and NheI sites of the pDEST26 vector.
ZCD2 cDNA was obtained from IMAGE clone 5105935 (Invitrogen). It was subcloned into the EcoRI and XhoI sites of the entry vector pENTR3C (Invitrogen), creating pENTR5105935. To create the fusion protein with the FLAG tag at the C-terminus, a ZCD2 cDNA without a stop codon was generated by PCR using the primers pENTR3C-F2 (tggcttttttgcgtttctac) and pogo3′no stop XhoR (gccgctcgaggctacttctttcttcttcagtattagtg), with use of pENTR5105935 as a template, and was cloned into the EcoRV site of pBluescript SK (Stratagene). The integrity of the clone was checked by sequencing, and then it was subcloned into the vector pENTR3C by use of EcoRI/XhoI sites.
The ZCD2 cDNA and the ZCD2 no-stop cDNA in the pENTR3C vectors were cloned into the N-terminus FLAG-tag pDEST26 vector and the C-terminus FLAG-tag pDEST26 vector, respectively, via the lamda-recombination reaction by use of the Gateway LR Clonase II kit (Invitrogen). The sequences of the expression constructs were confirmed by sequencing. pN-FLAG ZCD2 or pC-FLAG ZCD2 was transfected into either P19 or HEK293 cells by use of Lipofectamine 2000 (Invitrogen) in accordance with the manufacturer’s instructions.
Confocal Immunofluorescence Microscopy
HEK293 and P19 cells were grown on 4-well chambered slides (Lab-Tek) overnight. The cells were transfected with either pN-FLAG ZCD2 or pC-FLAG ZCD2 and were incubated overnight. Cells were fixed in 4% paraformaldehyde in PBS for 20 min at room temperature. After washing once with PBS, cells were permeabilized with 0.2% Triton X-100 in PBS for 10 min at room temperature. Then, cells were blocked with 1% BSA in PBS for 30 min at room temperature. Cells were then incubated with either anti-calnexin antibody (Santa Cruz), anti-FLAG M2 monoclonal antibody (SIGMA), or both at a concentration of 1 μg/250 μl in 1% BSA in PBS for 1 h at room temperature. After washing twice in 1% BSA in PBS, cells were incubated with either Alexa Fluor 488 anti-mouse (Invitrogen), Alexa Fluor 568 anti-rabbit (Invitrogen), or both at a dilution of 1:250 in 5% BSA in PBS for 30 min at room temperature. Cells were washed three times in PBS and were equilibrated in the pH Equilibration solution from SlowFade Antifade Kit with 4′,6-diamidino-2-phenylindole (DAPI) (Invitrogen) for 10 min. DAPI mounting medium with SlowFade was added to the cells. Confocal microscopy images were obtained using a Leica TCS SP2 AOBS confocal microscope with LCS software.
Western-Blot Analysis and Immunoprecipitation
Transfected HEK293 cells and P19 cell pellets were harvested and then were lysed with radioimmunoprecipitation (RIPA) buffer (50 mM Tris-HCl [pH 7.4], 10 mM NaCl2, 3 mM MgCl2, 1% NP-40, and 0.25% Na-deoxycholate) with 40 μl/ml protease inhibitor cocktail (Sigma). The lysed cells were pelleted, and SDS loading buffer was added in a 1:1 ratio. The lysate was denatured at 55°C for 30 min before loading onto 10% polyacrylamide gels and then was electroblotted onto a polyvinylidene fluoride membrane (BioRad). Primary antibodies were added at a dilution of 1:1,000 and 1:2,000 for mouse anti-FLAG (SIGMA) and rabbit anti-WFS1 (NOVUS), respectively. The appropriate secondary antibody was then added at a dilution of 1:10,000 for the anti-mouse antibody and 1:20,000 for the anti-rabbit antibody. Visualization of the immunoreactive proteins was performed using Western Lightning chemiluminescence reagent plus (PerkinElmer).
For immunoprecipitation, 5 μg/ml of primary antibody was added to the whole-cell lysate and was left on ice for 2 h, with periodic agitation. Then, 50 μl of prewashed protein A sepharose beads (Amersham) was added to the whole-cell lysate and was incubated overnight at 4°C with agitation. Beads were pelleted, and the supernatant was discarded. The beads were then washed with RIPA buffer twice, were resuspended in 18 μl of 2× SDS loading buffer, and were denatured at 55°C for 30 min. The proteins were then analyzed by western blot.
Ca2+ Studies
Lymphoblastoid cells at a concentration of 1×106 cells/ml were plated on coverslips treated with 0.1% gelatin for 48 h before measurement of Ca2+. Ca2+ measurements and calculations were performed as described elsewhere19 on single lymphoblastoid cells. In brief, intracellular Ca2+ ([Ca2+]i) was measured using a Ca2+-sensitive dye, fura-2-AM (Molecular Probes). Ca2+ values were determined from the fluorescence ratio of Ca2+-bound fura-2 (excited at 340 nm) to unbound fura-2 (excited at 380 nm) in blinded tests. For basal Ca2+ measurements, 30–37 cells were measured. A subset of cells (n=8) was treated with 2 μM thapsigargin (TG). The mean SE was calculated using unpaired two-tailed t tests with 95% CIs.
Results
Clinical Update
Family WS-2
Individual V:2 in the original pedigree,12 who was affected, committed suicide ∼5 years before this study, after battling with depression for a long time. Individual V:9 in the original pedigree12 developed high-frequency sensorineural hearing loss, although it is not as severe as that in his siblings.
Family WS-3
Individual II:3 died ∼7 years before this study (after the publication of the original linkage article12) from complications of dialysis. He developed renal failure and bleeding during dialysis. Individual II:9 died 5 years before this study, and the cause of death is unknown.
Family WS-4
All affected family members now have diabetes mellitus, optic atrophy, and high-frequency sensorineural hearing loss. The diabetes mellitus is mild and easily corrected with low doses of insulin. The hearing loss is currently symptomatic in all four affected individuals and not only during hearing tests.
Mutation Detection
The critical region for the WFS2 gene, between markers D4S591 and D4S3240, is a large region representing 7.1 cM. To narrow the critical gene region, additional markers within the region were genotyped in the families. These include markers D4S2961, D4S3043, D4S1572, Wpoly2, D4S2913, D4S411, and D4S1531 at the proximal end and D4S2917, Wpoly5, Wpoly6, and Wpoly7 at the distal end. The Wpoly markers are new microsatellite polymorphisms identified by Sputnik, a program that scans genomic DNA, identifies microsatellite stretches within the DNA, and evaluates their polymorphic potential. These markers were either uninformative in the families or did not cross the recombination breakpoint; thus, the critical gene region could not be reduced (data not shown).
The critical gene interval contains 62 transcripts, including known genes, full-length cDNAs, ESTs, and predicted proteins. To reduce the gene list, predicted proteins without a start codon and stop codon were not studied. In addition, homology searches of the translated sequences by use of BLAST were also performed, which eliminated additional putative genes. This left 42 transcripts in the region to be studied, including an excellent candidate gene, BANK1, known to induce Ca2+ mobilization in B cells.20
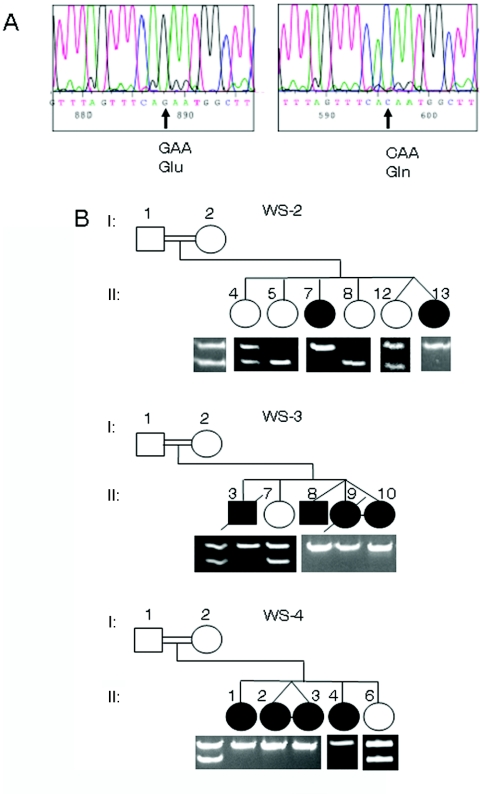
All the genes in the region were screened either by direct sequencing or by SSCP analysis, starting with known genes and full-length cDNAs. Various polymorphisms were identified, but only one nucleotide change in a novel predicted gene, ZCD2, was found to be a WFS2 mutation in these families. A G→C transversion in nucleotide 109 is predicted to change amino acid 37 from glutamic acid to glutamine (E37Q) (fig. 1A). This same mutation is found in all three families. The families are not known to be related, but haplotype analysis had revealed a common haplotype in the linked region,12 which would indicate a common ancestor for this mutation. This change disrupts an XmnI restriction-enzyme site that was used to genotype the family members. The mutation completely segregates with the phenotype in all three Jordanian families (fig. 1B). The mothers from all three pedigrees are shown to be obligate carriers, and the same is assumed for the fathers, although no samples were ever obtained from those individuals. Of the 20 original samples obtained from the children in the three families, only 16 were still currently available to test. The original blood spots of the four individuals not tested did not yield any appreciable DNA, and new samples were not able to be obtained.
Figure 1. .
Identification of a single base-pair change causative of WFS2. A, Sequencing results from a control individual (left panel) with a G at nucleotide 109 in exon 2 of the ZCD2 gene, which codes for glutamic acid, and sequence from an affected individual (right panel) with a homozygous C at the same position, which changes the amino acid to glutamine. B, Truncated pedigrees of WFS2-affected families. Genotyping of the mutation is shown below the pedigrees. All pedigrees represent consanguineous matings. Only children with DNA samples are shown. Children are numbered using designations from the original published pedigree.12 The mutation disrupts an XmnI restriction-enzyme site (GAANNNNTTC), and homozygous affected individuals show a 249-bp fragment, whereas the wild-type allele is cut into a 210-bp fragment and a 39-bp fragment (not shown).
The amplified region containing the mutation was sequenced in at least one affected individual in each family, and all showed the 109G→C mutation. This change is not found in the SNP database (dbSNP). Custom TaqMan SNP genotyping for the mutation was used to screen for this change in a large number of control individuals from various populations. Only one chromosome was identified to have the change in the ethnically matched control population. One individual from the affected Jordanian families is part of the Jordanian control panel. Since the panel is anonymous, it is unknown which individual from the families is included in the control panel. The allele frequency of this change is 0.08% in the Jordanian population, and it was not found in other populations after assessment of 2,004 chromosomes. Since this is an autosomal recessive condition, a very small portion of the population may carry this change in a heterozygous state, although, in our panel, the single sample carrying the change could be a carrier from the families studied.
Protein Analysis
The ZCD2 gene codes for a very small, 135-aa protein, with a molecular weight estimated to be 15,278 Da (fig. 2). It has a predicted transmembrane domain and a single zinc-finger domain of the CDGSH type, located close to the carboxy terminus. This zinc finger is similar to domains in the protein encoded by another human gene (ZCD1) and to those in proteins in many single-cell organisms, all of unknown function. Protein homologues from dog, mouse, rat, chicken, two frog species, two fish species, and Drosophila melanogaster have been identified. The protein is highly conserved and has 70% identity and 82% similarity across all the vertebrate species (fig. 2). The human and Drosophila proteins are 46% identical and 68% similar. Several serine and/or threonine and one tyrosine phosphorylation sites were identified in the human protein, although only four serine and/or threonine sites are conserved in all species, including Drosophila (fig. 2). The protein has a single β-adrenergic receptor kinase site at amino acids 25–29 ([E/D][S/T]XXX),21 two PKA/PKC kinase motifs at amino acids 32–34 and 67–69 ([R/K]X[S/T]),22 and a single CK2 (formerly, casein kinase 2) phosphorylation site ([S/T]XX[D/E])23 at amino acids 34–37, which encompasses the mutation in the last position.
Figure 2. .
Comparison of amino acid sequence of ERIS in nine species. The human sequence is shown, and identical amino acids in other species are represented with a dash. An asterisk (*) represents a missing amino acid in that species. The bold amino acids signify predicted conserved phosphorylation sites (β-adrenergic receptor kinase substrate at 25–29; PKA/PKC kinase substrate motif at 32–34 and 67–69; and CK2 substrate motif at 34–37). The glutamic acid that is mutated in ERIS in the Jordanian families is indicated by an arrow. The underlined amino acids represent a putative transmembrane domain. The double-underlined amino acids highlight the zinc-finger domain of the protein. The unblackened arrowheads indicate the region corresponding to splice junctions in the cDNA.
Hearing-Loss Mutation Screen
Although WFS is a recessive disorder, dominant mutations in the WFS1 gene have been associated with nonsyndromic hearing loss and optic atrophy with hearing impairment.9–11 Six families with WFS1 mutations have low-frequency sensorineural hearing loss,9 and one additional family has progressive hearing loss, with mild hearing loss in the low-frequency range but advanced-to-moderate hearing loss across all frequencies.10 Hearing loss in WFS mostly involves high frequencies, which is also observed in WFS2-affected families.3,12 To determine whether ZCD2 is involved in deafness in a general hearing-loss–affected population, we screened 377 probands for mutations in ZCD2. No mutations were identified in this cohort, so it seems that ZCD2 is not commonly involved in nonsyndromic deafness. Only one proband had low-frequency sensorineural hearing loss, so it remains to be determined whether ZCD2 is involved in this subset of hearing loss.
Expression Studies
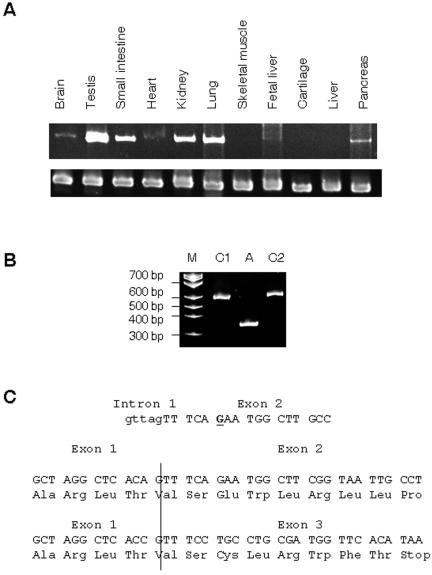
By use of RT-PCR on RNA from a variety of human tissues, expression was observed in the majority of tissues, including brain and pancreas. Expression was not observed in cartilage, fetal liver, and skeletal muscle (fig. 3A).
Figure 3. .
ZCD2 expression and WFS2 disease pathogenesis. A, RT-PCR of the WFS2 gene, ZCD2, in a variety of tissues (top row) and the GADPH control (bottom row). B, RT-PCR of lymphoblastoid RNA from two control individuals (C1 and C2) and an affected individual (A), by use of human ZCD2 cDNA primers. The primers amplify the full-length cDNA; the large band is 551 bp, and the smaller band is 336 bp. Lane M is the molecular marker. C, Genomic sequence of the intron-exon boundary shows that the mutation (bold and underlined) is within 6 bp of the 3′ splice-acceptor site. Below, cDNA and amino acid sequences are shown at the boundary between exon 1 and exon 2, compared with the premature stop codon that occurs when exon 1 and exon 3 are spliced together in an affected individual.
Disease Pathogenesis
The mutated glutamic acid is conserved in all species except mouse, in which there is an aspartic acid residue in its place (fig. 2). The replacement of the glutamic acid residue at position 37 with aspartic acid does not disrupt the CK2 phosphorylation site. This site is also found in Drosophila, although the sequence is divergent (SFKD). The mutation changes the conserved glutamic acid from the single conserved CK2 site, which would abolish phosphorylation at this site. It would be logical to conclude that the disruption of a functional domain would be indicative of disease pathogenesis, but, in actuality, the mutation also creates a splice-site mutation.
The mature cDNA is derived from three exons. The mutated base is located close to the 5′ end of exon 2, 6 bases from the intron-exon junction. Performance of RT-PCR with primers that amplify the full-length cDNA on templates derived from lymphoblastoid RNA from an affected individual and two controls showed a 336-bp product in the affected individual and a 551-bp product of expected size in controls (fig. 3B). Sequencing the RT-PCR products showed that the 336-bp product is the result of exon 2 being skipped in the final transcript (fig. 3C). This smaller RT-PCR product is not observed in RNA from other tissue types and is likely not a splice variant (data not shown).
The aberrant splice product would code for the 34 aa from exon 1, but the frameshifts in exon 3 would create a premature stop codon after 8 additional aa. This would eliminate 75% of the protein, including the transmembrane and zinc-finger domains. This particular nucleotide change does not reside in any known splice-acceptor or -donor regions; thus, it is likely that this change disrupts an exonic splice enhancer (ESE).
ERIS Cellular Localization
To determine the cellular localization of ZCD2, the human gene was tagged with FLAG on the amino (N-) or carboxy (C-) terminus and was expressed in mouse P19 and human HEK293 cells. N-FLAG protein is localized to the ER, colocalizing with calnexin, a known ER marker24 (fig. 4A). Because of the protein’s predicted domains and cellular localization, we have named the ZCD2-encoded protein “ERIS” (endoplasmic reticulum intermembrane small protein).
Figure 4. .
Localization and immunoprecipitation of ERIS. A, P19 cells transfected with pN-FLAG ZCD2. In panel 1, the transfected cells were stained with DAPI. Panel 2 shows the ER marker calnexin (red). Panel 3 shows pN-FLAG ZCD2 (green). Only a proportion of the cells that were transfected showed pN-FLAG ZCD2 expression. Panel 4 shows the merged picture, with pN-FLAG ZCD2 colocalizing with calnexin in the ER. B, Cell lysates from HEK293 cells transfected with pC-FLAG ZCD2, immunoprecipitated using anti-WFS1 antibodies (lane 1) and anti-FLAG antibodies (lane 2). The top half of the blot was probed with anti-WFS1 antibodies, and the bottom half with anti-FLAG antibodies. Lane 1 shows a strong Wolframin band at 100 kDa but does not show pC-FLAG ZCD2 (at 17.5 kDa). pC-FLAG ZCD2 is observed in lane 2, with no Wolframin. The 19-kDa band represents immunoglobulin G, whereas other bands represent nonspecific binding of the antibodies. The same results were observed when the experiment was performed with pN-FLAG ZCD2 (data not shown).
No Interaction of ERIS with Wolframin
To determine whether ERIS interacts directly with the WFS1-encoded protein, Wolframin, coimmunoprecipitation studies were performed in cell lines expressing N-FLAG and C-FLAG ERIS. Wolframin did not coprecipitate with ERIS (fig. 4B).
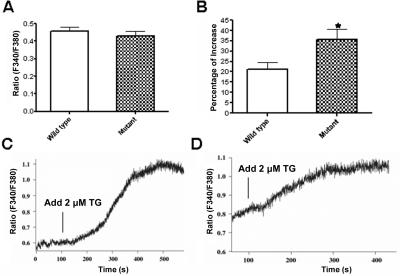
Ca2+ Measurements
Resting [Ca2+]i levels were not significantly different in a lymphoblastoid cell line derived from an affected individual compared with those in a cell line from an unaffected control (fig. 5A). When stimulated by TG, a known stimulator of ER Ca2+ release, there was significantly more intracellular Ca2+ release in the affected cell line than in the unaffected cell line (fig. 4B–4D).
Figure 5. .
Intracellular Ca2+ measurements. A, No difference in the mean basal [Ca2+]i levels observed between wild-type and mutant cells (n=30–37; P=.49). B, The mean [Ca2+]i increase, significantly greater in mutant cells than in wild-type cells when stimulated with 2 mM TG (n=8; the asterisk [*] indicates significance [P=.03]). Also shown are representative single-cell measurements of [Ca2+]i changes in mutant (C) and wild-type (D) cells when stimulated by TG.
Discussion
The majority of WFS cases seem to be caused by mutations in the WFS1 gene, but, in many studies, there has been evidence of genetic heterogeneity.12,25–27 We have shown that a mutation in a zinc-finger gene, ZCD2, also causes WFS. The same missense mutation was identified in all three unrelated, consanguineous Jordanian families studied. Through haplotype analysis, it is clear that these families share a common ancestor.12 Although the missense mutation occurs at a conserved amino acid, the actual pathogenesis of the nucleotide change has been shown to affect mRNA splicing. The mutation causes exon 2 to be skipped, which results in a frameshift leading to a premature stop codon, resulting in >75% of the protein to be missing, including a putative transmembrane domain and the zinc-finger domain. Although the mutation functionally affects splicing, the 10 bp surrounding the mutation site does not demonstrate any consensus for ESEs. Some of the best-defined ESEs share one of two features. One class of ESEs shows high purine content (⩾80%), although specific sequences within that region are known to be important; the second class of ESEs is A/C rich.28 ZCD2 encodes 60% purines in the sequence adjacent to the mutation, and the region is not particularly AC rich. In fact, the mutation replaces a G with a C. It is well known that a large number of sequences can function as ESEs but do not conform to any specific sequence. We have identified a functional ESE in ZCD2 that does not conform to a known class of ESEs.
We did not identify mutations in ZCD2 in a large cohort of 377 individuals from multiplex families with nonsyndromic deafness. Dominant mutations in the WFS1 gene have been reported in DFNA6/14-affected families with low-frequency sensorineural hearing loss or progressive hearing loss.9,10 Of the 377 probands, 240 had autosomal recessive inheritance, 84 had dominant hearing loss, and 53 were heterozygous for a single change in the GJB2 gene. Although the majority had severe-to-profound hearing loss, 14 had high-frequency hearing loss but only 1 had low-frequency hearing loss. ZCD2 does not seem to be generally associated with nonsyndromic hearing loss, but it remains to be determined whether it is specifically associated with low-frequency hearing loss, similar to the WFS1 gene.
ZCD2 has a fairly wide expression profile, including the pancreas and brain, two tissues affected by the disorder. Since the transcript could be amplified from RNA derived from lymphoblastoid cells, it may also be expressed in blood cell lineages. Expression studies using mass spectrometry listed in the Human Protein Reference Database (accession number 17413) show the presence of ZCD2 transcripts in platelets.17 This would correlate with the bleeding phenotype described in these families.
Although we did not perform in situ hybridization on tissue sections, these experiments were performed on sagittal sections with the mouse Zcd2 transcript (accession number 1500009M05Rik) as part of the Allen Brain Atlas project.29 Expression of the gene in adult mouse brain is observed in a wide variety of structures, including the cerebral cortex, piriform area, pyramidal cells of the hippocampus, the central amygdalar nucleus, the medulla, pons, thalamus, inferior and superior colliculus of the brainstem, and the Purkinje cell layer and dentate nucleus of the cerebellum. Interestingly, there is substantial overlap of expression of Zcd2 in mice and Wfs1 in rat brain.7 These brain regions primarily control memory, emotions, and motor skills. Affected individuals with WFS are known to have neurological complications, such as cerebellar ataxia and myoclonus, as well as psychiatric illness.4
Magnetic resonance imaging studies of individuals with WFS show degeneration of the brainstem, cerebellum, optic nerve, tracts, and chiasm.30 A small number of WFS-affected brains have been studied postmortem. These studies show the degeneration of specific brainstem structures and also show a much wider pathology.31,32 Neural degeneration of many other areas where Zcd2 is expressed is also observed. These include the superior and inferior colliculi and the thalamus (brainstem), as well as the pontine nuclei, Purkinje cells, and pons. Loss of the cochlear nerves and mild loss of cochlear nuclei has also been observed. In general, it appears that neurodegeneration in WFS is correlated with the spatial expression of ZCD2. Because the loss of sensory neurons, pancreatic β cells, or absence or hypoplasia of the hypothalamus could explain the main clinical symptoms, ZCD2 expression in these tissues needs to be examined further.
ERIS is an extremely small but well-conserved protein. Indirect immunofluorescence studies show that the protein is localized to the ER. ERIS does not have a predicted ER signal sequence at the amino terminus but has a single transmembrane domain. Transmembrane proteins have been shown to use their first transmembrane domain to be directed into the ER.33 The other major predicted domain is a zinc-finger domain of the CDGSH type with a CCHH finger. Zinc-finger domains are now known to be able to bind nucleic acids, lipids, and proteins.34 Wolframin coimmunoprecipitation studies performed in cell lines expressing N- or C-FLAG ERIS revealed that Wolframin does not coprecipitate or interact with ERIS.
Wolframin has been well studied and has been shown to localize to the ER and play a role in modulating [Ca2+]i. Although [Ca2+]i levels between WFS2-affected and control lymphoblastoid cells were not significantly different, a significantly greater increase in [Ca2+]i in affected cells was observed when they were stimulated with TG. TG is a known inhibitor of the sarco(endo)plasmic reticulum Ca2+ATPase, which results in ER Ca2+ efflux.35 This is consistent with a role for ERIS in [Ca2+]i homeostasis in cells expressing the gene. Recently, it has been shown that knockdown of WFS1 in HEK293 cells showed lower increases in Ca2+ when TG was added to the media, suggesting lower levels of Ca2+ in the ER ([Ca2+]ER) in cells without Wolframin.36 Measuring the rate at which Ca2+ moves back into the ER showed that this rate was decreased, indicating a role for Wolframin in ER Ca2+ uptake.36 Additional studies need to be undertaken to determine whether greater TG-stimulated Ca2+ release in WFS2-affected cells is because of higher [Ca2+]ER.
The symptoms of this disorder are caused by degeneration of neurons and pancreatic β cells. [Ca2+]ER is important not only for signaling but also for the folding and processing of newly synthesized proteins, which have shown to be Ca2+-dependent reactions.37,38 Cell lines and animals with WFS1 insufficiency have shown ER stress, which triggers the unfolded protein response (UPR), impairs cell-cycle progression, and increases apoptosis in pancreatic β cells.39,40 Wolcott-Rallison syndrome (MIM 226980), a rare autosomal recessive disorder that has infancy-onset diabetes as one of its symptoms, is caused by mutations in EIF2AK3, encoding translation initiation factor 2-alpha kinase 3, a kinase important in the UPR.41 Folding and processing of newly synthesized proteins require high Ca2+ levels; thus, higher-than-normal [Ca2+]ER may not impair protein folding and trigger the UPR.
High [Ca2+]ER has been shown to have a number of different cellular effects. It can increase the Ca2+ release rate constant in cardiomyocytes42,43; can affect the sensitivity of the ryanodine receptor, a major ER Ca2+ release channel44,45; and can inhibit Ca2+ uptake and affect the velocity of uptake in mouse pancreatic acinar cells and sensory neurons from dorsal root ganglia of rats.46,47 Interestingly, knock-in of presenilin-1 Alzheimer mutations in mice shows increased [Ca2+]ER.48 In hippocampal neurons from these mice, glutamate-induced oxidative stress and mitochondrial dysfunction is enhanced, resulting in a lower threshold for excitotoxic neuronal necrotic cell death.49 The mitochondria are also an important component in Ca2+ signaling, since they can rapidly sequester Ca2+ after an initial Ca2+ pulse and slowly release it during the recovery phase, through a permeability transition pore (PTP).50,51 Overloading the ER with Ca2+ enhances the sensitivity of ceramide-induced apoptosis, through the overload of Ca2+ in the mitochondria, and the release of cytochrome c, through the formation of the PTP.52,53 Disturbing Ca2+ homeostasis seems to predispose cells to multiple forms of cell death.
WFS has now been shown to be caused by at least two different genes, WFS1 and ZCD2. They both encode ER proteins and seem to affect cellular Ca2+ homeostasis. WFS1 has also been implicated in nonsyndromic hearing loss and optic atrophy. ZCD2 could be important in isolated cases of these disorders. It remains to be determined whether the two genes reside in the same pathway and what specific mechanism each gene uses that leads to nerve and pancreatic degeneration.
Acknowledgments
We acknowledge the technical assistance of Dr. Lin Li and Dr. B. Todd Webb. We are thankful for the families who participated in this study. We are grateful to the staff of the National Center for Diabetes, Endocrinology, and Genetics, for facilitating the collection of samples and cell lines. Confocal microscopy was performed at the Virginia Commonwealth University Department of Neurobiology and Anatomy Microscope Facility, supported in part by National Institutes of Health (NIH)–National Institute of Neurological Disorders and Stroke Center core grant 5P3ONSO47463. K.A. and H.E.-S. were funded by the National Center for Diabetes, Endocrinology, and Genetics and by the Higher Council for Science and Technology; L.S.S. and M.Z. were supported by NIH grant RO1 DK46409RS; and C.H. and R.S. were supported by American Diabetes Association grant 1-03-RA-80.
Web Resources
The accession number and URLs for data presented herein are as follows:
- Allen Brain Atlas, http://www.brain-map.org/welcome.do
- BLAST: tBLASTn, http://www.ncbi.nlm.nih.gov/BLAST/Blast.cgi?PAGE=Translations&PROGRAM=tblastn&BLAST_PROGRAMS=tblastn&PAGE_TYPE=BlastSearch&SHOW_DEFAULTS=on
- dbSNP, http://www.ncbi.nlm.nih.gov/SNP/
- Human Protein Reference Database, http://www.hprd.org/ (for accession number 17413)
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for WFS, WFS2, and Wolcott-Rallison syndrome)
- Prosite, http://www.expasy.org/prosite/
- Sputnik, http://espressosoftware.com/pages/sputnik.jsp
References
- 1.Wolfram DJ, Wagener HP (1938) Diabetes mellitus and simple optic atrophy among siblings: report of four cases. Mayo Clin Proc 13:715–718 [Google Scholar]
- 2.Blasi C, Pierelli F, Rispoli E, Saponara M, Vingolo E, Andreani D (1986) Wolfram’s syndrome: a clinical, diagnostic, and interpretative contribution. Diabetes Care 9:521–528 10.2337/diacare.9.5.521 [DOI] [PubMed] [Google Scholar]
- 3.Cremers CWRJ, Wijdeveld PGAB, Pinckers AJLG (1977) Juvenile diabetes mellitus, optic atrophy, hearing loss, diabetes insipidus, atonia of the urinary tract and bladder, and other abnormalities (Wolfram syndrome): a review of 88 cases from the literature with personal observations on 3 new patients. Acta Paediatr Scand Suppl 264:1–16 [DOI] [PubMed] [Google Scholar]
- 4.Minton JAL, Rainbow LA, Ricketts C, Barrett TG (2003) Wolfram syndrome. Rev Endocr Metab Disord 4:53–59 10.1023/A:1021875403463 [DOI] [PubMed] [Google Scholar]
- 5.Inoue H, Tanizawa Y, Wasson J, Behn P, Kalidas K, Bernal-Mizrachi B, Mueckler M, Marshall H, Donis-Keller H, Crock P, et al (1998) A gene encoding a transmembrane protein is mutated in patients with diabetes mellitus and optic atrophy (Wolfram syndrome). Nat Genet 20:143–148 10.1038/2441 [DOI] [PubMed] [Google Scholar]
- 6.Strom TM, Hörtnagel K, Hofmann S, Gekeler F, Scharfe C, Rabl W, Gerbitz KD, Meitinger T (1998) Diabetes insipidus, diabetes mellitus, optic atrophy and deafness (DIDMOAD) caused by mutations in a novel gene (wolframin) coding for a predicted transmembrane protein. Hum Mol Genet 7:2021–2028 10.1093/hmg/7.13.2021 [DOI] [PubMed] [Google Scholar]
- 7.Takeda K, Hiroshi I, Tanizawa Y, Matsuzaki Y, Oba J, Watanabe Y, Shinoda K, Oka Y (2001) WFS1 (Wolfram syndrome 1) gene product: predominant subcellular localization to endoplasmic reticulum in cultured cells and neuronal expression in rat brain. Hum Mol Genet 10:477–484 10.1093/hmg/10.5.477 [DOI] [PubMed] [Google Scholar]
- 8.Osman AA, Saito M, Makepeace C, Permutt MA, Schlesinger P, Mueckler M (2003) Wolframin expression induces novel ion channel activity in endoplasmic reticulum membranes and increases intracellular calcium. J Biol Chem 278:52755–52762 10.1074/jbc.M310331200 [DOI] [PubMed] [Google Scholar]
- 9.Bespalova IN, Van Camp G, Bom SJH, Brown DJ, Cryns K, DeWan AT, Erson AE, Flothmann K, Kunst HPM, Kurnool P, et al (2001) Mutations in the Wolfram syndrome 1 gene (WFS1) are a common cause of low frequency sensorineural hearing loss. Hum Mol Genet 10:2501–2508 10.1093/hmg/10.22.2501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Young T-L, Ives E, Lynch E, Person R, Snook S, MacLaren L, Cator T, Griffin A, Fernanadez B, Lee MK, et al (2001) Non-syndromic progressive hearing loss DFNA38 is caused by heterozygous missense mutation in the Wolfram syndrome gene WFS1. Hum Mol Genet 10:2509–2514 10.1093/hmg/10.22.2509 [DOI] [PubMed] [Google Scholar]
- 11.Eiberg H, Hansen L, Kjer B, Hansen T, Pederson O, Bille M, Rosenberg T, Tranebjaerg L (2006) Autosomal dominant optic atrophy associated with hearing impairment and impaired glucose regulation caused by a missense mutation in the WFS1 gene. J Med Genet 43:435–440 10.1136/jmg.2005.034892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.El-Shanti H, Lidral AC, Jarrah N, Druhan L, Ajlouni K (2000) Homozygosity mapping identifies an additional locus for Wolfram syndrome on chromosome 4q. Am J Hum Genet 66:1229–1236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Al-Sheyyab M, Jarrah N, Younis E, Shennak MM, Hadidi A, Awidi A, El-Shanti H, Ajlouni K (2001) Bleeding tendency in Wolfram syndrome: a newly identified feature with phenotype genotype correlation. Eur J Pediatr 160:243–246 10.1007/s004310000704 [DOI] [PubMed] [Google Scholar]
- 14.Ajlouni K, Jarrah N, El-Khateeb M, El-Zaheri M, El Shanti H, Lidral A (2002) Wolfram syndrome: identification of a phenotypic and genotypic variant from Jordan. Am J Med Genet 115:61–65 10.1002/ajmg.10345 [DOI] [PubMed] [Google Scholar]
- 15.Cann HM, de Toma C, Cazes L, Legrand MF, Morel V, Piouffre L, Bodmer J, Bodmer WF, Bonne-Tamir B, Cambon-Thomsen A, et al (2002) A human genome diversity cell line panel. Science 296:261–262 10.1126/science.296.5566.261b [DOI] [PubMed] [Google Scholar]
- 16.Hulo N, Bairoch A, Bulliard V, Cerutti L, De Castro E, Langendijk-Genevaux PS, Pagni M, Sigrist CJA (2006) The PROSITE database. Nucleic Acids Res 34:D227-D230 10.1093/nar/gkj063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Peri S, Navarro JD, Amanchy R, Kristiansen TZ, Jonnalagadda CK, Surendranath V, Niranjan V, Muthusamy B, Gandhi TK, Gronborg M, et al (2003) Development of Human Protein Reference Database as an initial platform for approaching systems biology in humans. Genome Res 13:2363–2371 10.1101/gr.1680803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410 [DOI] [PubMed] [Google Scholar]
- 19.Zhang M, Goforth P, Bertram R, Sherman A, Satin L (2003) The Ca2+ dynamics of isolated mouse β-cells and islets: implications for mathematical models. Biophys J 84:2852–2870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yokoyama K, Su IH, Tezuka T, Yasuda T, Mikoshiba K, Tarakhovsky A, Yamamoto T (2002) BANK regulates BCR-induced calcium mobilization by promoting tyrosine phosphorylation of IP3 receptor. EMBO J 21:83–92 10.1093/emboj/21.1.83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Onorato JJ, Palczewski K, Regan JW, Caron MG, Lefkowitz RJ, Benovic JL (1991) Role of acidic amino acids in peptide substrates of the beta-adrenergic receptor kinase and rhodopsin kinase. Biochemistry 30:5118–5125 10.1021/bi00235a002 [DOI] [PubMed] [Google Scholar]
- 22.Pearson RB, Kemp BE (1991) Protein kinase phosphorylation site sequences and consensus specificity motifs: tabulations. Methods Enzymol 200:62–81 [DOI] [PubMed] [Google Scholar]
- 23.Kuenzel EA, Mulligan JA, Sommercorn J, Krebs EG (1987) Substrate specificity determinants for casein kinase II as deduced from studies with synthetic peptides. J Biol Chem 262:9136–9140 [PubMed] [Google Scholar]
- 24.David V, Hochstenbach F, Rajagopalan S, Brenner MB (1993) Interaction with newly synthesized and retained proteins in the endoplasmic reticulum suggests a chaperone function for human integral membrane protein IP90 (calnexin). J Biol Chem 268:9585–9592 [PubMed] [Google Scholar]
- 25.Collier DA, Barrett TG, Curtis D, Macleod A, Arranz MJ, Maassen JA, Bundey S (1996) Linkage of Wolfram syndrome to chromosome 4p16.1 and evidence for heterogeneity. Am J Hum Genet 59:855–863 [PMC free article] [PubMed] [Google Scholar]
- 26.Gómez-Zaera M, Strom TM, Rodríguez Estivill X, Meitinger T, Nunes V (2001) Presence of a major WFS1 mutation in Spanish Wolfram syndrome pedigrees. Mol Genet Metab 72:72–81 10.1006/mgme.2000.3107 [DOI] [PubMed] [Google Scholar]
- 27.Khanim F, Kirk J, Latif F, Barrett TG (2001) WFS1/Wolframin mutations, Wolfram syndrome and associated diseases. Hum Mutat 17:357–367 10.1002/humu.1110 [DOI] [PubMed] [Google Scholar]
- 28.Cooper TA, Mattox W (1997) The regulation of splice-site selection and its role in human disease. Am J Hum Genet 61:259–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lein ES, Hawrylycz MJ, Ao N, Ayres M, Bensinger A, Bernard A, Boe AF, Boguski MS, Brockway KS, Byrnes EJ, et al (2007) Genome-wide atlas of gene expression in the adult mouse brain. Nature 445:168–176 10.1038/nature05453 [DOI] [PubMed] [Google Scholar]
- 30.Barrett TG, Bundey SE, Fielder AR, Good PA (1997) Optic atrophy in Wolfram (DIDMOAD) syndrome. Eye 11:882–888 [DOI] [PubMed] [Google Scholar]
- 31.Carson MJ, Slager UT, Steinberg RM (1977) Simultaneous occurrence of diabetes mellitus, diabetes insipidus, and optic atrophy in a brother and sister. Am J Dis Child 131:1382–1385 [DOI] [PubMed] [Google Scholar]
- 32.Genís D, Dávalos A, Molins A, Ferrer I (1997) Wolfram syndrome: a neuropathological study. Acta Neuropathol 93:426–429 10.1007/s004010050635 [DOI] [PubMed] [Google Scholar]
- 33.Kanner EM, Klein IK, Friedlander M, Simon SM (2002) The amino terminus of opsin translocates “posttranslationally” as efficiently as cotranslationally. Biochemistry 41:7707–7715 10.1021/bi0256882 [DOI] [PubMed] [Google Scholar]
- 34.Mackay JP, Crossley M (1998) Zinc fingers are sticking together. Trends Biochem Sci 23:1–4 10.1016/S0968-0004(97)01168-7 [DOI] [PubMed] [Google Scholar]
- 35.Lytton J, Westlin M, Hanley MR (1991) Thapsigargin inhibits the sarcoplasmic or endoplasmic reticulum Ca-ATPase family of calcium pumps. J Biol Chem 266:17067–17071 [PubMed] [Google Scholar]
- 36.Takei D, Ishihara H, Yamaguchi S, Yamada T, Tamura A, Katagiri H, Maruyama Y, Oka Y (2006) WFS1 protein modulates the free Ca2+ concentration in the endoplasmic reticulum. FEBS Lett 580:5635–5640 10.1016/j.febslet.2006.09.007 [DOI] [PubMed] [Google Scholar]
- 37.Kuznetsov G, Brostrom MA, Brostrom CO (1992) Demonstration of a calcium requirement for secretory protein processing and export: differential effects of calcium and dithiothreitol. J Biol Chem 267:3932–3939 [PubMed] [Google Scholar]
- 38.Lodish HF, Kong N, Wikstrom L (1992) Calcium is required for folding of newly made subunits of the asialoglycoprotein receptor within the endoplasmic reticulum. J Biol Chem 267:12753–12760 [PubMed] [Google Scholar]
- 39.Fonseca SG, Fukuma M, Lipson KL, Nguyen LX, Allen JR, Oka Y, Urano F (2005) WFS1 is a novel component of the unfolded protein response and maintains homeostasis of the endoplasmic reticulum in pancreatic β-cells. J Biol Chem 280:39609–39615 10.1074/jbc.M507426200 [DOI] [PubMed] [Google Scholar]
- 40.Yamada T, Ishihara H, Tamura A, Takahashi R, Yamaguchi S, Takei D, Tokita A, Satake C, Tashiro F, Katagiri H, et al (2006) WFS1-deficiency increases endoplasmic reticulum stress, impairs cell cycle progression and triggers the apoptotic pathway specifically in pancreatic β-cells. Hum Mol Genet 15:1600–1609 10.1093/hmg/ddl081 [DOI] [PubMed] [Google Scholar]
- 41.Delepine M, Nicolino M, Barrett T, Golamaully M, Lathrop GM, Julier C (2000) EIF2AK3, encoding translation initiation factor 2-α kinase 3, is mutated in patients with Wolcott-Rallison syndrome. Nat Genet 25:406–409 10.1038/78085 [DOI] [PubMed] [Google Scholar]
- 42.Han S, Schiefer A, Isenberg G (1994) Ca2+ load of guinea-pig ventricular myocytes determines efficacy of brief Ca2+ currents as trigger for Ca2+ release. J Physiol 480:411–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bassani JW, Yuan W, Bers DM (1995) Fractional SR Ca release is regulated by trigger Ca and SR Ca content in cardiac myocytes. Am J Physiol 268:C1313-C1319 [DOI] [PubMed] [Google Scholar]
- 44.Gyorke I, Gyorke S (1998) Regulation of the cardiac ryanodine receptor channel by luminal Ca2+ involves luminal Ca2+ sensing sites. Biophys J 75:2801–2810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gyorke I, Hester N, Jones LR, Gyorke S (2004) The role of calsequestrin, triadin, and junctin in conferring cardiac ryanodine receptor responsiveness to luminal calcium. Biophys J 86:2121–2128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mogami H, Tepikin AV, Petersen OH (1998) Terminatin of cytosolic Ca2+ signals: Ca2+ reuptake into intracellular stores is regulated by the free Ca2+ concentration in the store lumen. EMBO J 17:435–442 10.1093/emboj/17.2.435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Solovyova N, Veselovsky N, Toescu EC, Verkhratsky A (2002) Ca2+ dynamics in the lumen of the endoplasmic reticulum in sensory neurons: direct visualization of Ca2+-induced Ca2+ release triggered by physiological Ca2+ entry. EMBO J 21:622–630 10.1093/emboj/21.4.622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Leissring MA, Akbari Y, Fanger CM, Cahalan MD, Mattson MP, LaFerla FM (2000) Capacitative calcium entry deficits and elevated luminal calcium content in mutant presenilin-1 knockin mice. J Cell Biol 149:793–797 10.1083/jcb.149.4.793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Guo Q, Fu W, Sopher BL, Miller MW, Ware CB, Martin GM, Mattson MP (1999) Increased vulnerability of hippocampal neurons to excitotoxic necrosis in presenilin-1 mutant knock-in mice. Nat Med 5:101–106 10.1038/4789 [DOI] [PubMed] [Google Scholar]
- 50.Jouaville LS, Ichas F, Holmuhamedor EL, Camacho P, Lechieiter JD (1995) Synchronization of calcium waves by mitochondrial substrates in Xenopus laevis oocytes. Nature 377:438–441 10.1038/377438a0 [DOI] [PubMed] [Google Scholar]
- 51.Collins TJ, Lipp P, Berridge MJ, Li W, Bootman MD (2000) Inositol 1,4,5-triphosphate-induced Ca2+ release is inhibited by mitochondrial depolarization. Biochem J 347:593–600 10.1042/0264-6021:3470593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shimizu S, Manita M, Tsujimoto Y (1999) Bcl-2 family proteins regulate the release of apoptogenic cytochrome c by the mitochondrial channel VDAC. Nature 399:483–487 10.1038/20959 [DOI] [PubMed] [Google Scholar]
- 53.Pinton P, Ferrari D, Papizzi E, Di Virgilio F, Pozzan T, Rizzuto R (2001) The Ca2+ concentration of the endoplasmic reticulum is a key determinant of ceramide-induced apoptosis: significance for the molecular mechanism of Bcl-2 action. EMBO J 20:2690–2701 10.1093/emboj/20.11.2690 [DOI] [PMC free article] [PubMed] [Google Scholar]