Abstract
Germline mutations of PTEN (phosphatase and tensin homolog deleted on chromosome 10) are associated with the multihamartomatous disorder Cowden syndrome (CS). Moreover, patients with CS with germline PTEN promoter mutations have aberrant PTEN protein expression and an increased frequency of breast cancer. Here, we examined the downstream effect of five PTEN promoter variants (−861G/T, −853C/G, −834C/T, −798G/C, and −764G/A) that are not within any known cis-acting regulatory elements. Clinically, all five of these patients have been given diagnoses of breast, thyroid, and/or endometrial cancer. We demonstrated that protein binding to the PTEN promoter (−893 to −755) was not altered in the five variants when compared with the wild-type (WT) promoter. However, reporter assays indicated that three of the variants (−861G/T, −853C/G, and −764G/A) demonstrated an ∼50% decrease in luciferase activity compared with the WT construct. PTEN messenger RNA (mRNA) levels were not altered in these variants, whereas secondary structure predictions indicated that different PTEN 5′ untranslated region transcript-folding patterns exist in three variants, suggesting an inhibition of protein translation. This was confirmed by PTEN protein analysis. These data indicate that variants causing large mRNA secondary structure alterations result in an inhibition of protein translation and a decrease in PTEN protein expression. These data emphasize the importance of PTEN promoter nucleotide variations and their ability to lead to CS progression by a novel regulatory mechanism. Importantly, these patients have a high prevalence of breast, thyroid, and endometrial malignancies; thus, understanding of the mechanism of PTEN dysfunction in these patients will lead to more-sensitive molecular diagnostic and predictive testing and, ultimately, to rational targeted therapies to treat or prevent malignancy.
Germline mutations in PTEN (phosphatase and tensin homolog deleted on chromosome 10), a tumor-suppressor gene on 10q23, occur in 85% of patients with Cowden syndrome (CS [MIM 158350]).1–5 This syndrome affects ∼1 in 200,000 individuals; however, this is generally thought to be an underestimation.6 CS is characterized by hamartomas of multiple organs and increased risks of neoplasia. Patients given diagnoses of CS have a 25%–50% lifetime risk of developing female breast cancer, whereas the risk in the general population is ∼13%.7,8 Patients with CS have an ∼10% lifetime risk of developing thyroid cancer, which tends to be follicular, compared with a <1% risk in the general population. Furthermore, there is an ∼5%–10% lifetime risk of endometrial cancer for patients with CS, compared with ∼2%–4% in the general population.3,7 Proper recognition and diagnosis of CS is crucial not only because of the increased risk of cancers observed in this syndrome but also because of the morbidity, and even mortality, associated with its nonmalignant features.
Germline PTEN mutations are also found in 65% of patients with Bannayan-Riley-Ravalcaba syndrome (BRRS [MIM 153480]), characterized by macrocephaly, lipomatosis, hemangiomatosis, and speckled penis.3,9 In addition to BRRS, a number of other syndromes, including Proteus syndrome (MIM 176920), Proteus-like syndrome,6 and autism,10 share germline PTEN mutations as an etiology and have been classified as PTEN hamartoma tumor syndromes (PHTS). It is recommended that they all be managed in a similar manner to CS if a pathogenic PTEN mutation is identified.2
Before 2003, PTEN mutations had been identified in 80% of patients meeting strict diagnostic criteria for CS.8,11 CS is believed to be monogenic; therefore, determining the cause of PTEN dysfunction in the remaining 20% of patients is vitally important to the practice of personalized genetic health care. Our laboratory began aggressively pursuing alternative mechanisms of PTEN inactivation, including interrogating the mutation status of its own promoter. We have identified mutations in the PTEN promoter in patients with CS and have shown that ∼10% of previously classified PTEN mutation–negative patients have nucleotide variants within the full-length promoter.3 Furthermore, these mutations resulted in both a decrease in PTEN protein expression and loss of function.3 Interestingly, in this initial study, eight (89%) of nine patients with germline PTEN promoter mutations had breast cancer, although the overall number of organs involved was generally low in these patients, suggesting that these mutations may preferentially confer very high penetrance for breast cancer.3
Previous investigations of the PTEN promoter, including our own, have focused on areas within known transcription-factor binding sites. However, we have now identified variants of unknown significance (VUSs) 3′ of any known cis-acting elements. On the basis of our previous work,12–14 we predict that these VUSs might be pathogenic. Given the dilemma faced by the clinical cancer geneticist and genetics counselor when counseling patients with VUSs, an understanding of the molecular consequences of these promoter VUSs has important implications for patient care. In this study, we investigated both transcriptional and translational downstream effects of PTEN promoter VUSs in patients with CS. Our data demonstrate that certain PTEN promoter VUSs in patients with CS result in decreased PTEN expression through dysfunctional translation, rather than through altered transcription.
Material and Methods
Patient Recruitment
Human subjects were recruited from multiple institutions throughout the United States. All samples were acquired with informed consent in accordance with protocol approved by the human subjects protection committees of their respective institutions. Patients with CS used in this study were classified in accordance with both the National Comprehensive Cancer Network and the International Cowden Consortium operational diagnostic criteria, and 186 healthy individuals served as controls. In our entire series, patients are ∼85% white, ∼5% black, and 10% other (including Hispanic, Asian, etc.). For this particular study, both cases and controls were whites of western and northern European origin.
DNA Isolation and Promoter-Mutation Analysis
Unaffected control and patient genomic DNA was isolated by the Genomic Medicine Biorepository of the Cleveland Clinic Genomic Medicine Institute. Primers were designed to amplify the full PTEN promoter between –1389 and –715 (forward 5′-GCGTGGTCACCTGGTCCTTT-3′, reverse 5′-GCTGCTCACAGGCGCTGA-3′), and DNA was PCR amplified in 20-μl reactions with the use of HotStar and Q Solution (Qiagen). The PCR conditions consisted of 30 cycles at the annealing temperature of 55°C. PCR products were treated with exonuclease I (New England Biolabs) and shrimp alkaline phosphatase (USB) and were analyzed on both strands by direct sequencing (ABI 3730xl DNA Analyzer [Genomics Core Facility, Lerner Research Institute]). Variants were detected by direct analysis compared with unaffected control sequence through Lasergene software (DNASTAR).
Cell Culture
The MCF-7 breast cancer and HeLa cell lines were maintained at 37°C with 5% CO2 in Dulbecco's modified Eagle medium containing 10% fetal bovine serum (FBS) and 100 units/ml each of penicillin and streptomycin. Clonal lymphoblastoid cell lines (LBCLs) were generated by the Genomic Medicine Biorepository from patients with CS and unaffected, healthy controls. LBCLs were cultured in RPMI medium supplemented with 20% FBS and 100 units/ml of penicillin and streptomycin.
Protein Isolation
Total protein from LBCLs was harvested using M-PER (Pierce) lysis buffer containing phenylmethanesulphonylfluoride (0.75 mg/ml), benzamidine hydrochloride (0.5 mg/ml), leupeptin (2 μg/ml), aprotinin (2 μg/ml), pepstatin (2 μg/ml), β-glycerophosphate (10 mM), NaOV (0.2 mM), and NaF (25 mM). Cells were incubated at room temperature with lysis buffer for 1 min before harvesting. Samples were centrifuged at 16,000 g for 10 min at 4°C, to remove cellular debris. The resulting supernatant was stored at −80°C. Nuclear protein was extracted according to Pierce’s isolation protocol. In brief, cells were collected by scraping with PBS and were washed several times. Cell pellets were incubated with the appropriate amount of Cer I buffer on ice to isolate the cytoplasmic fraction. The remaining extract was incubated with the appropriate amount of Ner I buffer on ice with several agitation steps to isolate the nuclear fraction. The resultant cytoplasmic and nuclear extracts were stored at −80°C. Protein concentration was determined using the bicotinic method, with the use of BSA as a standard.15
Electromobility Shift Assay
For study of the VUSs in a heterozygous state, the PTEN promoter sequence (−893 to −755) was isolated through PCR amplification from either normal genomic DNA or from genomic DNA obtained from a patient with CS who has an identified variant. The DNA was PCR amplified for 30 cycles at the annealing temperature of 55°C in 20-μl reactions, with the use of HotStar and Q Solution (Qiagen) (forward 5′-ATGCGCTGCGGCAGGATAC-3′, reverse 5′-CTCATCTCCCTCGCCTGA-3′). To study the VUSs in a homozygous state, the above PCR products were cloned into TOPO-TA vectors, and product sequences were verified by direct DNA sequencing (ABI 3730xl DNA Analyzer). Nested PCR was subsequently performed with the above primers, for isolation of only the PTEN VUSs. Each of the above products was radiolabeled with 32P-γATP via T4 kinase. For examination of DNA-protein interaction, 1 ng of radiolabeled probe was incubated with 2 μg of nuclear protein extract for 20 min at room temperature with binding buffer containing 10 mM HEPES (pH 7.5), 2.5 mM MgCl2, 50 mM NaCl, 0.5 mM dithiothreitol, 4% glycerol, 1 μg/ml BSA, and 2 μg poly dI/dC. Unlabeled probe in 5× molar excess was used as the specific competitor, whereas a random oligonucleotide sequence was used as the nonspecific competitor. DNA-protein complexes were resolved on a 4% nondenaturing PAGE gel at 150 V for 3.5 h at 4°C and were visualized using a Phospho-Imager (Amersham Biosciences).
Reporter Assay
PTEN (−893 to −1) was PCR amplified from control genomic DNA or from genomic DNA from patients with CS (forward 5′-GCGTGGTCACCTGGTCCTTT-3′, reverse 5′-GCTGCTCACAGGCGCTGA-3′) and was subsequently cloned into a TOPO-TA vector. DNA was PCR amplified using 55°C as the annealing temperature for 30 cycles. All PCR-amplification products were verified by direct DNA sequencing (ABI 3730xl DNA Analyzer), and positive clones were subcloned into a pGL3.1-Basic vector (Promega). In the event that positive variant clones were not obtained, site-directed mutagenesis was performed, as described by the manufacturer’s protocol (GeneTailor Site-Directed Mutagenesis System [Invitrogen]). Site-directed mutagenesis was performed on the wild-type (WT) PTEN promoter in a TOPO-TA vector.
To determine promoter activity, 6-well plates of MCF-7 or HeLa cells were cotransfected with 1 μg/well of a PGL3_PTEN construct, and 50 ng/well Renilla luciferase control plasmid with 3 μl/well of FuGene (Roche), as described by the manufacturer. After 48 h, cells were harvested with 1× passive luciferase lysis buffer (Dual-Luciferase Reporter Assay System [Promega]) and were analyzed on a luminometer (LMax 11384 [Molecular Devices]) with the use of Renilla luciferase as an internal transfection control.
Western Blot
Fifteen micrograms of protein was prepared by the Laemelli method,16 was separated on 10% SDS-PAGE gel, and was electrophoretically transferred onto nitrocellulose. Equal protein loading between conditions was confirmed by staining with Ponceau S solution. Nonspecific binding was blocked by incubation of the nitrocellulose blots with 5% milk in Tris-buffered saline–Tween (TBS-T) (100 mM Tris [pH 7.5], 1 M NaCL, and 1% Tween-20) for 1 h at room temperature. Blots were then incubated with the primary antibody, either anti-PTEN (Cascade Bioscience) or anti-actin (Sigma), at a dilution of 1:1,000 in 3% BSA for 2 h at room temperature. After the primary incubation, the blots were washed with TBS-T for 1 h, with frequent changes of buffer. Blots were then incubated with the appropriate secondary antibody conjugated with horseradish peroxidase (Promega) at a dilution of 1:2,500 in 5% milk overnight at 4°C and were washed with TBS-T for 1 h. Protein bands were visualized using enhanced chemiluminescence, as described by the manufacturer (Amersham Pharmacia).
RT-PCR and Real-Time PCR
HeLa and MCF-7 cells, transfected with the above-described PGL3_PTEN –893 to –1 construct, and LBCLs were collected and were subsequently washed three times with PBS, through centrifugation. Total RNA was extracted from cells, in accordance with the Gentra Versagene RNA Purification System Protocol, and was then converted to cDNA by Superscript II Reverse Transcriptase after DNase treatment. The resultant cDNA was subjected to multiplex PCR amplification with the use of primers specific to luciferase (forward 5′-TCAAAGAGGCGAACTGTGTG-3′, reverse 5′-GGTGTTGGAGCAAGATGGAT-3′), PTEN exons 3 and 5 (forward 5′-TGGATTCAAAGCATAAAAACCA-3′, reverse 5′-AAAAGGATATTGTGCAACTCTGC-3′), and β-actin (Quantum RNA β-actin [Ambion]). Primers were allowed to anneal at 55°C for 28 cycles. The products from the PCRs were run on a 1% agarose gel containing ethidum bromide and were visualized under a UV light.
Real-time PCR was performed using the ABI 7500 real-time PCR system (ABI/Perkin Elmer) with the use of a SYBR Green–based assay, as described elsewhere.17 Primers were designed to amplify cDNA incorporating a portion of the PTEN transcript encoded by exons 7 and 8 (forward 5′-CCACAAACAGAACAAGATG-3′, reverse 5′-CTGGTCCTGGTATGAAGAAT-3′). Primers amplifying a portion of GAPDH were used as the control (forward 5′-CCATCTTCCAGGAGCGAGA-3′, reverse 5′-AAATGAGCCCCAGCCTTCT-3′). The thermal cycling conditions were 50°C for 2 min and 95°C for 10 min, followed by 40 cycles of denaturing at 95°C for 15 s and annealing and extension at 60°C for 1 min. All the reactions were performed in triplicate, and the comparative CT method was used for the quantification of the expression for each segment, by use of GAPDH as a normalization control. Each PCR generated only the expected amplicon, as shown by the melting temperature profiles of the final products and by gel electrophoresis.
Statistical Analysis
Statistical analysis was done using Student’s t test. Data are means ±SDs of three independent experiments and are normalized to a control. P<.05 is considered statistically significant.
Results
PTEN Promoter VUSs Demonstrate Similar Transcription-Factor Binding
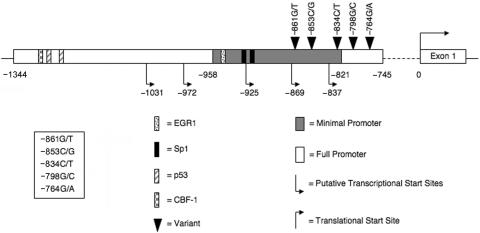
Despite the clear significance of PTEN dysfunction in the development of many types of cancers, the mechanisms that regulate PTEN’s promoter and 5′ UTR remain largely unknown. Figure 1 illustrates that the generally recognized full-length PTEN promoter is localized to positions −1344 to −745, and the minimal promoter to −958 to −821, with +1 representing the ATG site.18 While scanning the full-length PTEN promoter for nucleotide variants, we identified five VUSs (−861G/T, −853C/G, −834C/T, −798G/C, and −764G/A) in five unrelated patients with a clinical diagnosis of CS, which were absent in 186 unaffected, healthy controls. All five patients with CS had benign or malignant breast disease, and, importantly, three (60%) of the five had two component malignancies associated with CS (table 1). Additionally, all five of the VUSs used in this study are located 3′ of any known PTEN transcription-factor binding site (fig. 1). Previous data from our laboratory indicate that mutations within regulatory elements or transcription-factor binding sites interfere with normal transcriptional activity.12–14 However, these five VUSs are not predicted to alter any known cis-acting regulatory elements; therefore, the functional significance of these variants is even more difficult to ascertain.
Figure 1. .
Schematic diagram of the PTEN promoter. PTEN’s full-length promoter lies between −1344 and −745 (white bar), and the minimal promoter lies between −958 and −821 (gray bar). Within this region, the five nucleotide variants (black triangles) −861G/T, −853C/G, −834C/T, −798G/C, and −764G/A have been identified in a subset of patients with classic CS. The nearest 5′ transcription factors are EGR1 (dotted bar) and Sp1 (black bar).
Table 1. .
Increased Cancer Frequency in Patients with CS with PTEN Promoter Variants
| Mutation | Malignant Breast Cancer | Benign Breast Neoplasm | Follicular Thyroid Cancer | Endometrial Cancer |
| −861G/T | No | Yes | Yes | Yes |
| −853C/G | Yes | No | Yes | No |
| −834C/T | Yes | No | No | No |
| −798G/C | No | Yes | Yes | Yes |
| −764G/A | No | Yes | No | Yes |
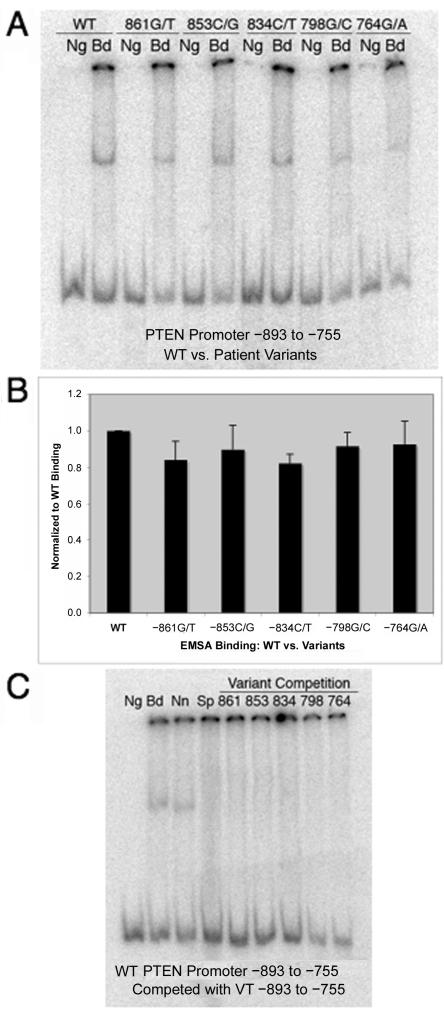
To determine what role these five VUSs may play in PTEN regulation, we performed electromobility shift assays (EMSAs) to find out if nuclear protein has the ability to associate with the promoter at this region. Interestingly, we found that nuclear protein does bind to the WT PTEN promoter from −893 to −755, suggesting that there is a yet-to-be-identified transcription-factor binding site in this region. We continued further to determine whether the nucleotide variants resulted in altered DNA-protein complex formation when compared with the WT PTEN promoter. EMSAs for each VUS were performed in either a homozygous state, where only the variant allele was present, or in a heterozygous state, where both the WT and variant allele were present. This allowed us to determine whether the variant allele has the ability to inhibit the WT allele’s function. As expected, no DNA-protein complex was observed when nuclear protein was not added to the reaction mixture (fig. 2A, Ng, lane 1), and nuclear protein does bind to the WT PTEN promoter from –893 to –755 (fig. 2A, Bd, lane 2). Unexpectedly, we observed that nuclear protein was also able to bind to the PTEN promoter VUSs in both a homozygous and a heterozygous state (fig. 2A, Bd, lanes 4, 6, 8, 10, and 12). Furthermore, these interactions did not differ significantly between the WT and VUS PTEN promoter probes (fig. 2B). In addition, the WT PTEN promoter DNA-protein complex could be competed with a cold unlabeled VUS probe (fig. 2C). Taken together, these data suggest that the DNA-protein interaction at this site is not affected by these specific promoter VUSs.
Figure 2. .
EMSA analysis probed with radiolabeled PTEN promoter from −893 to −755, either WT or variant. A, Each promoter probe, incubated in either the presence (Bd) or absence (Ng) of nuclear protein. The WT PTEN promoter (lanes 1 and 2), −861G/T (lanes 3 and 4), −853C/G (lanes 5 and 6), −834C/T (lanes 7 and 8), −798G/C (lanes 9 and 10), and −764G/A (lanes 11 and 12) demonstrate nuclear-protein binding. A representative blot from three individual experiments is displayed. B, Quantification of nuclear-protein binding to the PTEN promoter, confirming insignificant differences between WT (lane 1) and the five variants (lanes 2–6; P>.050). EMSA results are depicted as fold change compared with WT PTEN promoter and are shown in graphical format (Student’s t test). C, Radiolabeled WT PTEN promoter from −893 to −755. Nuclear protein either was not incubated with the probe (Ng) or was incubated with the probe (Bd) to test binding. Competition assays were performed with a nonspecific competitor (Nn), unlabeled WT promoter probe (Sp), and cold PTEN promoter variant probes (lanes 5–9). A representative blot from three individual experiments is displayed.
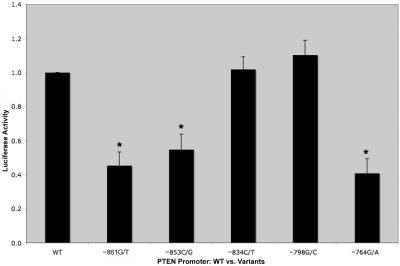
PTEN Promoter–Reporter Assays Demonstrate That VUSs Significantly Inhibit Luciferase Activity
The above data suggest that transcription-factor binding at positions −893 to −755 is not altered in patients who harbor VUSs within this region and that formation of this complex may not directly contribute to disease pathogenesis. To gain more insight into the potential mechanism(s) of PTEN dysfunction manifested by these variants, we examined the ability of each VUS to promote PTEN transcription, via reporter assays. We observed that the WT PTEN (positions −893 to −1) had a basal level of luciferase activity (fig. 3, WT). Two of the patient promoter VUSs, −834C/T and −798G/C, had similar luciferase read-outs (fig. 3). In contrast, three of the VUSs showed an inhibition in luciferase activity when compared with the WT PTEN construct. We found that the −861G/T and −764G/A VUSs had the greatest inhibition of luciferase activity, ∼50% less than that of the WT construct (fig. 3) (P<.001). Additionally, the −853C/G VUS resulted in a 40% decrease in luciferase activity, compared with WT PTEN (fig. 3) (P<.001).
Figure 3. .
Luciferase activity of PTEN 5′ UTR altered in three VUSs. MCF-7 cells were transfected with PGL3_PTEN 5′ UTR from −893 to −1, either WT or variant, as described in the “Material and Methods” section. After 48 h of treatment, cells were harvested, and luciferase activity was measured. Each bar represents a mean ±SEM of three individual experiments. An asterisk (*) indicates P<.001 (Student’s t test).
Promoter VUSs Do Not Inhibit PTEN Transcription
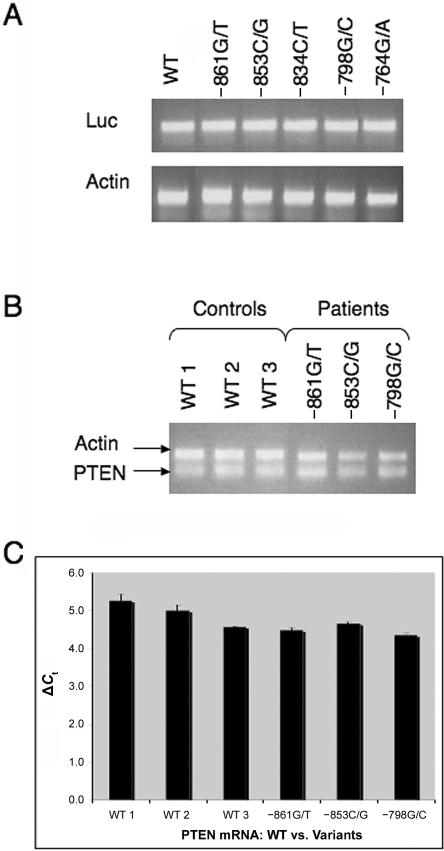
The above experiment indicates that WT PTEN induces more luciferase activity than do three of the patient-derived promoter VUSs—specifically, the −861G/T, −764G/A, and −853C/G variants. However, this experiment does not indicate whether this effect is due to a decrease in transcription or to an inhibition of protein translation. To differentiate the two scenarios, we analyzed luciferase mRNA levels in the reporter-assay samples described above. If the promoter VUSs are affecting transcription, we would expect to see changes in luciferase mRNA levels in those VUSs that caused significant decreases in luciferase activity. Unexpectedly, we found that luciferase mRNA was equally expressed in the cells transfected with the WT PTEN construct compared with cells transfected with the patient-derived VUS promoters (fig. 4A), suggesting that transcription efficiency and mRNA stability are not compromised in these five VUSs.
Figure 4. .
WT and variant PTEN promoters demonstrating equal mRNA expression. A, HeLa or MCF-7 cells transfected with a PGL3_PTEN −893 to −1 construct, either WT or containing a VUS. Cells were harvested 48 h after transfection, and total RNA was extracted. Luciferase (top panel) and actin (bottom panel) mRNA levels were measured by RT-PCR. P>.050 (represents a mean ±SEM of three individual experiments). B, LBCLs from unaffected, healthy controls that were either PTEN WT (lanes 1–3) or patient derived with a promoter VUS (−861G/T [lane 4], −853C/G [lane 5], and −798G/C [lane 6]). LBCLs were harvested, and total RNA was extracted. PTEN (bottom band) and actin (top band) mRNA levels were measured by RT-PCR. P>.050 (represents a mean ±SEM of three individual experiments). C, LBCLs from unaffected, healthy controls that were either PTEN WT (bars 1–3) or patient derived with a promoter VUS (−861G/T [bar 4], −853C/G [bar 5], and −798G/C [bar 6]). LBCLs were harvested, and total RNA was extracted. Real-time measured PTEN and GAPDH mRNA levels are shown, with quantification of PTEN mRNA normalized to GAPDH levels. Real-time RT-PCR results are depicted as ΔCt changes and are shown in graphical format. Each bar represents a mean ±SEM of three individual experiments. P>.050 (Student’s t test).
We next examined the VUSs ex vivo by assessing PTEN mRNA levels, using total RNA isolated from LBCLs from patients with promoter VUSs. We were able to assess mRNA levels in three cell lines derived from the patients with the following VUSs: −861G/T, −853C/G, and −798G/C. We were unable to obtain LBCLs from the patients with the remaining two VUSs (−834C/T and −764G/A). By semiquantitative RT-PCR, we demonstrated that there was equivalent PTEN mRNA expression in LBCLs harboring promoter VUSs compared with those obtained from unaffected, healthy controls (fig. 4B) (P>.05). To confirm these results, we performed a quantitative real-time PCR assay. Similarly, no differences were observed between PTEN mRNA levels in unaffected, healthy controls compared with patient-derived promoter VUSs (fig. 4C) (P>.05).
PTEN mRNA Secondary Structure Is Altered in VUSs versus PTEN WT Promoter
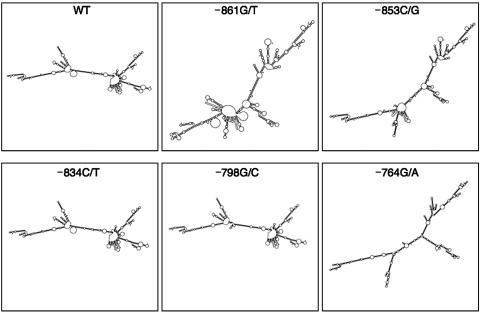
As the above data suggest, these PTEN VUSs do not result in altered transcription. Therefore, we next focused on whether they result in abnormal translation, by studying the PTEN mRNA transcript in more detail. Several laboratories have previously suggested a number of putative PTEN transcriptional start sites between −1031 and −9318–21; however, comparison of the human and mouse PTEN cDNA sequences suggests that the transcript begins around −925.20 In agreement with these results, we performed PTEN RT-PCR from −869 to exon 9 and verified that all five VUSs are included in the resulting transcript (data not shown). Therefore, we hypothesized that the inclusion of these nucleotide variants within the transcript causes an alteration of the normal mRNA secondary structure and, consequently, an inhibiting protein translation. To determine if the mRNA secondary structure is different in the VUSs compared with WT PTEN, we used the MFOLD software program.22 We analyzed the PTEN 5′ UTR from positions −893 to −1, using both the WT sequence and sequences containing each of the VUSs. Several potential secondary structures for each sequence were predicted, with the most-stable structures illustrated in figure 5.
Figure 5. .
MFOLD-predicted secondary structures resulting from the five VUSs in patients with CS. The most-stable mRNA secondary structures predicted by MFOLD are illustrated here.
The major secondary structure predicted for the WT PTEN promoter is Y shaped with multiple loops (fig. 5, WT). This shape is consistent with that predicted for the −834C/T and −798G/C VUSs (fig. 5). In contrast, the −861G/T, −853C/G, and −764G/A VUSs have different predicted secondary structures compared with the WT PTEN 5′ UTR. Whereas the −853C/G VUS maintains the general Y shape, the loop structures are altered to create a new branching arrangement (fig. 5, −853C/G). Similarly, the −764G/A VUS maintains a Y shape that is similar to the WT 5′ UTR; however, it has lost almost all of its loops, creating more of a sticklike structure (fig. 5, –764G/A). Finally, the –861G/T VUS is structurally the most different from WT PTEN (fig. 5, –861G/T). The general Y-shape appearance has been lost and has been replaced with a detailed, intricate looping and branching configuration. These structures suggest that PTEN translation may be altered or inhibited in patients with these promoter VUSs, particularly those that have the greatest deviation from the WT structure, such as –861G/T.
Altered PTEN Protein Expression in Select VUSs
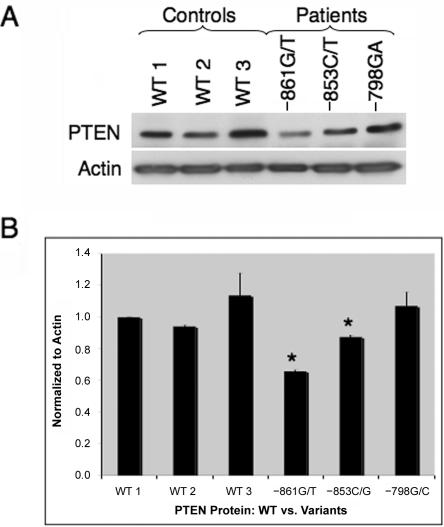
To determine whether these patients demonstrate altered protein translation due to nucleotide variants within the promoter region, we examined PTEN protein expression from the previously mentioned patient-derived LBCLs. We found that protein isolated from unaffected, healthy controls had similar PTEN protein expression (fig. 6A and 6B, lanes 1–3). Comparable to our control samples, normal PTEN protein expression was observed from patient-derived LBCLs with VUS –798G/C (fig. 6A and 6B, lane 6). Protein lysate derived from patient LBCLs with the −853C/G VUS had an ∼15% decrease in PTEN expression (P=.046) (fig. 6A and 6B, lane 5), whereas cells derived from VUS −861G/T had the largest decrease in PTEN levels, at ∼40% of control protein (P<.001) (fig. 6A and 6B, lane 4). Interestingly, these protein results can be seen as concurrent with the structural alterations observed with the MFOLD software. The secondary structure of the VUS −798G/C 5′ UTR does not predict a large change compared with WT PTEN; therefore, one would predict that PTEN protein levels would be comparable to WT controls, as we observed here. In contrast, the secondary structure predicted with the VUS −853C/G 5′ UTR predicted some alterations that correlate with the slight decrease in PTEN protein levels observed in this patient. Finally, the VUS promoter with the greatest structural change, −861G/T, demonstrated the largest alteration in PTEN levels. Taken together, these data indicate that these predicted alterations to the secondary structures of the 5′ UTR, including the PTEN promoter, which are consequent to the VUSs in our patients with CS, inhibit normal translation of PTEN.
Figure 6. .
PTEN protein expression decreased in promoter variants with the greatest mRNA secondary structure alterations. A, LBCLs from unaffected, healthy controls that were either PTEN WT (lanes 1–3) or patient derived with a promoter VUS (−861G/T [lane 4], −853C/G [lane 5], and −798G/C [lane 6]). LBCLs were harvested, and total protein was extracted. PTEN (top panel) and actin (bottom panel) protein levels were measured by western-blot analysis. B, Quantification of PTEN protein normalized to actin levels. Western-blot results are depicted as fold change and are shown in graphical format: −861G/T (P<.001), −853C/G (P=.046), and −798G/C (P>.050). Each bar represents a mean ±SEM of three individual experiments. An asterisk (*) indicates P<.050 (Student’s t test).
Discussion
Recently, the importance of gene regulation in the pathogenesis of hereditary cancer-predisposition syndromes has been advanced though the interrogation of promoter variation as a mechanism of disease development. PTEN promoter variants and their consequences have been only minimally studied; however, promoters within a few select genes, such as the baculoviral IAP repeat-containing 5 gene (BIRC5, also known as survivin), have been examined more extensively.23 Yet the majority of these studies have looked specifically at known regulatory regions and/or consensus sites within the promoter of interest. We hypothesized that novel nontraditional regulatory mechanisms within the PTEN promoter play an important role in gene regulation. This indicates that variants within unknown regulatory elements may be as significant as those in known cis-acting regions. To test this hypothesis, we studied five VUSs within the PTEN promoter that are not within a known cis-acting element. The culmination of our data reveals that protein translation is altered within a subset of patients with CS who lack traditional exonic or splice-site PTEN mutations but who harbor PTEN promoter variants, particularly those resulting in large mRNA structural changes compared with WT PTEN mRNA. These data also demonstrate abnormal protein translation as a novel mechanism of CS pathogenesis.
To date, analysis of PTEN's promoter has identified eight regulatory factors that have been implicated in modulating PTEN’s transcription: early growth response-1 (EGR1),24 nuclear factor–kappa B (NF-κB),21 Sp1,3,19 CBF-1,25 p53,20 USF1,12 peroxisome proliferator-activated receptor gamma (PPARγ),26–28 and c-Jun15 (fig. 1). The five VUSs (−861G/T, −853C/G, −834C/T, −798G/C, and −764G/A) used in our studies reside in the full-length PTEN promoter region between −893 and −755 but lie more 3′ of any of these known transcription-factor binding motifs. Our EMSA results, with use of PTEN −893 to −755 as bait, demonstrated nuclear-protein binding (fig. 2), thus suggesting that there may be a novel transcription-factor binding site contained within this region. Several potential transcription factors are anticipated to bind to this region, but only two were predicted by multiple prediction software programs: Sp1 (TESS,29 and Alibaba,30) and c-Myb (TESS and TFSEARCH,31). Sp1 is currently thought to be a putative PTEN transcription factor because the full-length PTEN promoter is very GC rich; however, research has yet to concretely show that it binds to any particular region of the promoter or has the ability to regulate its transcription.3,18 c-Myb has been shown to be up-regulated within tumors when PTEN expression is decreased,32 indicating that it may be acting as a PTEN transcriptional repressor; however, the pathway connecting the two has yet to be determined. On the basis of these prediction models, both Sp1 and c-Myb can be postulated as regulators of PTEN expression; however, more in-depth studies are necessary to determine the identity of this novel PTEN transcription factor.
In our initial EMSA results, we expected to observe a difference in this novel transcription factor’s ability to bind to the PTEN promoter, since previous data from our laboratory have shown that PTEN promoter alterations within the p53 (−1190 to −1157)13 and USF1 (−2237 and −2058)12 binding sites inhibit both normal PTEN mRNA expression and protein function. In contrast, data presented herein indicated that protein-binding inhibition is not the primary mechanism of PTEN alterations (fig. 2). Interestingly, our reporter-assay results indicated that several of the VUSs had a decrease in luciferase activity (fig. 3), whereas, seemingly paradoxically, PTEN mRNA was equally expressed relative to WT (fig. 4). This suggests that normal protein translation is disrupted by these alterations, whereas conventional mRNA transcription remains unaffected. In 2001, Signori and colleagues observed a similar effect caused by a variant located 3 nt upstream of the ATG site, thus lying within the Kozak consensus sequence of the 5′ UTR within BRCA1.33 This nucleotide variant is thought to have weakened the Kozak sequence enough to inhibit normal protein translation. In contrast to Signori et al.,33 the variants discussed in this publication did not immediately indicate this mechanism, and more intricate analyses were necessary.
PTEN has a number of putative transcription start sites, whose analysis reveals five potential Kozak translational start sites. However, none of these start sites perfectly fit the mammalian Kozak consensus sequence, GCCRCCATGG, where the −3 and +4 positions are the most conserved.34 Analyses of these ATG sites, 5′→3′, indicate that they would produce 28-aa, 5-aa, 4-aa, 46-aa, and 403-aa proteins. PTEN is just one of many genes that has the potential to produce upstream ORFs and may not follow the “first ATG” rule. Moreover, this tends to lead to leaky scanning by the 40S ribosome, thus allowing for translation of one or several of the 5′ ORFs. In this situation, the ribosomes do not fall off the transcript but do proceed to scan 3′ to the true ATG site, thus allowing for the production of the correct mRNA. This suggests that PTEN’s long 5′ UTR with potential ORFs and a weak Kozak consensus sequence make it more prone to influences from nucleotide variants, which can subsequently decrease its translation efficiency.35
Another mechanism that can confound normal protein-translation efficiency is through aberrant mRNA secondary structures. To determine whether modified PTEN mRNA structure was the cause of altered protein translation in our patients, we used the MFOLD software program to compare the WT PTEN promoter with the five VUSs. Our results demonstrate that some of these VUSs contribute to the mRNA structure, thus creating a significantly different configuration compared with WT PTEN (fig. 5). This is important because normal mRNA primary and secondary structures are essential for the ability of mRNA-binding proteins to both accurately bind and initiate and, subsequently, to affect protein translation.35 Recently, Saxena and colleagues identified an 11-bp deletion in MeCP2, 103 nt upstream of the ATG site.36 Through the use of mRNA structural prediction models, they concluded that this deletion disrupted normal protein translation without affecting MeCP2 transcription.
Studies of the nucleotide alterations isolated within both BRCA1 and MeCP2, which specifically inhibit normal translation and not gene transcription, are currently the only known ones to demonstrate that promoter VUSs can affect protein expression through such a mechanism.33,36 However, there have been a few recent examples of SNPs within protein-coding regions that also alter the normal protein outcome through translation. In both of these examples, it is hypothesized that synonymous SNPs induce structural changes in mRNA structure, which ultimately lead to the protein’s dysregulation. Kimchi-Sarfaty et al.37 report that specific MDR1 haplotypes, generated from silent SNPs, inhibit normal protein translation and, subsequently, its function. This is thought to occur through the slowing of ribosomal scanning at these specific codons.37,38 Furthermore, Nackley and colleagues39 described three major haplotypes formed by four SNPs within the human catechol-O-methyltransferase (COMT) gene, whose MFOLD predictions indicated that the haplotypes result in altered mRNA secondary structures. More interestingly, a decrease in both protein expression and enzymatic activity was produced from a haplotype containing two synonymous SNPs.39 These data suggest that nucleotide changes that may initially seem insignificant can disrupt normal protein translation through alterations of mRNA secondary structure.
As stated above, our reporter-assay, MFOLD-analysis, and western-blot results are all in agreement with regard to the −861C/G, −853C/G, and −789G/C VUSs. Because of unavailability, we were unable to directly study the −834C/T and −764G/A VUSs; however, one can speculate on the likely outcome on the basis of the above results. Our data indicate that the −834C/T VUS, which displays no secondary structural differences compared with WT PTEN mRNA, and which had only a slight decrease in luciferase activity, would not have a decrease in PTEN protein expression. In contrast, one can hypothesize that the −764G/A variant would have a significant decrease in PTEN protein expression. Similar to the −861C/G VUS, the −764G/A VUS demonstrated a significant decrease in luciferase activity and a large alteration in mRNA secondary structure, as predicted by MFOLD, when compared with WT PTEN 5′ UTR.
It is thought that the decrease in translation efficiency of these mRNA structures can be compensated for by more-efficient translation through the regulation of the eukaryotic translation initiation factor (eIF4F) complex. This complex, which is composed of eIF4A, eIF4B, and eIF4H, is involved in unwinding mRNA secondary structures to induce protein translation.40 These data suggest that one could potentially modulate one or several of these translation factors as a personal therapeutic target for patients with PTEN promoter VUSs.
In 2003, our laboratory was the first to demonstrate the pathogenicity of PTEN promoter variants in patients with CS.3 These genetic alterations within PTEN’s promoter appear to correlate with a high prevalence of breast cancer in this subset of patients. In agreement with these previous data, the patients included in the current analysis all harbor promoter VUSs and exhibit a high prevalence of breast neoplasia, as well as follicular thyroid and endometrial cancer. All five patients with the PTEN promoter VUSs interrogated in this study developed breast tumors (table 1). Two of these five patients were given diagnoses of breast cancer, whereas the remaining three patients were given diagnoses of benign breast neoplasms. In addition to breast cancer, three of the five patients developed follicular thyroid cancer, and three of the five patients had endometrial cancer, suggesting that these VUSs are associated with neoplastic risk. All five patients ultimately were given diagnoses of at least one component malignancy, and three (60%) were diagnosed with two component malignancies.
Through more-detailed functional analysis of the VUSs located within the PTEN 5′ UTR, we have now elucidated this mechanism in three of five patients with CS, each of whom harbors a previously uncharacterized promoter VUS (specifically, −861G/T, −853C/G, and −764G/C). However, our data indicate that aberrant protein translation likely is not the primary mechanism of CS development in patients with the −834C/T and −798G/C variants. One can speculate that, within the patients that have these two VUSs, a specific haplotype may be playing a key role14 or the PTEN protein function may be altered.41 This suggests that the region upstream of PTEN plays an important role in the development of CS; however, the precise pathomechanism(s) of these mutations remains to be elucidated.
Our data reinforce the importance of PTEN promoter nucleotide variations and their ability to lead to CS progression through protein-translation inhibition. As discussed, patients with CS with promoter mutations have a high prevalence of breast, thyroid, and endometrial malignancies, and an understanding of the mechanism of PTEN dysfunction in these patients may lead to rational targeted therapies to treat or prevent malignancy. Our data suggest that a therapeutic tool that can regulate its transcription and/or translation, such as Lovastatin28 or an eIF4F target, could be highly effective for patients with germline nucleotide alterations within this region or in sporadic tumors with somatic 5′-UTR VUSs. Moreover, our data also reiterate the importance of looking for variants within the PTEN promoter and even elsewhere in the 5′ UTR of patients who have CS features yet do not have a detectable mutation within its ORF. This approach should increase the frequency of finding germline PTEN mutations in PHTS, thus increasing the sensitivity of molecular diagnosis, and hence broadening those families amenable to predictive testing. Furthermore, this knowledge can be extended to other diseases and to their respective susceptibility genes. Currently, promoters are rarely analyzed in the clinical setting; therefore, it is very likely that nucleotide changes will be identified in many other genes and that these patients may also benefit from personalized treatment, given their promoter-mutation status.
Acknowledgments
This work was funded in part by American Cancer Society grant RSG-02–151–01-CCE (to C.E.). R.E.T. and M.G.P. are predoctoral fellows of the Cleveland Clinic Genomic Medicine Institute and are graduate students of the Integrated Biomedical Sciences Graduate Program of The Ohio State University (Columbus). K.M.Z. is a Crile Fellow of the Cleveland Clinic. C.E. is a recipient of the Doris Duke Distinguished Clinical Scientist Award. The authors thank Mr. Todd Romigh (C.E.'s lab) for constructing the vectors, Dr. X. P. Zhou for technical assistance during the early phases of this work, and, especially, Drs. Jodi Bubenik, Donna Driscoll, Don Luse, and Kwaku Dayie for their insight and critical discussions. R.E.T. also acknowledges Dr. Yufang Tang, Ms. Pat Kessler, Mr. Robert Pilarski, and Ms. Jennifer Stein for helpful discussions.
Web Resources
Accession numbers and URLs for data presented herein are as follows:
- Alibaba, http://www.gene-regulation.com/pub/programs/alibaba2/index.html
- Genomic Medicine Biorepository of the Cleveland Clinic Genomic Medicine Institute, http://www.lerner.ccf.org/gmi/gmb/methods.php
- MFOLD, http://bioweb.pasteur.fr/seqanal/interfaces/mfold-simple.html
- National Comprehensive Cancer Network, http://www.nccn.org/
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for CS, BRRS, and Proteus syndrome)
- TESS, http://www.cbil.upenn.edu/cgi-bin/tess/tess
- TFSEARCH, http://www.cbrc.jp/research/db/TFSEARCH.html
References
- 1.Liaw D, Marsh DJ, Li J, Dahia PL, Wang SI, Zheng Z, Bose S, Call KM, Tsou HC, Peacocke M, et al (1997) Germline mutations of the PTEN gene in Cowden disease, an inherited breast and thyroid cancer syndrome. Nat Genet 16:64–67 10.1038/ng0597-64 [DOI] [PubMed] [Google Scholar]
- 2.Marsh DJ, Kum JB, Lunetta KL, Bennett MJ, Gorlin RJ, Ahmed SF, Bodurtha J, Crowe C, Curtis MA, Dasouki M, et al (1999) PTEN mutation spectrum and genotype-phenotype correlations in Bannayan-Riley-Ruvalcaba syndrome suggest a single entity with Cowden syndrome. Hum Mol Genet 8:1461–1472 10.1093/hmg/8.8.1461 [DOI] [PubMed] [Google Scholar]
- 3.Zhou XP, Waite KA, Pilarski R, Hampel H, Fernandez MJ, Bos C, Dasouki M, Feldman GL, Greenberg LA, Ivanovich J, et al (2003) Germline PTEN promoter mutations and deletions in Cowden/Bannayan-Riley-Ruvalcaba syndrome result in aberrant PTEN protein and dysregulation of the phosphoinositol-3-kinase/Akt pathway. Am J Hum Genet 73:404–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Witman PM (2006) More than just a bump: the hamartoma syndromes. Adv Dermatol 22:157–180 10.1016/j.yadr.2006.08.002 [DOI] [PubMed] [Google Scholar]
- 5.Zbuk KM, Eng C (2007) Cancer phenomics: RET and PTEN as illustrative models. Nat Rev Cancer 7:35–45 10.1038/nrc2037 [DOI] [PubMed] [Google Scholar]
- 6.Eng C (2003) PTEN: one gene, many syndromes. Hum Mutat 22:183–198 10.1002/humu.10257 [DOI] [PubMed] [Google Scholar]
- 7.Ries LAG, Eigne EM, Kosary CL, Hankey BF, Miller BA, Clegg L, Mariotto A, Fay MP, Feuer EJ, Edwards BK (2003) SEER cancer statistics review, 1975–2000. National Cancer Institute, Bethesda, MD [Google Scholar]
- 8.Pilarski R, Eng C (2004) Will the real Cowden syndrome please stand up (again)? Expanding mutational and clinical spectra of the PTEN hamartoma tumour syndrome. J Med Genet 41:323–326 10.1136/jmg.2004.018036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gorlin RJ, Cohen MM Jr, Condon LM, Burke BA (1992) Bannayan-Riley-Ruvalcaba syndrome. Am J Med Genet 44:307–314 10.1002/ajmg.1320440309 [DOI] [PubMed] [Google Scholar]
- 10.Butler MG, Dasouki MJ, Zhou XP, Talebizadeh Z, Brown M, Takahashi TN, Miles JH, Wang CH, Stratton R, Pilarski R, et al (2005) Subset of individuals with autism spectrum disorders and extreme macrocephaly associated with germline PTEN tumour suppressor gene mutations. J Med Genet 42:318–321 10.1136/jmg.2004.024646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eng C (2000) Will the real Cowden syndrome please stand up: revised diagnostic criteria. J Med Genet 37:828–830 10.1136/jmg.37.11.828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pezzolesi MG, Zbuk KM, Waite KA, Eng C (2007) Comparative genomic and functional analyses reveal a novel cis-acting PTEN regulatory element as a highly conserved functional E-box motif deleted in Cowden syndrome. Hum Mol Genet 16:1058–1071 10.1093/hmg/ddm053 [DOI] [PubMed] [Google Scholar]
- 13.Tang Y, Eng C (2006) p53 Down-regulates phosphatase and tensin homologue deleted on chromosome 10 protein stability partially through caspase-mediated degradation in cells with proteasome dysfunction. Cancer Res 66:6139–6148 10.1158/0008-5472.CAN-06-0772 [DOI] [PubMed] [Google Scholar]
- 14.Pezzolesi MG, Li Y, Zhou XP, Pilarski R, Shen L, Eng C (2006) Mutation-positive and mutation-negative patients with Cowden and Bannayan-Riley-Ruvalcaba syndromes associated with distinct 10q haplotypes. Am J Hum Genet 79:923–934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hettinger K, Vikhanskaya F, Poh MK, Lee MK, de Belle I, Zhang JT, Reddy SA, Sabapathy K (2007) c-Jun promotes cellular survival by suppression of PTEN. Cell Death Differ 14:218–229 10.1038/sj.cdd.4401946 [DOI] [PubMed] [Google Scholar]
- 16.Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685 10.1038/227680a0 [DOI] [PubMed] [Google Scholar]
- 17.Sarquis MS, Agrawal S, Shen L, Pilarski R, Zhou XP, Eng C (2006) Distinct expression profiles for PTEN transcript and its splice variants in Cowden syndrome and Bannayan-Riley-Ruvalcaba syndrome. Am J Hum Genet 79:23–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sheng X, Koul D, Liu JL, Liu TJ, Yung WK (2002) Promoter analysis of tumor suppressor gene PTEN: identification of minimum promoter region. Biochem Biophys Res Commun 292:422–426 10.1006/bbrc.2002.6662 [DOI] [PubMed] [Google Scholar]
- 19.Han B, Dong Z, Liu Y, Chen Q, Hashimoto K, Zhang JT (2003) Regulation of constitutive expression of mouse PTEN by the 5′-untranslated region. Oncogene 22:5325–5337 10.1038/sj.onc.1206783 [DOI] [PubMed] [Google Scholar]
- 20.Stambolic V, MacPherson D, Sas D, Lin Y, Snow B, Jang Y, Benchimol S, Mak TW (2001) Regulation of PTEN transcription by p53. Mol Cell 8:317–325 10.1016/S1097-2765(01)00323-9 [DOI] [PubMed] [Google Scholar]
- 21.Vasudevan KM, Gurumurthy S, Rangnekar VM (2004) Suppression of PTEN expression by NF-kappa B prevents apoptosis. Mol Cell Biol 24:1007–1021 10.1128/MCB.24.3.1007-1021.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zuker M (1989) On finding all suboptimal foldings of an RNA molecule. Science 244:48–52 10.1126/science.2468181 [DOI] [PubMed] [Google Scholar]
- 23.Jiang Y, Saavedra HI, Holloway MP, Leone G, Altura RA (2004) Aberrant regulation of survivin by the RB/E2F family of proteins. J Biol Chem 279:40511–40520 10.1074/jbc.M404496200 [DOI] [PubMed] [Google Scholar]
- 24.Virolle T, Adamson ED, Baron V, Birle D, Mercola D, Mustelin T, de Belle I (2001) The Egr-1 transcription factor directly activates PTEN during irradiation-induced signalling. Nat Cell Biol 3:1124–1128 10.1038/ncb1201-1124 [DOI] [PubMed] [Google Scholar]
- 25.Whelan JT, Forbes SL, Bertrand FE (2007) CBF-1 (RBP-Jkappa) binds to the PTEN promoter and regulates PTEN gene expression. Cell Cycle 6:80–84 [DOI] [PubMed] [Google Scholar]
- 26.Bonofiglio D, Gabriele S, Aquila S, Catalano S, Gentile M, Middea E, Giordano F, Ando S (2005) Estrogen receptor alpha binds to peroxisome proliferator-activated receptor response element and negatively interferes with peroxisome proliferator-activated receptor gamma signaling in breast cancer cells. Clin Cancer Res 11:6139–6147 10.1158/1078-0432.CCR-04-2453 [DOI] [PubMed] [Google Scholar]
- 27.Patel L, Pass I, Coxon P, Downes CP, Smith SA, Macphee CH (2001) Tumor suppressor and anti-inflammatory actions of PPARgamma agonists are mediated via upregulation of PTEN. Curr Biol 11:764–768 10.1016/S0960-9822(01)00225-1 [DOI] [PubMed] [Google Scholar]
- 28.Teresi RE, Shaiu CW, Chen CS, Chatterjee VK, Waite KA, Eng C (2006) Increased PTEN expression due to transcriptional activation of PPARgamma by Lovastatin and Rosiglitazone. Int J Cancer 118:2390–2398 10.1002/ijc.21799 [DOI] [PubMed] [Google Scholar]
- 29.Schung J, Overton GC (1997) Technical report CBIL-TR-1997-1001-v0.0. School of Medicine, University of Pennsylvania, Philadelphia [Google Scholar]
- 30.Grabe N (2002) AliBaba2: context specific identification of transcription factor binding sites. In Silico Biol 2:S1–S15 [PubMed] [Google Scholar]
- 31.Heinemeyer T, Wingender E, Reuter I, Hermjakob H, Kel AE, Kel OV, Ignatieva EV, Ananko EA, Podkolodnaya OA, Kolpakov FA, et al (1998) Databases on transcriptional regulation: TRANSFAC, TRRD and COMPEL. Nucleic Acids Res 26:362–367 10.1093/nar/26.1.362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Williams K, Fernandez S, Stien X, Ishii K, Love HD, Lau YF, Roberts RL, Hayward SW (2005) Unopposed c-MYC expression in benign prostatic epithelium causes a cancer phenotype. Prostate 63:369–384 10.1002/pros.20200 [DOI] [PubMed] [Google Scholar]
- 33.Signori E, Bagni C, Papa S, Primerano B, Rinaldi M, Amaldi F, Fazio VM (2001) A somatic mutation in the 5′UTR of BRCA1 gene in sporadic breast cancer causes down-modulation of translation efficiency. Oncogene 20:4596–4600 10.1038/sj.onc.1204620 [DOI] [PubMed] [Google Scholar]
- 34.Kozak M (1986) Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Cell 44:283–292 10.1016/0092-8674(86)90762-2 [DOI] [PubMed] [Google Scholar]
- 35.Mignone F, Gissi C, Liuni S, Pesole G (2002) Untranslated regions of mRNAs. Genome Biol 3:REVIEWS0004 10.1186/gb-2002-3-3-reviews0004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Saxena A, de Lagarde D, Leonard H, Williamson SL, Vasudevan V, Christodoulou J, Thompson E, MacLeod P, Ravine D (2006) Lost in translation: translational interference from a recurrent mutation in exon 1 of MECP2. J Med Genet 43:470–477 10.1136/jmg.2005.036244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kimchi-Sarfaty C, Oh JM, Kim IW, Sauna ZE, Calcagno AM, Ambudkar SV, Gottesman MM (2007) A “silent” polymorphism in the MDR1 gene changes substrate specificity. Science 315:525–528 10.1126/science.1135308 [DOI] [PubMed] [Google Scholar]
- 38.Komar AA (2007) Genetics: SNPs, silent but not invisible. Science 315:466–467 10.1126/science.1138239 [DOI] [PubMed] [Google Scholar]
- 39.Nackley AG, Shabalina SA, Tchivileva IE, Satterfield K, Korchynskyi O, Makarov SS, Maixner W, Diatchenko L (2006) Human catechol-O-methyltransferase haplotypes modulate protein expression by altering mRNA secondary structure. Science 314:1930–1933 10.1126/science.1131262 [DOI] [PubMed] [Google Scholar]
- 40.Svitkin YV, Pause A, Haghighat A, Pyronnet S, Witherell G, Belsham GJ, Sonenberg N (2001) The requirement for eukaryotic initiation factor 4A (elF4A) in translation is in direct proportion to the degree of mRNA 5′ secondary structure. RNA 7:382–394 10.1017/S135583820100108X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chung JH, Eng C (2005) Nuclear-cytoplasmic partitioning of phosphatase and tensin homologue deleted on chromosome 10 (PTEN) differentially regulates the cell cycle and apoptosis. Cancer Res 65:8096–8100 10.1158/0008-5472.CAN-05-1888 [DOI] [PubMed] [Google Scholar]