Abstract
The ADH1B Arg47His polymorphism has been convincingly associated with alcoholism in numerous studies of several populations in Asia and Europe. In a review of literature from the past 30 years, we have identified studies that report allele frequencies of this polymorphism for 131 population samples from many different parts of the world. The derived ADH1B*47His allele reaches high frequencies only in western and eastern Asia. To pursue this pattern, we report here new frequency data for 37 populations. Most of our data are from South and Southeast Asia and confirm that there is a low frequency of this allele in the region between eastern and western Asia. The distribution suggests that the derived allele increased in frequency independently in western and eastern Asia after humans had spread across Eurasia.
It has been >30 years since variation in alcohol dehydrogenase (ADH) was shown to be associated with alcoholism.1 The ADH1B (MIM 103720) and ADH1C (MIM 103730) genes, which encode the primary ADH enzymes for alcohol metabolism in the liver, both harbor polymorphisms resulting in functional differences in their respective enzymes. The evolutionarily derived ADH1B*47His allele (previously called “ADH2*2”) results in enhanced catalytic activity, increased blood levels of acetaldehyde, flushing, and protection from alcoholism.2–5 These phenotypic changes are also associated with some other polymorphisms in the ADH region, but the associations are weaker or are not consistently replicated.6,7 Variants at ADH1C may be responsible for weak effects in Europeans, and linkage disequilibrium between variants in either gene may explain these results. In any case, the ADH1B Arg47His polymorphism (rs1229984) is the SNP generally regarded as the most important with respect to alcoholism (or alcoholism protection) in the ADH gene family in Asia.
The frequency of the evolutionarily derived ADH1B*47His allele is particularly high in eastern Asian populations (often exceeding 80%), but the allele is almost absent in sub-Saharan, European, and Native American populations. The high frequency of the derived allele in Asia could have resulted from either of two possible evolutionary processes: (1) a selective advantage existing only in eastern Asia for the allele or (2) random genetic drift increasing the frequency of the allele in eastern Asia. Indeed, we recently showed8 that positive selection led to the high frequency in some eastern Asian populations, most especially in Japanese and Koreans. However, we have also observed that there is another region of relatively high frequency (>40%) of the ADH1B*47His allele in southwestern Asia and in populations deriving from southwestern Asia.9 However, the few population samples we have studied from central Asia lacked or had very low frequencies of ADH1B*47His. To learn more about the distribution of this allele and to present a more detailed global view of this polymorphism (rs1229984), we typed 31 new population samples and reviewed the literature for population studies. In our review, we found 131 normal (nonalcoholic) population samples that have been studied for this polymorphism; populations studied, allele frequencies, and references for each study are available in ALFRED.
We studied 1,449 individuals from 37 population samples not previously studied (table 1). The majority of these population samples come from South and Southeast Asia, except for the Sandawe from Tanzania and Hungarians from Europe. Two of the population samples were collected in Pakistan but represent migrant individuals from Africa (Somali) or descendants from eastern Africans but with the strong possibility of some admixture with other Pakistanis (Negroid Makrani). Three new South Asian populations are included: the Mohannas are indigenous fishermen from southern Pakistan, the Hazaras are from the northeastern border of Pakistan and Afghanistan and claim descent from Genghis Khan’s army, and the Keralites are a Dravidian group from southwestern India. The 23 Laotian populations are among the most ethnically diverse in Southeast Asia and include many Mon-Khmer and Daic groups. Linguistic subfamilies such as Bahnaric, Katuic, Viet-Muong, Khmuic, and Palaungic are included as parts of the Mon-Khmer family; Tai-Sek and Kam-Sui subfamilies are parts of the Daic family.10 The remaining seven population samples are from southwestern China, a region ethnically related to Southeast Asia under the same linguistic families, Mon-Khmer and Daic. All individuals provided informed consent approved by the relevant human subjects committees of the relevant countries and collaborating institutions. With all 37 population samples, the ADH1B Arg47His polymorphism (rs1229984) was typed by the TaqMan method with use of an assay we designed and had synthesized by Shanghai GeneCore BioTechnologies (probe 1, FAM-AATCTGTCACACAGATGA-MGB; probe 2, TET-TCTGTCGCACAGATG-MGB [bold font indicates polymorphic nucleotide site]; forward primer, 5′-TCTTTTCTGAATCTGAACAGCTTCTCT-3′; reverse primer, 5′-GGGTCACCAGGTTGCCACTA-3′).
Table 1. .
Populations Sampled in This Study
| Population Numbera |
Country | Population | ISO639-3b | 2N |
ADH1B*47His Frequency (%) |
Longitude | Latitude |
| 3 | Mozambique | Negroid Makrani | VMW | 56 | 1.8 | 30–41 E | 10.5–27 S |
| 4 | Tanzania | Sandawe | SAD | 80 | .0 | 35–38 E | 4–7 S |
| 17 | Somalia | Somali | SOM | 36 | 2.8 | 40–52 E | 12N–2 S |
| 40 | Hungary | Hungarian | HUN | 178 | 10.1 | 16–23 E | 48.5–45.5 N |
| 61 | Pakistan | Mohanna | SBN | 104 | 7.7 | 67–68 E | 27–25 N |
| 62 | India | Keralite | MAL | 58 | .0 | 76–77 E | 10–8 N |
| 71 | Pakistan | Hazara | HAZ | 58 | 24.1 | 70 E | 33 N |
| 74 | Laos | Oy | OYB | 100 | 48.0 | 106.4 E | 14.4 N |
| 75 | Laos | Brao | BRB | 100 | 20.0 | 106.5 E | 15.0 N |
| 76 | Laos | Talieng | TDF | 96 | 38.5 | 106.6 E | 15.2 N |
| 77 | Laos | Alak | ALK | 92 | 35.9 | 106.5 E | 15.2 N |
| 78 | Laos | Jeh | JEH | 100 | 34.0 | 107.0 E | 15.3 N |
| 79 | Laos | Ngeq | NGT | 124 | 39.5 | 106.6 E | 15.3 N |
| 80 | Laos | Taoih | TTH | 10 | 50.0 | 106.4 E | 15.4 N |
| 81 | Laos | Kataang | KGD | 92 | 54.3 | 106.5 E | 15.5 N |
| 82 | Laos | Suy | KDT | 104 | 26.0 | 106.4 E | 15.6 N |
| 83 | Laos | Inh | IRR | 98 | 56.1 | 106.2 E | 15.6 N |
| 84 | Laos | So | SSS | 108 | 25.9 | 105.1 E | 17.4 N |
| 85 | Laos | Phuthai | PHT | 100 | 14.0 | 105.0 E | 17.5 N |
| 86 | Laos | Aheu | THM | 90 | 31.1 | 105.0 E | 18.0 N |
| 87 | Laos | Bo | BGL | 104 | 31.7 | 105.1 E | 18.2 N |
| 88 | Laos | Tai Mene | TMP | 104 | 35.6 | 104.4 E | 18.2 N |
| 89 | Laos | Phuan | PHU | 106 | 39.6 | 103.1 E | 19.2 N |
| 90 | Laos | Rien | RIE | 100 | 26.0 | 101.3 E | 19.1 N |
| 91 | Laos | Mal | MIF | 100 | 21.0 | 101.3 E | 19.3 N |
| 92 | Laos | Kang | KYP | 36 | 36.1 | 104.3 E | 19.5 N |
| 93 | Laos | Tai Deang | TYR | 96 | 41.7 | 103.6 E | 20.0 N |
| 94 | Laos | Tai Dam | BLT | 102 | 33.3 | 104.2 E | 20.3 N |
| 95 | Laos | Puoc | PUO | 82 | 25.6 | 104.0 E | 20.5 N |
| 96 | Laos | Bit | BGK | 86 | 10.5 | 101.3 E | 20.5 N |
| 97 | China | Bugan | BBH | 46 | 87.0 | 104.6 E | 23.4 N |
| 98 | China | Lachi | LBT | 40 | 100.0 | 104.2 E | 22.5 N |
| 99 | China | Yerong | YRN | 14 | 42.9 | 105.6 E | 23.2 N |
| 100 | China | Cao Lan | MLC | 12 | 91.7 | 107.3 E | 21.4 N |
| 101 | China | Pahng | PHA | 20 | 100.0 | 108.6 E | 25.3 N |
| 104 | China | Pou | BYK | 18 | 83.3 | 111.5 E | 23.4 N |
| 105 | China | Tujia | TJI | 48 | 60.4 | 109.1 E | 29.5 N |
The population numbers are the same as those in figure 1.
ISO639-3 is the international standard devised to enable the uniform identification of all known languages in a wide range of applications, particularly including information systems. For our sample populations, these linguistic tags can be used to search Ethnologue for information on the populations, although some African-derived populations in Pakistan no longer speak their original languages.
The allele frequencies for these 37 new population samples are given in table 1. The frequencies of the derived ADH1B*47His allele were very low in the African samples as well as in the Keralites. Low to moderate frequencies were found in the two Pakistani populations, with the moderate frequency occurring in the Hazaras (24%), consistent with some eastern Asian ancestry. Frequencies <15% occurred in two of the Laotian populations, the Phuthai and Bit. The other Laotian populations had frequencies ranging from 20% to 56%. With the exception of the Yerong (42.9%), the Chinese populations had frequencies higher than those of any Laotian population, ranging from 60% to 100%.
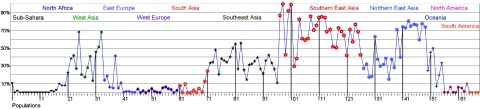
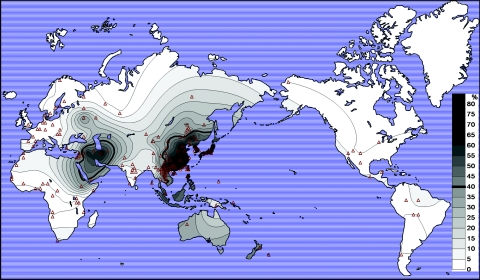
For a global analysis, we combined our new data with previously published data, for a total of 168 population samples. The frequency data are displayed graphically in figure 1, with the population samples arranged “geographically” from Africa to South America. Clearly, the frequencies are high in western and eastern Asia and are lower in between. Moderate frequencies exist in Southeast Asia and North Africa—that is, the geographic regions surrounding those populations with the highest frequencies. In allele-frequency contours plotted geographically (fig. 2), a clinal pattern is quite obvious. The Moscow Russian sample appears anomalous with a fairly high frequency of ADH1B*47His; however, the sample is said in the original publication11 to be admixed with eastern Asians. It is generally accepted that the Polynesians originated in Southeast Asia12; they also have a moderate frequency of ADH1B*47His. In the rest of the world, the ADH1B*47His allele is almost absent.
Figure 1. .
ADH1B*47His frequencies in populations from different regions of the world. Descriptions of the specific samples, full references, and allele frequencies are available in ALFRED. The geographic-region labels are colored the same as the corresponding population values. “Southeast Asia” corresponds primarily to populations in peninsular eastern Asia and island regions; “Southern East Asia” corresponds primarily to populations in southern China. The numbers on the X-axis correspond to the following populations: 1, Western Cape; 2, !Kung San; 3, Negroid Makrani; 4, Sandawe; 5, Chagga; 6, Biaka; 7, Mbuti; 8, southwestern Bantu; 9, Hausa; 10, Ibo; 11, Yoruba; 12, Yoruba; 13, Masai; 14, African; 15, African American; 16, African American; 17, Somali; 18, Saharans; 19, northern Moroccans; 20, central Moroccans; 21, black American; 22, Ethiopian Jews; 23, Yemenite Jews; 24, Druze; 25, Samaritans; 26, non-Russian Ashkenazi; 27, Russian Ashkenazi; 28, Ashkenazi Jews; 29, Sephardic Jews; 30, Turks (Turkey); 31, Turks (Iran); 32, Turkomans; 33, Persian Zoroastrians; 34, Adygei; 35, Chuvash; 36, Russian (Vologda); 37, Russian (Archangelsk); 38, Russian (Moscow); 39, Hungarians; 40, Hungarian; 41, Finns; 42, Finns; 43, Lapps; 44, Khanty; 45, Komi; 46, Swedes (Stockholm); 47, Swedes; 48, Poles (Krakow); 49, Danes; 50, German; 51, Germans; 52, German (Heidelberg); 53, French (Bordeaux); 54, Irish; 55, Spanish (Tarragona); 56, Spanish (Catalans); 57, Basque; 58, European American; 59, European American; 60, white American; 61, Mohanna; 62, Keralite; 63, Indians; 64, Bhil; 65, Gond; 66, Sahariya; 67, Korku; 68, Pattapu; 69, Onge; 70, Kachari; 71, Hazara; 72, Thais; 73, Cambodian; 74, Oy; 75, Brao; 76, Talieng; 77, Alak; 78, Jeh; 79, Ngeq; 80, Taoih; 81, Kataang; 82, Suy; 83, Inh; 84, So; 85, Phuthai; 86, Aheu; 87, Bo; 88, Tai Mene; 89, Phuan; 90, Rien; 91, Mal; 92, Kang; 93, Tai Deang; 94, Tai Dam; 95, Puoc; 96, Bit; 97, Bugan; 98, Lachi; 99, Yerong; 100, Cao Lan; 101, Pahng; 102, Bouyei (Yunnan); 103, Hui (Guizhou); 104, Pou; 105, Tujia; 106, Malays; 107, Filipinos; 108, Paiwan; 109, Ami; 110, Ami; 111, Bunun; 112, Atayal; 113, Atayal; 114, Chinese; 115, Han (Canton); 116, Han (Fujian); 117, Han (South Taiwan); 118, Han (Taiwan); 119, Han (Hakka); 120, Han (Jiangsu); 121, Han (Sichuan); 122, Han (Shandong); 123, Han (Henan); 124, Han (Beijing); 125, Han (Jilin); 126, Han (Xinjiang); 127, Uygur (Urumqi); 128, Xibo (Ili); 129, Kazak (Ili); 130, Yakut; 131, Mongolian (Baotou); 132, Manchu; 133, Daur; 134, Mongol (Hulunbel); 135, Ewenk (Hulunbel); 136, Oroqen (Hulunbel); 137, Elunchun (Mohe); 138, Hezhen; 139, Korean (Changchun); 140, Korean (Yanbian); 141, Korean; 142, Korean (Incheon); 143, Korean; 144, Japanese (Gunma); 145, Japanese (Kyoto); 146, Japanese; 147, Japanese; 148, Japanese (Aichi); 149, Japanese (Tokyo); 150, Micronesian; 151, Maori; 152, Polynesian; 153, Papua New Guineans; 154, Nasioi; 155, Australia Aborigines; 156, Eskimos; 157, Cheyenne; 158, Mexican American; 159, Pima (Arizona); 160, southwestern Amerindian; 161, Maya (Yucatan); 162, Mestizos; 163, Pima (Mexico); 164, Caboclos; 165, Karitiana; 166, Surui; 167, Ticuna; and 168, Aurocanians.
Figure 2. .
Global contour plot of the ADH1B*47His allele frequency drawn by the Surface Mapping System program Surfer 8.00 developed by Golden Software. The frequency data are the same as in figure 1, with the use of the geographic coordinates in table 1 and in ALFRED.
A discontinuity between two high-frequency regions, western Asia and eastern Asia, appears in South Asia and the western part of Southeast Asia. The route from western Asia, via South Asia and Southeast Asia, to eastern Asia is one of the supposed routes of modern human expansion in Asia.13 The observed gap supports an argument against maintenance of the high western Asian frequency throughout this expansion. Another supposed route of expansion is via central Asia. Although there are few samples in the region and the frequencies are somewhat higher than those in South Asia, the frequencies are still much lower than those to the west and the east. Historic events such as travel along the Silk Road, Ghengis Khan’s conquests, and recent migrations—such as the movement of the Han Chinese to Xinjiang and the Xibo population from northeastern China to Xinjiang during the Qing Dynasty—could all have contributed to these somewhat higher central Asian frequencies. It is noteworthy that the derived allele frequencies in two central Asian populations, the Uygur and Kazakh, are not high. Similarly, the Khanty from western Siberia lack the allele. These data strongly support the argument that a significantly lower frequency exists in central Asia, with two separate flanking regions of high frequency of the ADH1B*47His allele.
Plausible explanations for the observed high frequencies of ADH1B*47His in eastern and western Asia but not in between involve separate increases in frequency in the two regions. Either drift or natural selection could have been the cause of these high frequencies. There is strong evidence that natural selection is responsible in Koreans, Japanese, and Han Chinese8 but not in other eastern Asian populations in that study. However, a selective force could have been too weak to be detected but still strong enough to have resulted in the widespread distribution of the ADH1B*47His allele in eastern Asia. Or, gene flow could have spread the allele into populations in which selection was not operating, since there seem to be no strongly detrimental fitness consequences of having the allele in areas where natural selection could not be detected. In this expanded study, which combines all known data from the literature with new data about many relevant populations, we have shown that the pattern of the ADH1B*47His distribution is more complicated than was previously apparent. In western Asia, the highest frequencies appear in the Persians, Turks, Samaritans, and Jews from a variety of regions. These populations belong to three different linguistic families—Indo-European (Iranian), Altaic (Turks), and Afro-Asiatic (Semitic). This linguistic difference provides an argument against a recent common origin of these groups, although their geographic proximity may have allowed considerable gene flow among them in more recent millennia.
At least three hypotheses may explain the observed allele-frequency pattern of ADH1B*47His: two independent mutations, loss of the allele from intervening populations, and local selective factors. We can confidently exclude independent mutation, because the haplotype phylogeny clearly shows overall strong linkage disequilibrium but a recombination event on one side of the Arg47His site that distinguishes the southwestern Asian from eastern Asian haplotypes.9,14,15 Hypothesizing the loss of the allele from central and South Asian populations requires explanation of several facts. Implicit is the assumption that the high frequency arose in southwestern Asia and was carried into eastern Asia. However, if the derived allele were always frequent, the western Asian haplotype should also be found in eastern Asia, because it would have been the common haplotype initially, and total replacement by the recombinant haplotype would have been very unlikely. The western Asian haplotype has never been observed in the several eastern Asian populations studied for the necessary polymorphisms.9 Also, there has never been evidence that ADH1B*47His itself was selected against and, therefore, no reason to speculate that selection eliminated the allele in South and central Asia. Local selection focused on the genomic region containing ADH1B*47His is already strongly supported for eastern Asian populations.8 We speculate that a low allele frequency had been maintained in different Asian populations after the settlements of western, central, South, and eastern Asia. Because the allele frequency was quite low, different haplotypes easily drifted to different frequencies, including complete loss, in different geographic regions. Then, at a relatively recent time, local selective factors on this allele or on one tightly linked increased the allele frequency in eastern Asia.8 The high frequency in western Asia might have resulted from local selection or might have simply resulted from random drift during population expansion but after human expansion into Europe. So far, there is no evidence of selection in western Asia, but the data are incomplete. In either case, the data presented here support the argument that the high allele frequency must have occurred independently.
We still lack data from multiple populations in central Asia and haplotype data from the majority of these 168 population samples. A clearer understanding of the complex geographic pattern of this important allele at a metabolically important gene will result only when more haplotype data exist for this locus from more population samples from regions already densely studied, as well as new allele- and haplotype-frequency data from central parts of Asia—for example, Afghanistan, Iran, Kazakhstan, and southern Siberia. Clearly, additional research is needed; there is still no last word on the ADH1B*47His story, which grows ever more complex.
Acknowledgments
This work was supported in part by U.S. Public Health Service grants AA009379 and GM057672 and by National Science Foundation grant BCS0096588. We especially thank the many individuals who volunteered to provide samples for this study.
Web Resources
The URLs for data presented herein are as follows:
- ALFRED, http://alfred.med.yale.edu/alfred/SiteTable1A_working.asp?siteuid=SI000229N
- Ethnologue, http://www.ethnologue.com/ (for ISO639-3 symbols and linguistic relationships) and http://www.ethnologue.com/show_country.asp?name=LA (for Laotian population information)
- Online Mendelian Inheritance in Man (OMIM) http://www.ncbi.nlm.nih.gov/Omim/ (for ADH1B and ADH1C)
References
- 1.Li TK, Bosron WF, Dafeldecker WP, Lange LG, Vallee BL (1977) Isolation of pi-alcohol dehydrogenase of human liver: is it a determinant of alcoholism? Proc Natl Acad Sci USA 74:4378–4381 10.1073/pnas.74.10.4378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thomasson HR, Edenberg HJ, Crabb DW, Mai XL, Jerome RE, Li TK, Wang SP, Lin YT, Lu RB, Yin SJ (1991) Alcohol and aldehyde dehydrogenase genotypes and alcoholism in Chinese men. Am J Hum Genet 48:677–681 [PMC free article] [PubMed] [Google Scholar]
- 3.Goedde HW, Agarwal DP, Fritze G, Meier-Tackmann D, Singh S, Beckmann G, Bhatia K, Chen LZ, Fang B, Lisker R (1992) Distribution of ADH2 and ALDH2 genotypes in different populations. Hum Genet 88:344–346 10.1007/BF00197271 [DOI] [PubMed] [Google Scholar]
- 4.Nakamura K, Iwahashi K, Matsuo Y, Miyatake R, Ichikawa Y, Suwaki H (1996) Characteristics of Japanese alcoholics with the atypical aldehyde dehydrogenase 2*2. I. A comparison of the genotypes of ALDH2, ADH2, ADH3, and cytochrome P-450E1 between alcoholics and nonalcoholics. Alcohol Clin Exp Res 20:52–55 10.1111/j.1530-0277.1996.tb01043.x [DOI] [PubMed] [Google Scholar]
- 5.Mulligan CJ, Robin RW, Osier MV, Sambuughin N, Goldfarb LG, Kittles RA, Hesselbrock D, Goldman D, Long JC (2003) Allelic variation at alcohol metabolism genes (ADH1B, ADH1C, ALDH2) and alcohol dependence in an American Indian population. Hum Genet 113:325–336 10.1007/s00439-003-0971-z [DOI] [PubMed] [Google Scholar]
- 6.Shen YC, Fan JH, Edenberg HJ, Li TK, Cui YH, Wang YF, Tian CH, Zhou CF, Zhou RL, Wang J (1997) Polymorphism of ADH and ALDH genes among four ethnic groups in China and effects upon the risk for alcoholism. Alcohol Clin Exp Res 21:1272–1277 [PubMed] [Google Scholar]
- 7.Chen WJ, Loh EW, Hsu YP, Cheng AT (1997) Alcohol dehydrogenase and aldehyde dehydrogenase genotypes and alcoholism among Taiwanese aborigines. Biol Psychiatry 41:703–709 10.1016/S0006-3223(96)00072-8 [DOI] [PubMed] [Google Scholar]
- 8.Han Y, Gu S, Oota H, Osier MV, Pakstis AJ, Speed WC, Kidd JR, Kidd KK (2007) Evidence of positive selection on a class I ADH locus. Am J Hum Genet 80:441–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Osier MV, Pakstis AJ, Soodyall H, Comas D, Goldman D, Odunsi K, Okonofua F, Parnas J, Schulz L, Bertranpetit J, et al (2002) A global perspective on genetic variation at the ADH genes reveals unusual patterns of linkage disequilibrium and diversity. Am J Hum Genet 71:84–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gordon RG Jr (2005) Ethnologue: languages of the world, 15th ed. SIL International, Dallas [Google Scholar]
- 11.Ogurtsov PP, Garmash IV, Miandina GI, Guschin AE, Itkes AV, Moiseev VS (2001) Alcohol dehydrogenase ADH2-1 and ADH2-2 allelic isoforms in the Russian population correlate with type of alcoholic disease. Addict Biol 6:377–383 10.1080/13556210020077109 [DOI] [PubMed] [Google Scholar]
- 12.Zhang F, Su B, Zhang YP, Jin L (2007) Genetic studies of human diversity in East Asia. Phil Trans R Soc B 362:987–995 10.1098/rstb.2007.2028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goebel T (2007) The missing years for modern humans. Science 315:194–196 10.1126/science.1137564 [DOI] [PubMed] [Google Scholar]
- 14.Whitfield JB (2002) Alcohol dehydrogenase and alcohol dependence: variation in genotype-associated risk between populations. Am J Hum Genet 71:1247–1250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kidd KK, Osier MV, Pakstis AJ, Kidd JR (2002) Reply to Whitfield. Am J Hum Genet 71:1250–1251 [Google Scholar]