Abstract
HLA-G is a nonclassic, class I HLA molecule that has important immunomodulatory properties. Previously, we identified HLA-G as an asthma-susceptibility gene and discovered that the risk of asthma in a child was determined by both the child’s HLA-G genotype and the mother’s affection status. Here we report a SNP in the 3′ untranslated region of HLA-G that influences the targeting of three microRNAs (miRNAs) to this gene, and we suggest that allele-specific targeting of these miRNAs accounts, at least in part, for our earlier observations on HLA-G and the risk of asthma.
The class Ib HLA gene, HLA-G (MIM 142871), is primarily expressed by fetal cells at the maternal-fetal interface, where it displays immunosuppressive properties, and is thought to contribute to the maternal tolerance of the genetically foreign fetus.1 We previously identified HLA-G as an asthma (MIM 600807) gene in a positional cloning study in white Chicago families.2 In the Chicago families and in a Dutch population, the risk genotype at HLA-G varied depending on whether the mother had asthma or bronchial hyperresponsiveness (BHR) or was unaffected. For example, in both populations, the GG genotype at SNP −964G/A in the promoter region of HLA-G was associated with asthma in children of affected mothers, whereas the AA genotype was associated with asthma in children of unaffected mothers. A similar interaction was observed with +1489C/T, the SNP that explained nearly the entire linkage signal in the Chicago families.2 We then focused our studies on the highly polymorphic promoter region and showed that the −964G/A SNP tagged two major haplotype clades with evidence of longstanding balancing selection,3 but we could not demonstrate functional differences between these two promoter clades by use of luciferase reporter assays.4 Last, we showed expression of soluble (s)HLA-G protein in bronchial epithelial cells in two individuals with asthma but not in one unaffected control individual, suggesting that this gene was up-regulated in the asthmatic lungs.2
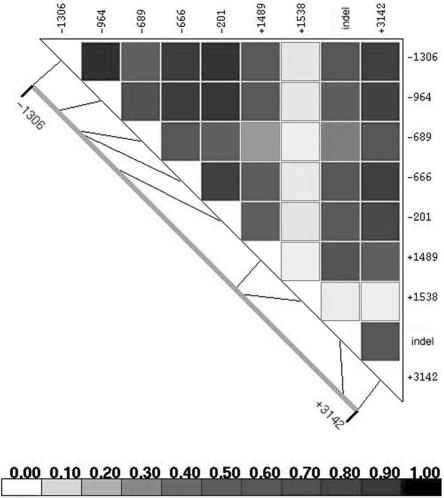
Because of the strong linkage disequilibrium (LD) between SNPs in the HLA-G gene (fig. 1), we turned our attention to other regions of the gene to identify variation that contributes to asthma risk. As part of these studies, we identified in the 3′ UTR of HLA-G a putative target site for three microRNAs (miRNAs): miR-148a, miR-148b, and miR-152. A C/G SNP at +3142 in the HLA-G mRNA is located in the seed region of this target site.
Figure 1. .
Pairwise linkage disequilibrium (r2) map of HLA-G in the Chicago families. The LD plot was made by LDPlotter.
miRNAs are a family of endogenous, small, noncoding RNAs, ∼22 nt in length, that negatively regulate gene expression by suppressing translation or degrading mRNA.5 They have important and diversified functions—including development and differentiation,6 apoptosis,7 hematopoiesis,8 and tumorigenesis,9 as well as insulin secretion10 and viral defense11—and have been shown to be involved in various human diseases, including cancer12 and fragile X syndrome.13 Although polymorphisms in miRNA genes may be scarce,14 polymorphisms in miRNA target sites are abundant. Some of these polymorphisms influence the binding of miRNAs and have been associated with muscularity in sheep and with Tourette syndrome.15,16 Here we demonstrate that +3142C/G influences miRNA targeting of HLA-G and is associated with risk of asthma.
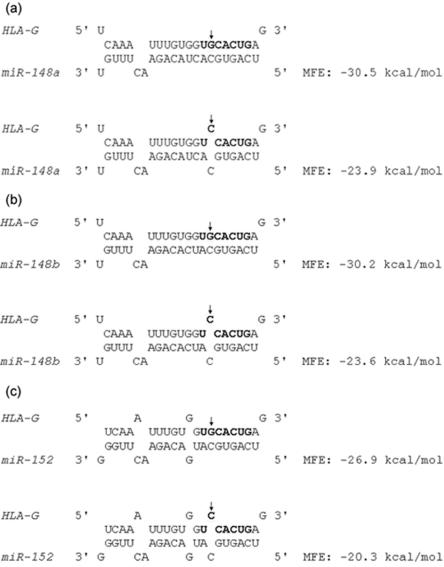
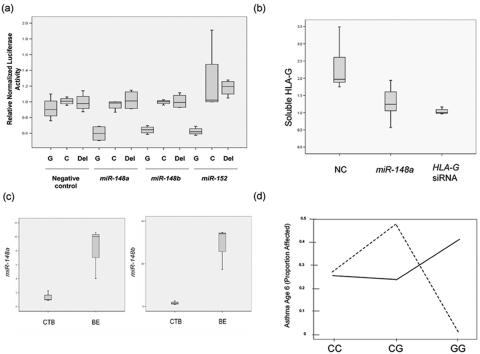
For miRNA target site predictions in HLA-G, we used MicroInspector,17 TargetScan,18 DIANA-microT,19 MiRanda,6 and PicTar,20 as well as our own algorithm, which is based on the criteria of Watson-Crick base-pair of the 5′ “2–8” nucleotides of miRNA to target sequence and more stable binding of miRNA to the target region than to the flanking region. On the basis of the results of those prediction programs, we selected three target sites for further study: target for miR-148a/148b/152, target for miR-143, and target for miR-524*. We used RNAhybrid,21 to analyze the targeting of miR-148a, miR-148b, and miR-152 to HLA-G. On the basis of the calculation of minimum free energy (MFE), the binding of miRNA to the +3142G allele is predicted to be more stable than binding to the +3142C allele (fig. 2). Thus, +3142 might influence the binding of miRNA to HLA-G, and therefore HLA-G protein expression, in an allele-specific manner. To test this hypothesis, we performed luciferase assays. We used firefly luciferase plasmid pLS-HX, which was constructed by inserting firefly luciferase ORF between SacI and XbaI of pCL-con,22 and renilla plasmid pRL, which was constructed by inserting renilla luciferase between SacI and EcoRI of pCL-con. A 365-bp region of HLA-G 3′ UTR, containing the insertion allele at the exon 8 insertion/deletion (indel),23 was amplified using forward (CTAGAAGCTTTGTGAAACAGCTGCCC) and reverse (CGCTAGTCTAGATGTCTCTCAAATTTC) primers, was digested with HindIII and XbaI, and was ligated into pLS-HX. Site-directed mutagenesis was performed using QuikChange II XL kit (Stratagene) to generate pluc-HLAG-G and pluc-HLAG-C that carry either C or G at HLA-G +3142, and pluc-HLA-G-Del, which has the miRNA target site (22 bp) deleted. We then cotransfected pluc-HLAG-G, pluc-HLAG-C, or pluc-HLA-Del and pRL with either miR-148a, miR-148b, miR-152, or a negative control miRNA (NC miRNA) (Dharmacon) into a human bronchial epithelial cell line, 16HBE14o-.24 For each well, 50 ng luciferase plasmid, 2 ng renilla plasmid, and 20 nM (final concentration) miRNA were transfected using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s protocol. Cells were lysed 40 h after transfection, and luciferase and renilla activities were determined by use of Dual-Luciferase Reporter kit (Promega) and Veritas Microplate Luminometer (Turner Biosystems). We found that, although normalized luciferase activity of either plasmid was not significantly different when cells were cotransfected with NC miRNA, the activity of pluc-HLAG-G in the presence of each of the three miRNAs was significantly lower than those of pluc-HLAG-C and pluc-HLA-G-Del. This indicates allele-specific targeting of the three miRNAs to HLA-G (fig. 3a). We were not able to confirm the target for miR-143 or the one for miR-524* by luciferase experimentation.
Figure 2. .
Predicted binding of miR-148a, miR-148b, and miR-152 to HLA-G. The seed region of the target site is shown in bold letters, and +3142C/G is indicated by an arrow. The minimum free energy (MFE) of the RNA duplex was analyzed by RNAhybrid.
Figure 3. .
HLA-G +3142C/G affects the targeting of miR-148a, miR-148b, and miR-152 to HLA-G and interacts with mother’s asthma status to determine risk of asthma in the child. a, Luciferase assays showing the allele-specific targeting of the three miRNAs to HLA-G in 16HBE14o- cells. pluc-HLAG-G (G), pluc-HLAG-C (C), or pluc-HLA-G-Del (Del) luciferase plasmid was cotransfected with negative control miRNA, miR-148a, miR-148b, or miR-152. At least six replicate assays were performed for each transfection. For each sample, luciferase activity was normalized by renilla activity and then normalized again by the median value of the pluc-HLAG-C plasmid within each miRNA transfection group. The P values for the difference in luciferase activity of the three plasmids are as follows. For NC miRNA transfection: G versus C, P>.05; G versus Del, P>.05. For miR-148a transfection: G versus C, P=.0002; G versus Del, P<.0001. For miR-148b transfection: G versus C, P<.0001; G versus Del, P<.0001. For miR-152 transfection: G versus C, P=.0002; G versus Del, P=.0016. b, Endogenous HLA-G expression is inhibited by miR-148a in JEG3 cells. JEG3 cells, which are +3142GG, were transfected with either negative control miRNA, miR-148a, or HLA-G siRNA as a positive control. A total of seven replicate assays were performed for each RNA. miR-148a and HLA-G siRNA significantly reduced the level of sHLA-G in JEG3 supernatant compared with the NC miRNA. sHLA-G level was normalized to the median of the HLA-G siRNA group. c, miR-148a and miR-148b levels in primary cytotrophoblast (CTB) cells from six individuals and primary bronchial epithelial (BE) cells from three individuals, determined by miRNA real-time PCR. miR-148a (P=.0016) and miR-148b (P=.0006) levels were significantly lower in CTB cells than in BE cells. d, +3142C/G interacts with maternal asthma status and is associated with asthma in the COAST children. Dashed lines are the children of mothers with asthma (N=58), and solid lines are the children of mothers without asthma (N=119). The interaction P=.0011.
To validate the presence of the miRNA target site in the endogenous HLA-G gene, we used a choriocarcinoma cell line JEG3 (ATCC), which constitutively expresses high levels of HLA-G and has the GG genotype at +3142. We transfected JEG3 cells with miR-148a, NC miRNA, or HLA-G small interfering RNA (siRNA) (positive control), and we measured soluble HLA-G level in cell supernatants 5 d after transfection by use of an HLA-G ELISA kit (Biovendor & Exbio). sHLA-G levels were significantly decreased by miR-148a and by HLA-G siRNA, compared with the NC miRNA (P=.019 and P=.0048, respectively), confirming the presence of a functional target site on the +3142G allele (fig. 3b). Next, we measured expression of the three miRNAs by real-time PCR in mRNA from primary cytotrophoblast (CTB) cells, which have high constitutive expression of HLA-G, and in primary bronchial epithelial cells,25 which have low constitutive expression of HLA-G. miRNA was extracted by mirVana miRNA isolation kit (Ambion), and miRNA real-time PCR was performed with 20 ng miRNA and mirVana miRNA detection kit and primer sets (Ambion). For normalization, U6 real-time PCR was also performed for these samples, and comparative Ct method was used to calculate the relative abundance of the miRNAs in these samples. The expression of miR-148a and miR-148b, but not of miR-152, was detected in bronchial epithelial cells and to a lesser extent in CTB cells (fig. 3c). The higher expression of the miRNAs in bronchial epithelial cells compared with cytotrophoblast cells is consistent with the inverse expression pattern of HLA-G, which is low in the former and high in the latter cells (unpublished data).
HLA-G +3142C/G is in high LD with the HLA-G promoter polymorphism that was associated with asthma in the Chicago and Dutch families (fig. 1). Thus, it is possible that +3142C/G could account for at least some of the previously observed association. To further explore this possibility, we studied a third population consisting of children participating in Childhood Origins of ASThma (COAST), a birth cohort study. Couples in which at least one parent had asthma or allergies were recruited into the COAST study in Madison, WI, during pregnancy.26 Their children are at high risk for developing asthma and have been followed from birth through age 6 years, at which time asthma was diagnosed. We genotyped 180 white COAST children for four polymorphisms in HLA-G: −1306G/A, which is in perfect LD with the promoter SNP (−964G/A) that was associated with asthma in the Chicago and Dutch families2 (fig. 1); +1489C/T, a synonymous SNP in exon 3 that explained most of the evidence for linkage in the Chicago families2; a 14-bp indel in the 3′ UTR that has been associated with mRNA stability,23 and +3142C/G. We tested for interaction effects between mother’s affection status (asthma vs. no asthma) and the child’s genotype on the development of asthma by age 6 years in this cohort by logistic regression, as described elsewhere.26 To assess the significance of this interaction, children’s asthma status was permuted 10,000 times, whereas mothers’ status and children’s genotypes were unchanged. For each generated data set, a log likelihood–ratio test was performed to compare the model with interaction to the one without interaction. Permuted P values (Pinteraction) were determined by the number of data sets that had smaller log likelihood P values than the original data set. Significant interactions (P<.01) were observed at all SNPs tested except the exon 3 synonymous +1489C/T SNP (table 1); however, the association with +3142C/G was the most significant (permutation Pinteraction=0.0011). In the COAST cohort, the GG genotype is protective against asthma among children of asthmatic mothers (0 of 20 asthmatic children were GG; P=.0036, Fisher’s exact test) but is associated with modest risk of asthma among children of nonasthmatic mothers (7 of 32 asthmatic children were GG; P=.39, Fisher’s exact test) (fig. 3d and table 1). Thus, consistent with our earlier report in two other white populations, the +3142G allele, or promoter alleles in strong LD with +3142G (r2=0.76 between +3142 and either −964 or −1306), is a susceptibility allele among children of asthmatic mothers, and the C allele may be a risk allele (albeit more modest) among children of nonasthmatic mothers. Interestingly, the three miRNA genes are located at regions that have been linked to asthma or IgE: 7p15 (miR-148a), 12q13 (miR-148b), and 17q21 (miR-152),27,28 raising the possibility that variation in the miRNA genes themselves might contribute to asthma risk, perhaps by interacting with HLA-G.
Table 1. .
Interaction of HLA-G Genotypes and Maternal Asthma Status on Asthma Risk in COAST Children[Note]
| No. of Children, by Affection Status |
|||||
| MothersAffected |
MothersUnaffected |
||||
| SNP and Genotype | Children Affected |
Children Unaffected |
Children Affected |
Children Unaffected |
Pinteraction |
| HLA-G −1306 (rs1736936): | .0045 | ||||
| GG | 6 | 10 | 14 | 35 | |
| AG | 14 | 15 | 11 | 43 | |
| AA | 0 | 11 | 5 | 9 | |
| HLA-G +1489 (rs1130356): | .0978 | ||||
| CC | 9 | 16 | 18 | 44 | |
| CT | 10 | 12 | 9 | 28 | |
| TT | 0 | 3 | 4 | 4 | |
| HLA-G exon 8 indel (rs16375): | .0142 | ||||
| DelDel | 6 | 15 | 14 | 38 | |
| InsDel | 14 | 14 | 14 | 44 | |
| InsIns | 0 | 9 | 4 | 8 | |
| HLA-G +3142 (rs1063320): | .0011 | ||||
| CC | 3 | 8 | 10 | 29 | |
| CG | 17 | 18 | 15 | 48 | |
| GG | 0 | 12 | 7 | 10 | |
Note.— The minor-allele frequencies at each polymorphism in the COAST children are 0.395 (−1306A), 0.293 (+1489T), 0.372 (exon 8 ins), and 0.461 (+3142G).
It has become evident in recent years that gene-environment interactions play an important role in the etiology of common diseases,29,30 and it has even been suggested that genes with main effects on disease risk are more the exception than the rule.31 Identification and characterization of such interactions is critical for both basic research and clinical practice by helping to explain some of the inconsistencies of disease-association studies and by identifying individuals with genotype-specific risks for detrimental environmental exposures. Here we observed that a functional polymorphism in HLA-G interacts with mother’s asthma status, which represents an “in utero” environmental exposure. This interaction has now been observed in three white populations ascertained using three different study designs: families ascertained through affected sib pairs in the Chicago study,2 families ascertained through an affected parent originally studied 20 years earlier in the Dutch study,32 and children participating in a birth cohort study.26 Thus, the finding of an interaction between maternal affection status and child’s HLA-G genotype seems robust to study design and is particularly intriguing for two reasons. First, maternal asthma remains the most significant and best replicated risk factor for asthma in children,26,33 and, second, HLA-G is most highly expressed during pregnancy, when it is thought to play a key role in modulating immune tolerance toward the genetically foreign fetus by enhancing the Th2 arm of the immune system.1 Asthma and allergic disease are also characterized by a skewing toward Th2 immunity. Our study suggests that maternal asthma influences children’s risk in an allele-specific manner and that the immunosuppressive (Th2-skewing) properties of HLA-G promote asthma pathogenesis.
Although the mechanism of this interaction is still unknown, the results of this study suggest that fetal miRNA regulation might differ in pregnancies of asthmatic and nonasthmatic mothers. The fetal lung is bathed in amniotic fluid that is swallowed by the fetus, and this could provide a direct route for maternal factors to modulate fetal gene expression in the lung as an epigenetic phenomenon. Regardless of mechanism, however, the discovery that a 3′ UTR polymorphism affects miRNA targeting reveals a novel functional mechanism by which noncoding polymorphisms can contribute to the risk of common disease. Because ∼30% of human genes are regulated by miRNAs,18 the involvement of miRNA regulation in human diseases may be quite prevalent. This might be particularly relevant to common diseases, to which many susceptibility loci with small effects on gene expression likely contribute. Our study suggests that allele-specific miRNA targeting may indeed be a common mechanism of human disease pathogenesis.
Acknowledgments
We thank Ligang Wu and Joel Belasco of New York University for providing luciferase and renilla plasmids pLS-HX and pRL and for giving valuable suggestions. We also thank Andrew M. Shon and Matthew G. Schwartz for technical assistance. This work was supported by National Institutes of Health grants HL72414, HL70831, and HD21244.
Web Resources
Accession numbers and URLs for data presented herein are as follows:
- dbSNP, http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?CMD=search&DB=snp (for HLA-G —1306A/G [accession number rs1736936], HLA-G +1489C/T [accession number rs1130356], HLA-G exon 8 indel [accession number rs16375], and HLA-G +3142C/G [accession number rs1063320])
- LDPlotter, http://innateimmunity.net/IIPGA2/Bioinformatics/
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for HLA-G and asthma)
- RNAhybrid, http://bibiserv.techfak.uni-bielefeld.de/rnahybrid/submission.html
References
- 1.Hunt JS, Petroff MG, McIntire RH, Ober C (2005) HLA-G and immune tolerance in pregnancy. FASEB J 19:681–693 10.1096/fj.04-2078rev [DOI] [PubMed] [Google Scholar]
- 2.Nicolae D, Cox NJ, Lester LA, Schneider D, Tan Z, Billstrand C, Kuldanek S, Donfack J, Kogut P, Patel NM, et al (2005) Fine mapping and positional candidate studies identify HLA-G as an asthma susceptibility gene on chromosome 6p21. Am J Hum Genet 76:349–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tan Z, Shon AM, Ober C (2005) Evidence of balancing selection at the HLA-G promoter region. Hum Mol Genet 14:3619–3628 10.1093/hmg/ddi389 [DOI] [PubMed] [Google Scholar]
- 4.Ober C, Billstrand C, Kuldanek S, Tan Z (2006) The miscarriage-associated HLA-G -725G allele influences transcription rates in JEG-3 cells. Hum Reprod 21:1743–1748 10.1093/humrep/del036 [DOI] [PubMed] [Google Scholar]
- 5.Bartel DP (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116:281–297 10.1016/S0092-8674(04)00045-5 [DOI] [PubMed] [Google Scholar]
- 6.Johnston RJ, Hobert O (2003) A microRNA controlling left/right neuronal asymmetry in Caenorhabditis elegans. Nature 426:845–849 10.1038/nature02255 [DOI] [PubMed] [Google Scholar]
- 7.Brennecke J, Hipfner DR, Stark A, Russell RB, Cohen SM (2003) bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell 113:25–36 10.1016/S0092-8674(03)00231-9 [DOI] [PubMed] [Google Scholar]
- 8.Chen CZ, Li L, Lodish HF, Bartel DP (2004) MicroRNAs modulate hematopoietic lineage differentiation. Science 303:83–86 10.1126/science.1091903 [DOI] [PubMed] [Google Scholar]
- 9.He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, Goodson S, Powers S, Cordon-Cardo C, Lowe SW, Hannon GJ, et al (2005) A microRNA polycistron as a potential human oncogene. Nature 435:828–833 10.1038/nature03552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Poy MN, Eliasson L, Krutzfeldt J, Kuwajima S, Ma X, Macdonald PE, Pfeffer S, Tuschl T, Rajewsky N, Rorsman P, et al (2004) A pancreatic islet-specific microRNA regulates insulin secretion. Nature 432:226–230 10.1038/nature03076 [DOI] [PubMed] [Google Scholar]
- 11.Lecellier CH, Dunoyer P, Arar K, Lehmann-Che J, Eyquem S, Himber C, Saib A, Voinnet O (2005) A cellular microRNA mediates antiviral defense in human cells. Science 308:557–560 10.1126/science.1108784 [DOI] [PubMed] [Google Scholar]
- 12.Calin GA, Croce CM (2006) MicroRNA signatures in human cancers. Nat Rev Cancer 6:857–866 10.1038/nrc1997 [DOI] [PubMed] [Google Scholar]
- 13.Jin P, Zarnescu DC, Ceman S, Nakamoto M, Mowrey J, Jongens TA, Nelson DL, Moses K, Warren ST (2004) Biochemical and genetic interaction between the fragile X mental retardation protein and the microRNA pathway. Nat Neurosci 7:113–117 10.1038/nn1174 [DOI] [PubMed] [Google Scholar]
- 14.Iwai N, Naraba H (2005) Polymorphisms in human pre-miRNAs. Biochem Biophys Res Commun 331:1439–1444 10.1016/j.bbrc.2005.04.051 [DOI] [PubMed] [Google Scholar]
- 15.Abelson JF, Kwan KY, O’Roak BJ, Baek DY, Stillman AA, Morgan TM, Mathews CA, Pauls DL, Rasin MR, Gunel M, et al (2005) Sequence variants in SLITRK1 are associated with Tourette’s syndrome. Science 310:317–320 10.1126/science.1116502 [DOI] [PubMed] [Google Scholar]
- 16.Clop A, Marcq F, Takeda H, Pirottin D, Tordoir X, Bibe B, Bouix J, Caiment F, Elsen JM, Eychenne F, et al (2006) A mutation creating a potential illegitimate microRNA target site in the myostatin gene affects muscularity in sheep. Nat Genet 38:813–818 10.1038/ng1810 [DOI] [PubMed] [Google Scholar]
- 17.Rusinov V, Baev V, Minkov IN, Tabler M (2005) MicroInspector: a web tool for detection of miRNA binding sites in an RNA sequence. Nucleic Acids Res 33:W696–700 10.1093/nar/gki364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lewis BP, Burge CB, Bartel DP (2005) Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120:15–20 10.1016/j.cell.2004.12.035 [DOI] [PubMed] [Google Scholar]
- 19.Kiriakidou M, Nelson PT, Kouranov A, Fitziev P, Bouyioukos C, Mourelatos Z, Hatzigeorgiou A (2004) A combined computational-experimental approach predicts human microRNA targets. Genes Dev 18:1165–1178 10.1101/gad.1184704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krek A, Grun D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel M, et al (2005) Combinatorial microRNA target predictions. Nat Genet 37:495–500 10.1038/ng1536 [DOI] [PubMed] [Google Scholar]
- 21.Rehmsmeier M, Steffen P, Hochsmann M, Giegerich R (2004) Fast and effective prediction of microRNA/target duplexes. RNA 10:1507–1517 10.1261/rna.5248604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu L, Belasco JG (2005) Micro-RNA regulation of the mammalian lin-28 gene during neuronal differentiation of embryonal carcinoma cells. Mol Cell Biol 25:9198–9208 10.1128/MCB.25.21.9198-9208.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rousseau P, Le Discorde M, Mouillot G, Marcou C, Carosella ED, Moreau P (2003) The 14 bp deletion-insertion polymorphism in the 3′ UT region of the HLA-G gene influences HLA-G mRNA stability. Hum Immunol 64:1005–1010 10.1016/j.humimm.2003.08.347 [DOI] [PubMed] [Google Scholar]
- 24.Cozens AL, Yezzi MJ, Kunzelmann K, Ohrui T, Chin L, Eng K, Finkbeiner WE, Widdicombe JH, Gruenert DC (1994) CFTR expression and chloride secretion in polarized immortal human bronchial epithelial cells. Am J Respir Cell Mol Biol 10:38–47 [DOI] [PubMed] [Google Scholar]
- 25.Schroth MK, Grimm E, Frindt P, Galagan DM, Konno SI, Love R, Gern JE (1999) Rhinovirus replication causes RANTES production in primary bronchial epithelial cells. Am J Respir Cell Mol Biol 20:1220–1228 [DOI] [PubMed] [Google Scholar]
- 26.Hoffjan S, Nicolae D, Ostrovnaya I, Roberg K, Evans M, Mirel DB, Steiner L, Walker K, Shult P, Gangnon RE, et al (2005) Gene-environment interaction effects on the development of immune responses in the 1st year of life. Am J Hum Genet 76:696–704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hoffjan S, Ober C (2002) Present status on the genetic studies of asthma. Curr Opin Immunol 14:709–717 10.1016/S0952-7915(02)00393-X [DOI] [PubMed] [Google Scholar]
- 28.Wills-Karp M, Ewart SL (2004) Time to draw breath: asthma-susceptibility genes are identified. Nat Rev Genet 5:376–387 10.1038/nrg1326 [DOI] [PubMed] [Google Scholar]
- 29.Hunter DJ (2005) Gene-environment interactions in human diseases. Nat Rev Genet 6:287–298 10.1038/nrg1578 [DOI] [PubMed] [Google Scholar]
- 30.Ober C, Thompson EE (2005) Rethinking genetic models of asthma: the role of environmental modifiers. Curr Opin Immunol 17:670–678 10.1016/j.coi.2005.09.009 [DOI] [PubMed] [Google Scholar]
- 31.Vercelli D, Martinez FD (2006) The Faustian bargain of genetic association studies: bigger might not be better, or at least it might not be good enough. J Allergy Clin Immunol 117:1303–1305 10.1016/j.jaci.2006.03.030 [DOI] [PubMed] [Google Scholar]
- 32.Koppelman GH, Stine OC, Xu J, Howard TD, Zheng SL, Kauffman HF, Bleecker ER, Meyers DA, Postma DS (2002) Genome-wide search for atopy susceptibility genes in Dutch families with asthma. J Allergy Clin Immunol 109:498–506 10.1067/mai.2002.122235 [DOI] [PubMed] [Google Scholar]
- 33.Wright AL (2004) The epidemiology of the atopic child: who is at risk for what? J Allergy Clin Immunol 113:S2–7 10.1016/j.jaci.2003.09.050 [DOI] [PubMed] [Google Scholar]