Abstract
Globozoospermia is a rare (incidence <0.1% in male infertile patients) form of teratozoospermia, mainly characterized by round-headed spermatozoa that lack an acrosome. It originates from a disturbed spermiogenesis, which is expected to be induced by a genetic factor. Several family cases and recessive mouse models with the same phenotype support this expectation. In this study, we present a consanguineous family with three affected brothers, in whom we have identified a homozygous mutation in the spermatogenesis-specific gene SPATA16. This is the first example of a nonsyndromic male infertility condition in humans caused by an autosomal gene defect, and it could also mean that the identification of other partners like SPATA16 could elucidate acrosome formation.
Approximately 15% of couples are confronted with the inability to conceive after 2 years of unprotected intercourse.1 In about half of these cases, infertility is due to the inability of the male partner to produce spermatozoa of sufficient number (oligozoospermia), adequate motility (asthenozoospermia), or normal morphology (teratozoospermia) or to combinations of these defects. Globozoospermia (MIM 102530) is a rare but severe teratozoospermia, characterized by ejaculates consisting completely of round-headed spermatozoa that lack an acrosome or, in partial globozoospermia, containing a variable proportion (20%-90%) of acrosomeless spermatozoa.2–4 Men that are affected with total globozoospermia are infertile, and even the application of intracytoplasmic sperm injection (ICSI) has met with disappointingly low success rates.2 Globozoospermia originates from a disturbed spermiogenesis, and, although the underlying cause is still unknown, a genetic contribution appears to be supported by several familial case reports5–7 and by three recessive mouse models involving CSNK2A2 (MIM 115442), HRB (MIM 600862), and GOPC (MIM 606845).8–10 However, no causative gene mutations have been identified in these orthologues or any other human genes to date.11,12 We describe a family with three affected brothers, in whom we have identified a homozygous mutation in the spermatogenesis-specific gene SPATA16 (MIM 609856). To our knowledge, this is the first example of a nonsyndromic male infertility condition in humans caused by a single gene defect.
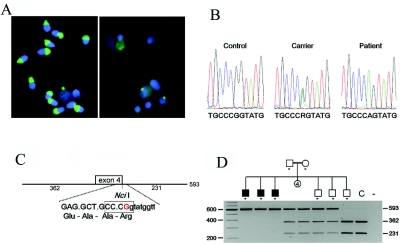
We investigated an Ashkenazi Jewish family with six brothers (three affected and three healthy) and four sisters (fig. 1D) that was identified at the Centre for Reproductive Medicine of the Dutch-Speaking Brussels Free University. The three unaffected brothers fathered seven, six, and five children, respectively, but the three affected brothers were childless and presented with a fertility disorder due to oligoasthenoteratozoospermia, showing the characteristics of total globozoospermia, such as roundheadedness and acrosomelessness, as shown by acrosin (MIM 102480) staining in figure 1A. No known consanguinity was reported, although the family belonged to an isolated Jewish population. A normal karyotype and no Y-chromosome microdeletion were found. In two brothers, ICSI was performed, but fertilization was poor, and no pregnancy occurred.
Figure 1. .
Family with globozoospermia and a mutation in the SPATA16 gene. A, Sperm morphology. Fluorescent acrosin (green) staining (with fluorescein) of acrosomes and 4′,6-diamidino-2-phenylindole (blue) staining of nuclei. On the left is a sample from a fertile control, in which the most important content of the acrosome (acrosin) is clearly and abundantly present; on the right is a sample from a patient. Sperm morphology and acrosome structures are severely disrupted in patient cells. Remnants of acrosin staining were observed for some deformed sperm cells, but most signals represent nonspecific acrosin staining in the leukocytes. B, Chromatograms of the mutation. Shown are the sequences from a control sample, the heterozygous father, and one of the patients. C, The NciI recognition site (5′-CCCGG-3′) is lost because of the G→A mutation at the last nucleotide of exon 4. (The HpaII recognition site is not shown but overlaps at 5′-CCGG-3′). This mutation predicts a R283Q amino acid substitution, as well as the disruption of the 5′ splice site of intron 4. D, Pedigree of the Ashkenazi Jewish family in this study. The order of the 10 siblings is arbitrary. The segregation of the mutation was studied by NciI digestion of a PCR amplification of exon 4 and its flanking sequences. The first lane is the marker lane. As asterisk (*) indicates tested individuals. The two parents and two siblings are heterozygous for the mutation. The three affected males are homozygous, and one unaffected male and a control (C) are not carriers of the mutation.
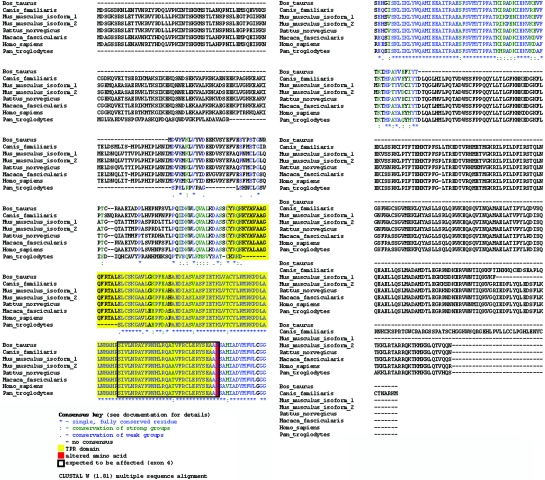
We performed a genomewide scan analysis of all six brothers, using a 10K SNP array (Affymetrix GeneChip). Regions of homozygosity were defined by the presence of >25 consecutive homozygous SNPs. Large regions of homozygosity were observed in all six individuals (tables 1 and 2), indicating consanguinity in the second or third degree in the family. Therefore, we considered this family to be consanguineous and expected the pathology to be autosomal recessive. We identified a unique region of haplotypic identical homozygosity shared by all affected brothers, in which the healthy brothers were heterozygous. The smallest region of overlap spanned 17 Mb of chromosome 3q26 (167054711–184087390). This region contains ∼50 known genes in the UCSC Genome Browser. We selected the SPATA16 gene (spermatogenesis-associated 16, also known as “NYD-SP12”) as the most plausible candidate gene, because recent studies showed that SPATA16 is specifically expressed in human testis and that the mouse ortholog is primarily expressed in the spermatocyte and spermatids.13 Localization in the Golgi apparatus and the shift with Golgi vesicles to the acrosome was observed in round and elongated spermatids by use of a SPATA19-GTP (green fluorescent protein) fusion protein, strongly suggesting a role for the SPATA16 protein in acrosome formation during spermiogenesis.14 SPATA16 is composed of 11 exons encoding a highly conserved protein of 65 kDa (569 aa), which contains a tetratricopeptide repeat (TPR [MIM 602259]) domain. Sequence alignment (by use of ClustalW 1.81 [SDSC Biology Workbench]) (fig. 2) shows that SPATA16 is highly conserved across mammals, exhibiting an identity rate varying from 77% (mouse) to 96% (chimpanzee) (NCBI Blast) (table 3). The conservation is even higher (92% and 98% in mouse and chimpanzee, respectively) for the TPR domain, a protein-protein interaction domain commonly but exclusively found in cochaperone proteins.15
Table 1. .
Results of the SNP Array for the Three Affected Brothers[Note]
| Genotype Call in Sibling |
||||
| Chromosome 3 Position |
SNP | 1 | 2 | 3 |
| 165727121 | SNP_A-1508753 | AA | AA | AA |
| 167054711 | SNP_A-1512676 | AB | AB | AB |
| 167631594 | SNP_A-1508627 | AA | AA | AA |
| 167801377 | SNP_A-1519252 | AA | AA | AA |
| 169050879 | SNP_A-1511126 | BB | BB | BB |
| 169140975 | SNP_A-1509483 | AA | AA | AA |
| 169748882 | SNP_A-1518417 | BB | BB | BB |
| 170222552 | SNP_A-1518965 | BB | BB | BB |
| 170229669 | SNP_A-1509719 | BB | BB | BB |
| 170266790 | SNP_A-1519387 | BB | BB | BB |
| 170341708 | SNP_A-1513150 | BB | BB | BB |
| 170440150 | SNP_A-1513458 | AA | AA | AA |
| 170815817 | SNP_A-1516975 | AA | AA | AA |
| 171062673 | SNP_A-1514614 | No Call | AA | No Call |
| 171141972 | SNP_A-1508020 | BB | BB | BB |
| 172368188 | SNP_A-1510308 | AA | AA | AA |
| 172643136 | SNP_A-1517656 | BB | BB | BB |
| 172771949 | SNP_A-1509479 | AA | AA | AA |
| 172933454 | SNP_A-1509435 | AA | AA | AA |
| 172933529 | SNP_A-1509382 | BB | BB | BB |
| 173575998 | SNP_A-1516388 | BB | BB | BB |
| 173580444 | SNP_A-1514288 | BB | BB | BB |
| 173580729 | SNP_A-1514241 | AA | AA | AA |
| 174671975 | SNP_A-1507368 | AA | AA | AA |
| 175782878 | SNP_A-1508795 | BB | BB | BB |
| 176537318 | SNP_A-1511779 | AA | AA | AA |
| 176610712 | SNP_A-1509801 | BB | BB | BB |
| 176827123 | SNP_A-1511813 | AA | AA | AA |
| 177333023 | SNP_A-1517824 | AA | AA | AA |
| 177621541 | SNP_A-1518682 | BB | BB | BB |
| 177708137 | SNP_A-1510834 | AA | AA | AA |
| 177723240 | SNP_A-1516780 | BB | BB | BB |
| 178105620 | SNP_A-1516425 | AA | AA | AA |
| 178105781 | SNP_A-1515959 | No Call | BB | No Call |
| 179324469 | SNP_A-1517000 | No Call | AA | No Call |
| 179597123 | SNP_A-1516215 | AA | AA | AA |
| 179597352 | SNP_A-1516746 | AA | AA | AA |
| 179636158 | SNP_A-1512810 | BB | BB | BB |
| 179706441 | SNP_A-1510950 | BB | BB | BB |
| 179839122 | SNP_A-1514173 | BB | BB | BB |
| 180413909 | SNP_A-1511671 | AA | AA | AA |
| 180729255 | SNP_A-1508156 | BB | BB | BB |
| 181028753 | SNP_A-1512457 | BB | BB | BB |
| 181180943 | SNP_A-1507868 | BB | BB | BB |
| 181251555 | SNP_A-1512336 | AA | AA | AA |
| 181298402 | SNP_A-1513747 | AA | AA | AA |
| 181506449 | SNP_A-1517808 | BB | BB | BB |
| 181552274 | SNP_A-1509494 | BB | BB | BB |
Note.— Several large areas of shared haplotype were identified, indicating consanguinity. The area of shared haplotype on which we concentrated is shown.
Table 2. .
Areas of Shared Haplotype and Shared Homozygosity[Note]
| Shared Haplotype | Shared Homozygosity | ||||||
| Chromosome and Start Position | Stop Position | Fragment Length (bp) |
No. of SNPsa | Start Position | Stop Position | Fragment Length (bp) |
No. of SNPsa |
| 1: | |||||||
| 33955677 | 36556426 | 2,600,749 | 15 | ||||
| 117399167 | 119296010 | 1,896,843 | 11 | 117866019 | 119296010 | 1,429,991 | 6 |
| 151489481 | 156376556 | 4,887,075 | 11 | 151489481 | 156376556 | 4,887,075 | 11 |
| 160643582 | 162290883 | 1,647,301 | 14 | ||||
| 194965564 | 198362174 | 3,396,610 | 10 | ||||
| 2: | |||||||
| 19716714 | 34386124 | 14,669,410 | 55 | ||||
| 54483803 | 57217291 | 2,733,488 | 14 | ||||
| 59155900 | 65801963 | 6,646,063 | 11 | ||||
| 65801963 | 107509848 | 41,707,885 | 91 | ||||
| 113469814 | 115959977 | 2,490,163 | 12 | ||||
| 212719583 | 215032622 | 2,313,039 | 10 | ||||
| 224103332 | 226044921 | 1,941,589 | 16 | ||||
| 234216839 | 239151790 | 4,934,951 | 14 | 234719101 | 239151790 | 4,432,689 | 8 |
| 3: | |||||||
| 653347 | 3558063 | 2,904,716 | 12 | ||||
| 100634082 | 103786422 | 3,152,340 | 10 | ||||
| 113839242 | 127152417 | 13,313,175 | 48 | ||||
| 165655532 | 184087390 | 18,431,858 | 49 | 167054711 | 184087390 | 17,032,679 | 47 |
| 184087390 | 190712906 | 6,625,516 | 26 | ||||
| 4: | |||||||
| 173190930 | 176925273 | 3,734,343 | 10 | 173530324 | 176925273 | 3,394,949 | 8 |
| 181334851 | 191091333 | 9,756,482 | 48 | ||||
| 5: | |||||||
| 120723042 | 122283900 | 1,560,858 | 11 | 121110284 | 122283900 | 1,173,616 | 7 |
| 134671015 | 139500740 | 4,829,725 | 15 | ||||
| 6: | |||||||
| 28766533 | 31094058 | 2,327,525 | 10 | 29479394 | 31094058 | 1,614,664 | 6 |
| 46333713 | 47986997 | 1,653,284 | 14 | 46824038 | 47986997 | 1,162,959 | 5 |
| 96879221 | 102207593 | 5,328,372 | 12 | ||||
| 105174572 | 108446020 | 3,271,448 | 10 | ||||
| 108446020 | 52383480 | 43,937,460 | 169 | ||||
| 7: | |||||||
| 77627148 | 131407468 | 53,780,320 | 169 | ||||
| 8: | |||||||
| 53838124 | 57959516 | 4,121,392 | 15 | ||||
| 72728798 | 76200122 | 3,471,324 | 10 | ||||
| 9: | |||||||
| 507715 | 13460671 | 12,952,956 | 85 | 12445236 | 13460671 | 1,015,435 | 6 |
| 72760108 | 75532057 | 2,771,949 | 10 | 73265909 | 75532057 | 2,266,148 | 7 |
| 75532057 | 90138831 | 14,606,774 | 56 | ||||
| 10: | |||||||
| 63884288 | 66164664 | 2,280,376 | 10 | ||||
| 66164664 | 68113302 | 1,948,638 | 10 | ||||
| 91501439 | 105835217 | 14,333,778 | 47 | ||||
| 14: | |||||||
| 19490525 | 22620727 | 3,130,202 | 20 | ||||
| 31508337 | 33265230 | 1,756,893 | 10 | ||||
| 36021565 | 37647284 | 1,625,719 | 10 | ||||
| 15: | |||||||
| 21490270 | 23325412 | 1,835,142 | 13 | ||||
| 23325412 | 59077153 | 35,751,741 | 127 | ||||
| 79723090 | 84387340 | 4,664,250 | 12 | ||||
| 16: | |||||||
| 22705353 | 50026393 | 27,321,040 | 24 | ||||
| 17: | |||||||
| 28942222 | 34693022 | 5,750,800 | 10 | 29183029 | 34693022 | 5,509,993 | 9 |
| 66657008 | 72151125 | 5,494,117 | 10 | ||||
| 18: | |||||||
| 63746898 | 66906214 | 3,159,316 | 13 | 64766169 | 66906214 | 2,140,045 | 9 |
| 20: | |||||||
| 38790879 | 43185196 | 4,394,317 | 10 | ||||
| 21: | |||||||
| 18764912 | 21113081 | 2,348,169 | 11 | 18764912 | 21113081 | 2,348,169 | 11 |
| 29925652 | 31842275 | 1,916,623 | 12 | ||||
| 36234195 | 37974748 | 1,740,553 | 10 | 36447405 | 37974748 | 1,527,343 | 7 |
Note.— In the left part of the table, a selection of the areas of shared haplotype (those with (>9 SNPs) is displayed. In the right part of the table, the regions of shared homozygosity (with >4 SNPs) that lie within are shown.
Number of SNPs that form the area of shared haplotype or homozygosity.
Figure 2. .
Sequence alignment of SPATA16
Table 3. .
Identity Rates among Species for the SPATA16 Sequence and the TPR Domain of the Protein
| Identity with Human(%) |
Positivesa(%) |
Gapsb(%) |
||||
| Species | SPATA16 | TPR Domain | SPATA16 | TPR Domain | SPATA16 | TPR Domain |
| Homo sapiens | 100 | 100 | 100 | 100 | 0 | 0 |
| Bos taurus | 87 | 94 | 92 | 97 | 0 | 0 |
| Canis familiaris | 83 | 97 | 89 | 97 | 0 | 0 |
| Macaca fascicularis | 95 | 97 | 97 | 97 | 0 | 0 |
| Mus musculus isoform 1 | 77 | 92 | 86 | 96 | 0 | 0 |
| M. musculus isoform 2 | 78 | 92 | 87 | 96 | 0 | 0 |
| Pan troglodytes | 96 | 98 | 98 | 98 | 0 | 0 |
| Rattus norvegicus | 80 | 93 | 87 | 96 | 0 | 0 |
Amino acid positive-match score.
Space introduced into an alignment to compensate for insertions and deletions in one sequence relative to another. In our search, the gaps did not exceed 0.5%.
Sequence analysis of one of the affected sons revealed a homozygous sequence variation in exon 4 (c.848G→A), which disrupts a NciI or an HpaII recognition site (fig. 1C). Restriction-enzyme analysis revealed that the three affected brothers are homozygous and that the two parents and two healthy brothers are heterozygous for the mutation. The third unaffected brother appeared to be homozygous for the wild-type sequence (fig. 1D). The c.848G→A nucleotide variation is not known in any SNP database and was not identified in 231 controls, including 151 random controls of both sexes and 80 fertile males.
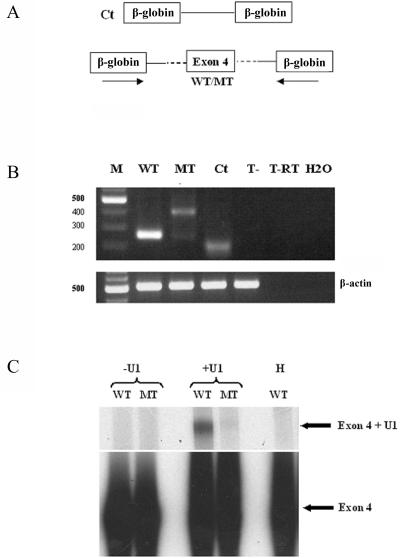
The mutation predicts an amino acid change of a highly conserved residue (p.R283Q) located at the C-terminal end of the highly conserved TPR domain. In addition, the c.848G→A mutation affects the last nucleotide of exon 4 (fig. 1C) and, therefore, may disrupt the 5′ splice site of intron 4. Three different splice-site prediction models predicted that the mutation disrupts this splice site (table 4). Unfortunately, the SPATA16 protein presents a testis-restricted expression, and we were not allowed to use fresh sperm cells or to perform a biopsy in these religious patients to verify the predicted aberrant splicing in vivo. Therefore, minigene constructs were made that consisted of two constitutive β-globin exons surrounding a 420-bp fragment containing either the wild-type or the mutated form of exon 4 and the flanking intronic sequences of SPATA16. These minigene constructs were transfected into COS1 or HeLa cells, and transcripts were analyzed by RT-PCR 24 h after the transfection (fig. 3A). As shown in figure 3B, wild-type exon 4 is invariably included in the final mRNA, as confirmed by the sequencing of the PCR product. In sharp contrast, the mutated exon gives rise to two aberrant splicing forms, as shown by cloning and sequencing of these PCR products. The most prominent, larger product is the result of the use of a splice site situated in the β-globin intron used for the minigene construct. The weaker, smaller product corresponds to the use of a cryptic splice site situated 18 bp upstream of the normal splice site. These aspecific products are likely the result of the very short intron sequences in the minigene construct. Such products are often seen in exon-trapping experiments in the absence of a bona fide splice site and are indicative of the occurrence of exon skipping due to the mutation.16,17 Importantly, we did not detect any transcript containing the correct junctions from the mutated exon 4, indicating that the mutation hinders normal splicing.
Table 4. .
Splice-Site Predictions from Three Web Sites
| Odds Ratio for Sequence |
||
| Web Sitea | Wild Type | Mutant |
| NetGene2 Server | .80 | <.50 |
| SpliceSiteFinder | .805 | .681 |
| Splice Site Prediction by Neural Network | .97 | <.40 |
See the Web Resources for URLs.
Figure 3. .
Mutated donor splice site of SPATA16 intron 4 and U1 SnRNP binding to wild-type (WT) and mutant (MT) donor splice sites. A, Overview of the used prediction sites. B, Minigene constructs used to test the splicing of exon 4. T- = construct without exon; T-RT = construct without exon in RT-PCR. C, Gel showing that wild-type exon 4 is invariably included in the final mRNA. In sharp contrast, the mutant exon gives rise to two aberrant splicing forms. M = marker lane. D, U1 SnRNP binding analyzed by psoralen-mediated UV-crosslinking experiments, revealing that mutant exon 4 is not recognized by the splicing machinery, whereas wild-type exon 4 is clearly recognized. The identity of the U1 SnRNP was confirmed by RNAse H treatment with use of an oligodeoxynucleotide complementary to nucleotide positions 1–15 of U1 SnRNA.
The first and critical step of exon inclusion is the binding of the U1 small nuclear ribonucleoprotein (SnRNP) splicing factor to the 5′ splice sites.18 To confirm that the mutated SPATA16 exon 4 is not recognized by the splicing machinery, we checked its binding to the U1 SnRNP by psoralen-mediated UV crosslinking. Whereas binding of U1 SnRNP to wild-type DNA was readily detected, this was not observed when DNA carrying the c.848G→A mutation was used as template (fig. 3C). The identity of the U1 SnRNP was confirmed by RNAse H treatment by use of an oligodeoxynucleotide complementary to positions 1–15 of U1 SnRNA.19 Therefore, the results of the bioinformatic prediction, the minigene, and the U1 binding strongly suggest that the c.848G→A mutation leads to inappropriate splicing of exon 4 and, therefore, the disruption of the TPR domain.
SPATA16 was also analyzed in 29 patients with globozoospermia, including 6 familial cases involving 14 patients. Of the 29 patients, 12 presented with total globozoospermia, and 17 with partial globozoospermia. None of them presented with any variation in the SPATA16 sequence, with the exception of three known polymorphisms and two point mutations that did not segregate with the disease (table 5).
Table 5. .
Polymorphisms[Note]
| Segregation in |
|||||
| Patient(s), Type of Globozoospermia, and Variation |
Exon | Known SNP | Parents | Siblings | Ethnicity |
| 1: | |||||
| Partial: | |||||
| c.232G→A (p.E78K) | 2 | Yes | … | … | European |
| c.397A→G (p.M133V) | 2 | Yes | … | … | European |
| 2: | |||||
| Partial: | |||||
| c.232G→A (p.E78K) | 2 | Yes | … | … | European |
| c.397A→G (p.M133V) | 2 | Yes | … | … | European |
| c.1526C→T (p.A509V) | 10 | No | Mother | Absent in affected brother | European |
| c.1577T→C (p.M526T) | 10 | No | Mother | Absent in affected brother | European |
| 3 and 4: | |||||
| Partial: | |||||
| c.232G→A (p.E78K) | 2 | Yes | … | … | European |
| c.397A→G (p.M133V) | 2 | Yes | … | … | European |
| 5: | |||||
| Partial: | |||||
| c.232G→A (p.E78K) | 2 | Yes | … | Absent in affected brother | North African |
| c.397A→G (p.M133V) | 2 | Yes | … | … | … |
| c.440G→A (p.G147E) | 2 | Yes | … | … | … |
| 7 and 8: | |||||
| Partial: | |||||
| c.232G→A (p.E78K) | 2 | Yes | … | Patients are brothers | European |
| c.397A→G (p.M133V) | 2 | Yes | … | … | … |
| c.440G→A (p.G147E) | 2 | Yes | … | … | … |
| 9: | |||||
| Total: | |||||
| c.397A→G (p.M133V) | 2 | Yes | … | … | European |
| 10: | |||||
| Total: | |||||
| c.232G→A (p.E78K) | 2 | Yes | … | … | North African |
| c.397A→G (p.M133V) | 2 | Yes | … | … | … |
| c.440G→A (p.G147E) | 2 | Yes | … | … | … |
Note.— All nonsynonymous coding variations that were found (in 9 of 28 patients) are shown. Three known SNPs were identified, located next to two unknown but nonsegregating variations. In patients 6 and 11–28, no nonsynonymous coding variations were identified (dbSNP database).
This is, to our knowledge, the first description of a gene involved in the pathogenesis of human globozoospermia. The data strongly suggest that the identified homozygous mutation in SPATA16 causes globozoospermia in three of six brothers in the family studied, which allows us to state that globozoospermia can be a genetic trait with an autosomal recessive mode of transmission. The SPATA16 protein localizes to the Golgi apparatus and to the proacrosomic vesicles that are transported to the acrosome in round and elongated spermatids during spermiogenesis. Our observations support the hypothesis of a crucial role for SPATA16 in acrosome formation.14 The strongest protein conservation is seen in the TPR domain, which is disrupted in these cases of globozoospermia. The TPR domain is known to mediate protein-protein interactions and assembly of multiprotein complexes. Study of the x-ray structure revealed that the TPR domain adopts a helix-turn-helix arrangement, with the ability to associate with other α-helical structures. A possible interacting protein may be from the gene GOPC, a Golgi-associated protein containing coiled-coil motif α-helices,10 or from the HIV-1 rev binding protein gene (HRB), which also localizes to the Golgi complex.9 Both these genes are involved in the pathogenesis of globozoospermia in mouse models. Finally, it is worth noting that SPATA16 contains six casein kinase II phosphorylation sites and that the casein kinase IIa′ is the most abundant casein kinase in the testis,8 for which the knockout model shows acrosome and other morphological defects.20 Moreover, the existence of several candidate genes8–10 suggests genetic heterogeneity in human globozoospermia, which could be a reason why we did not find other patients with a gene alteration in SPATA16. Noteworthy as well is the fact that the heterozygous mouse models show no sperm abnormalities. This indicates that mutation carriers should have normal fertility. In this family, this seems to be the case, since the father and two heterozygous brothers have fathered 10, 7, and 6 children, respectively. In two of the affected brothers, ICSI was performed to induce fertilization and pregnancy, but without success. This is in accordance with the literature, which shows that ICSI enables oocyte fertilization, but with low fertilization rates in about half of the cases.
Since male infertility does not respect the canonical rule of genetics, the determination of inheritance patterns and the elucidation of genetic causes are complicated. Several genetic factors have been described that affect male fertility,21 but these give rise to more complex phenotypes. However, the patients in this study did not show any mental or physical abnormalities—in particular, no andrological abnormalities—in addition to their aberrant semen analysis. Thus, the mutation in SPATA16 that we found in this study appears to present a human gene in which mutations give rise to male infertility without any associated other anomalies.
Further studies of other patients may help to identify other participant genes involved of the formation of the acrosome, allowing the fine dissection of the mechanisms involved in the setup of such a specialized cellular organelle. SPATA16 defects influence spermiogenesis, whereas meiosis is not disturbed. Thus, modulation of SPATA16 function or that of other components in the same pathway could offer an innovative, reversible approach to male contraception that is not based on controlling the hormonal pathway of sperm production.
Acknowledgments
We thank N. Dondaine, M. Jochem, J.-C. Nicod, and L. Ramos for precious technical assistance. We are grateful to the Institute of Genetics and Molecular and Cellular Biology Services and the Department of Human Genetics for their invaluable assistance. This work was supported by the French Centre National de la Recherche Scientifique, the Institut National de la Santé et de la Recherche Médicale, the Ministère de l’Education Nationale, the l’Enseignement Supérieur et de la Recherche, the Louis Pasteur University of Strasbourg, and the Radboud University Nijmegen Medical Centre.
Web Resources
Accession numbers and URLs for data presented herein are as follows:
- BLAST, http://www.ncbi.nlm.nih.gov/BLAST/Blast.cgi?
- dbSNP, http://www.ncbi.nlm.nih.gov/SNP/ (for exon 10 c.1526C→T [accession number ss73688634], exon 10 c.1577T→C [accession number ss73688636], and exon 4 c.848G→A [accession number ss73688635])
- NetGene2 Server, http://www.cbs.dtu.dk/services/NetGene2/
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for globozoospermia, CSNK2A2, HRB, GOPC, SPATA16, acrosin, and TPR)
- SDSC Biology Workbench, http://workbench.sdsc.edu/ (for Biology Workbench 3.2 and ClustalW 1.81)
- SpliceSiteFinder, http://www.genet.sickkids.on.ca/~ali/splicesitefinder.html
- Splice Site Prediction by Neural Network, http://www.fruitfly.org/seq_tools/splice.html
- UCSC Genome Browser, http://genome.cse.ucsc.edu/ (for March 2006 version)
References
- 1.World Health Organization (1999) WHO laboratory manual for the examination of human semen and sperm–cervical mucus interaction. 4th ed. Cambridge University Press, Cambridge, United Kingdom [Google Scholar]
- 2.Dam AH, Feenstra I, Westphal JR, Ramos L, van Golde RJ, Kremer JA (2007) Globozoospermia revisited. Hum Reprod Update 13:63–75 10.1093/humupd/dml047 [DOI] [PubMed] [Google Scholar]
- 3.Holstein AF, Schirren CG, Schirren C, Mauss J (1973) [Round headed spermatozoa: a cause of male infertility.] Dtsch Med Wochenschr 98:61–62 [DOI] [PubMed] [Google Scholar]
- 4.Schirren CG, Holstein AF, Schirren C (1971) Uber die Morphogenese rundkopfiger Spermatozoen des Menschen. Andrologie 3:117–125 [Google Scholar]
- 5.Dale B, Iaccarino M, Fortunato A, Gragnaniello G, Kyozuka K, Tosti E (1994) A morphological and functional study of fusibility in round-headed spermatozoa in the human. Fertil Steril 61:336–340 [DOI] [PubMed] [Google Scholar]
- 6.Florke-Gerloff S, Topfer-Petersen E, Muller-Esterl W, Mansouri A, Schatz R, Schirren C, Schill W, Engel W (1984) Biochemical and genetic investigation of round-headed spermatozoa in infertile men including two brothers and their father. Andrologia 16:187–202 [DOI] [PubMed] [Google Scholar]
- 7.Kilani Z, Ismail R, Ghunaim S, Mohamed H, Hughes D, Brewis I, Barratt CL (2004) Evaluation and treatment of familial globozoospermia in five brothers. Fertil Steril 82:1436–1439 10.1016/j.fertnstert.2004.03.064 [DOI] [PubMed] [Google Scholar]
- 8.Xu X, Toselli PA, Russell LD, Seldin DC (1999) Globozoospermia in mice lacking the casein kinase II α′ catalytic subunit. Nat Genet 23:118–121 10.1038/12729 [DOI] [PubMed] [Google Scholar]
- 9.Kang-Decker N, Mantchev GT, Juneja SC, McNiven MA, Van Deursen JM (2001) Lack of acrosome formation in Hrb-deficient mice. Science 294:1531–1533 10.1126/science.1063665 [DOI] [PubMed] [Google Scholar]
- 10.Yao R, Ito C, Natsume Y, Sugitani Y, Yamanaka H, Kuretake S, Yanagida K, Sato A, Toshimori K, Noda T (2002) Lack of acrosome formation in mice lacking a Golgi protein, GOPC. Proc Natl Acad Sci USA 99:11211–11216 10.1073/pnas.162027899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Christensen GL, Ivanov IP, Atkins JF, Campbell B, Carrell DT (2006) Identification of polymorphisms in the Hrb, GOPC, and Csnk2a2 genes in two men with globozoospermia. J Androl 27:11–15 10.2164/jandrol.05087 [DOI] [PubMed] [Google Scholar]
- 12.Pirrello O, Machev N, Schimdt F, Terriou P, Menezo Y, Viville S (2005) Search for mutations involved in human globozoospermia. Hum Reprod 20:1314–1318 10.1093/humrep/deh799 [DOI] [PubMed] [Google Scholar]
- 13.Xu M, Xiao J, Chen J, Li J, Yin L, Zhu H, Zhou Z, Sha J (2003) Identification and characterization of a novel human testis-specific Golgi protein, NYD-SP12. Mol Hum Reprod 9:9–17 10.1093/molehr/gag005 [DOI] [PubMed] [Google Scholar]
- 14.Lu L, Lin M, Xu M, Zhou ZM, Sha JH (2006) Gene functional research using polyethylenimine-mediated in vivo gene transfection into mouse spermatogenic cells. Asian J Androl 8:53–59 10.1111/j.1745-7262.2006.00089.x [DOI] [PubMed] [Google Scholar]
- 15.Smith DF (2004) Tetratricopeptide repeat cochaperones in steroid receptor complexes. Cell Stress Chaperones 9:109–121 10.1379/CSC-31.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wieringa B, Meyer F, Reiser J, Weissmann C (1983) Unusual splice sites revealed by mutagenic inactivation of an authentic splice site of the rabbit beta-globin gene. Nature 301:38–43 10.1038/301038a0 [DOI] [PubMed] [Google Scholar]
- 17.Treisman R, Orkin SH, Maniatis T (1983) Specific transcription and RNA splicing defects in five cloned beta-thalassaemia genes. Nature 302:591–596 10.1038/302591a0 [DOI] [PubMed] [Google Scholar]
- 18.Baralle D, Baralle M (2005) Splicing in action: assessing disease causing sequence changes. J Med Genet 42:737–748 10.1136/jmg.2004.029538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Forch P, Puig O, Kedersha N, Martinez C, Granneman S, Seraphin B, Anderson P, Valcarcel J (2000) The apoptosis-promoting factor TIA-1 is a regulator of alternative pre-mRNA splicing. Mol Cell 6:1089–1098 10.1016/S1097-2765(00)00107-6 [DOI] [PubMed] [Google Scholar]
- 20.Escalier D, Silvius D, Xu X (2003) Spermatogenesis of mice lacking CK2α′: failure of germ cell survival and characteristic modifications of the spermatid nucleus. Mol Reprod Dev 66:190–201 10.1002/mrd.10346 [DOI] [PubMed] [Google Scholar]
- 21.Matzuk MM, Lamb DJ (2002) Genetic dissection of mammalian fertility pathways. Nat Cell Biol Suppl 4:s41–s49 [DOI] [PubMed] [Google Scholar]