Abstract
Background
We investigated if global gene expression and transcription networks in B-lymphocytes of siblings with multiple sclerosis (MS) were different from healthy siblings.
Results
Using virus-transformed immortalized B cells and human whole genome bioarrays with validation using RT-qPCR, we found that in siblings with MS, genes for CXCL10, serpin B1 and FUT4 were up regulated whereas CDC5L, TNFRSF19 and HLA-DR were down regulated, among others; transcription analysis showed two intersecting clusters of transcriptional factors - the larger, governed by the upregulated transcription factor 2 (TCF2) and the smaller network regulated by the downregulated CDC5L.
Conclusion
No study has linked TCF2 to MS and to better understand the role of TCF2 in MS, studies in larger cohorts are required.
Background
MS is a complex genetic disease associated with inflammation predominantly in the white matter of brain and spinal cord. It is thought to be mediated by autoreactive T cells [1,2]. Susceptibility to MS is determined by both inherited and non-inherited factors [3]. Approximately 15-20% of MS patients have a family history of MS, but large extended pedigrees are uncommon. Studies in twins [4,5] and conjugal pairs [6] show that much of the familial clustering is the result of shared genetic risk factors.
MS susceptibility is linked to HLA-DR2 [7]. Increased risk of MS in women has been detected with interleukin-1 receptor antagonist (IL-1RA) allele 2 [8], 5G5G genotype of plasminogen activator inhibitor 1 (PAI-1) gene [9] and interaction between estrogen receptor 1 (ESR1) and HLA-DR2 [10]. B cells are implicated in MS and have been found in the cerebrospinal fluid (CSF) of MS patients [11]. Additionally, oligoclonal bands identified in CSF point to the role of B cells in MS pathogenesis [12]. Furthermore, antibody-secreting B cells contribute to tissue injury [13]. We hypothesized that B cells derived from MS patients could harbor genes that confer a higher MS risk as compared to B cell gene expression in healthy siblings.
Human B-cells have a receptor for Epstein-Barr virus (EBV) and can become immortalized after in vitro infection with EBV. Additionally, the link between EBV and MS is highly impressive [14] though inconclusive; we hypothesized that analysis of gene expression and transcription networks in EBV-transformed B cells between siblings with and without MS could yield important clues to understanding the pathology of MS.
Large-scale analyses of transcripts from peripheral blood cells or brain lesions from MS patients have created possibilities for therapeutics [15] and global gene expression analysis using microarrays is a sensitive method to investigate molecular heterogeneity [16]. In this study, we tested a new software tool that is in development, to map transcription networks in the microarray data.
Our objectives were to i) to determine if gene expression and transcription networks in B-lymphocytes of siblings with MS were different from healthy siblings in EBV-transformed B cells and ii) to validate data using qPCR techniques.
Methods
EBV-transformed B cell lines from Coriell Institute for medical research (Camden, NJ, USA) and the National Institute of General Medical Sciences (NIGMS, Bethesda, MD) were obtained for our study. As shown in Table 1, B cells were harvested from one family (# 2108, proband, affected sister) and an unaffected brother (control); cells from another family (# 2112, proband, affected brother) and three unaffected brothers (controls). Cells from the third family (# 2102) comprised of proband and an affected sister but no unaffected controls. None of the patients were on immunomodulatory agents (IMAs) to treat MS.
Table 1.
Cell lines obtained from Corielle for our study.
| Sample ID | Relation | Family | Affected | Sex | Age |
| GM8923 | Proband | 2108 | Yes | Male | 38 |
| GM8922A | Sister | 2108 | Yes | Female | 43 |
| GM8830 | Proband | 2102 | Yes | Female | 47 |
| GM8839 | Sister | 2102 | Yes | Female | 39 |
| GM9013 | Proband | 2112 | Yes | Male | 35 |
| GM9016 | Brother | 2112 | Yes | Male | 38 |
| GM9018 | Brother | 2112 | No | Male | 42 |
| GM9017 | Brother | 2112 | No | Male | 45 |
| GM9023 | Brother | 2112 | No | Male | 47 |
| GM8921A | Brother | 2108 | No | Male | 46 |
Cells were harvested in 4 ml of Tri-Reagent (Molecular research center, Inc) and the total RNA was isolated using standard protocols. Briefly, B-cells were incubated at 37°C and 5% CO2 overnight, the day after cells were counted. Cells were expanded in RPMI 1640 with 2 mM L-glutamine and 15% fetal bovine serum following Coriell's lymphoblast line maintenance protocols. 2 vials were frozen and 1 × 107 cells were spun down and resuspended in 2 ml of Tri-reagent. The cell mixture was frozen at -80°C until further analysis. The RNA was isolated using the standard Tri-reagent protocol and further cleaned up using RNeasy kits (Qiagen) using a protocol that included an on column Dnase step. RNA samples were checked for quality on a Bioanalyzer and a nanodrop spectrophotometer. Samples with 28s/18s ratio > 1.8 were deemed acceptable for the microarray experiment. The total RNA (1 ug) was first converted to double stranded cDNA using the standard Codelink protocol. This was then purified on Qiaquick PCR columns and in vitro transcribed to labeled cRNA using biotin-11-UTP (Perkin Elmer). The labeled cRNA was purified on Qiagen RNeasy Mini columns and 10 ug was fragmented as per the Codelink protocol following which it was hybridized to the Codelink Human Whole Genome bioarrays for 18 h at 37°C. The arrays were subsequently washed, stained with Cy5-Streptavidin (GE Amersham) and scanned on an Axon Genepix 4000B scanner. The data were collected using Codelink Expression Analysis software and analyzed using Genespring software version 7.3.1 (Agilent). All microarray data are deposited at Gene Expression Omnibus as GSE 10064 and linked via [17].
Microarray data analysis
Values below 0.01 were set to 0.01. Each measurement was divided by the 50th percentile of all measurements in that sample. All samples were normalized to the median of the control samples. Each measurement for each gene in those specific samples was divided by the median of that gene's measurements in the corresponding control samples. The gene list (54,902 transcripts) was filtered using the cross gene error model for replicates to select genes with good signal-to-noise ratios (31, 168 genes). This gene list was subjected to a student's t-test (p-value < 0.05) and yielded 1, 417 genes with significant differences between the two conditions. This list was further filtered for confidence using a Benjamini-Hochberg false discovery rate correction to obtain 1405 genes. A 1.5-fold cut-off filter was then applied to identify genes that were preferentially upregulated (260) or downregulated (452) genes in samples with disease as compared to controls. These 712 genes were further filtered to only select those which were consistently 'absent" or "present" in 3 of 4 controls and 5 of 6 diseased samples. The final gene list had 705 genes; 259 upregulated and 446 downregulated genes while using 2-fold cut off resulted in 84 upregulated and 111 downregulated genes (Tables 2 &3).
Table 2.
Representative upregulated genes (out of 259 total).
| Probe ID | Fold Change | Common | Genbank | Description |
| Upregulated genes | ||||
| GE53598 | 5.88 | TINAG | NM_014464 | tubulointerstitial nephritis antigen, complete cds |
| GE59642 | 5.77 | CXCL10 | NM_001565 | gamma-interferon inducible early response gene (with homology to platelet proteins) |
| GE57550 | 4.74 | CRYM | NM_001888 | mu-crystallin mRNA, complete cds |
| GE518123 | 4.27 | IGLL1 | NM_020070 | immunoglobulin lambda-like polypeptide 1 (IGLL1), transcript variant 1, mRNA |
| GE58736 | 4.20 | DACT1 | NM_016651 | dapper homolog 1, antagonist of beta-catenin (xenopus) (DACT1), mRNA |
| GE79033 | 4.15 | CD9 | NM_001769 | CD9 antigen (p24) (CD9), mRNA |
| GE55346 | 3.57 | LRRC16 | NM_017640 | leucine rich repeat containing 16 (LRRC16), mRNA |
| GE61239 | 3.50 | SERPINB1 | NM_030666 | serine (or cysteine) proteinase inhibitor, clade B (ovalbumin), member 1 (SERPINB1), mRNA |
| GE62781 | 3.50 | MT1F | NM_005949 | metallothionein 1F (functional) (MT1F), mRNA |
| GE87501 | 3.49 | FXYD2 | NM_001680 | FXYD domain containing ion transport regulator 2 (FXYD2), transcript variant a, mRNA |
| GE61946 | 3.26 | ARHGAP6 | NM_001174 | Rho GTPase activating protein 6 (ARHGAP6), transcript variant 2, mRNA |
| GE56932 | 3.22 | EGLN3 | NM_022073 | egl nine homolog 3 (C. elegans) (EGLN3), mRNA |
| GE61093 | 3.18 | ACTN1 | NM_001102 | actinin, alpha 1 (ACTN1), mRNA |
| GE87626 | 2.97 | OSR2 | NM_053001 | odd-skipped-related 2A protein (OSR2), mRNA |
| GE59655 | 2.93 | ATP1B1 | NM_001677 | ATPase, Na+/K+ transporting, beta 1 polypeptide (ATP1B1), mRNA |
| GE58575 | 2.91 | PTPLA | NM_014241 | protein tyrosine phosphatase-like (proline instead of catalytic arginine), member a (PTPLA), mRNA |
| GE57767 | 2.84 | COL9A3 | NM_001853 | collagen, type IX, alpha 3 (COL9A3), mRNA |
| GE57621 | 2.83 | LGALS3BP | NM_005567 | lectin, galactoside-binding, soluble, 3 binding protein (LGALS3BP), mRNA |
| GE81389 | 2.82 | CCR6 | NM_004367 | chemokine (C-C motif) receptor 6 (CCR6), transcript variant 1, mRNA |
| GE79898 | 2.81 | LYPLA3 | NM_012320 | lysophospholipase 3 (lysosomal phospholipase A2) (LYPLA3), mRNA |
| GE62292 | 2.79 | CPXM | NM_019609 | carboxypeptidase X (M14 family) (CPXM), mRNA |
| GE57547 | 2.66 | TCN2 | NM_000355 | transcobalamin II; macrocytic anemia (TCN2), mRNA |
| GE81525 | 2.52 | IFI27 | NM_005532 | ISG12 protein |
| GE79641 | 2.48 | FUT4 | NM_002033 | fucosyltransferase 4 (alpha (1,3) fucosyltransferase, myeloid-specific) (FUT4), mRNA |
| GE81272 | 2.47 | VIM | NM_003380 | vimentin (VIM), mRNA |
| GE57737 | 2.45 | ENPP2 | NM_006209 | autotaxin mRNA, complete cds |
| GE80150 | 2.38 | ELF3 | NM_004433 | E74-like factor 3 (ets domain transcription factor, epithelial-specific) (ELF3), mRNA |
| GE58207 | 2.34 | ENG | NM_000118 | endoglin (Osler-Rendu-Weber syndrome 1) (ENG), mRNA |
| GE57048 | 2.33 | CCR1 | NM_001295 | chemokine (C-C motif) receptor 1 (CCR1), mRNA |
| GE57883 | 2.28 | SELL | NM_000655 | selectin L (lymphocyte adhesion molecule 1) (SELL), mRNA |
| GE60214 | 2.27 | NRGN | NM_006176 | neurogranin (protein kinase C substrate, RC3) (NRGN), mRNA |
| GE498904 | 2.22 | DACT1 | NM_016651 | dapper homolog 1, antagonist of beta-catenin (xenopus) (DACT1), mRNA |
| GE81340 | 2.17 | PSTPIP1 | NM_003978 | proline-serine-threonine phosphatase interacting protein 1 (PSTPIP1), mRNA |
| GE58931 | 2.16 | TCF8 | U12170 | zinc finger homeodomain protein mRNA, complete cds |
| GE61464 | 2.15 | SERPINB9 | NM_004155 | serine (or cysteine) proteinase inhibitor, clade B (ovalbumin), member 9 (SERPINB9), mRNA |
| GE82947 | 2.12 | SNX22 | NM_024798 | sorting nexin 22 (SNX22), mRNA |
| GE60134 | 2.09 | UPP1 | NM_003364 | uridine phosphorylase (UP), transcript variant 1, mRNA |
| GE54152 | 2.07 | APOL1 | NM_003661 | apolipoprotein L, 1 (APOL1), transcript variant 1, mRNA |
| GE56786 | 2.02 | DUSP4 | NM_001394 | dual specificity phosphatase 4 (DUSP4), transcript variant 1, mRNA |
| GE54528 | 2.02 | TNFSF12; | NM_003809 | tumor necrosis factor (ligand) superfamily, member 12 (TNFSF12), transcript variant 1, mRNA |
| GE61789 | 2.02 | TDE2L | NM_178865 | tumor differentially expressed protein 2 (TDE2), mRNA |
| GE61399 | 2.01 | LLT1 | NM_013269 | lectin-like NK cell receptor (LLT1), mRNA |
Fold change = diseased/normal
Table 3.
Representative downregulated genes (out of 446 total)
| Probe ID | Fold change | Common | Genbank | Description |
| Down regulated genes | ||||
| GE54030 | 0.09 | EIF1AY | NM_004681 | eukaryotic translation initiation factor 1A, Y-linked (EIF1AY), mRNA |
| GE79500 | 0.1 | RPS4Y | NM_001008 | ribosomal protein S4, Y-linked (RPS4Y), mRNA |
| GE54029 | 0.12 | DDX3Y | NM_004660 | DEAD/H (Asp-Glu-Ala-Asp/His) box polypeptide, Y chromosome (DBY), mRNA |
| GE59227 | 0.12 | JARID1D | NM_004653 | Smcy homolog, Y-linked (mouse) (SMCY), mRNA |
| GE55836 | 0.15 | MLAT4 | NM_018192 | myxoid liposarcoma associated protein 4 (MLAT4), mRNA |
| GE54974 | 0.19 | TNFRSF19 | NM_148957 | tumor necrosis factor receptor superfamily, member 19 (TNFRSF19), transcript variant 2, mRNA |
| GE788416 | 0.28 | KRTAP4-9 | AJ406941 | partial mRNA for keratin associated protein 49 (KRTAP4.9 gene) |
| GE489986 | 0.32 | ZFY | NM_003411 | zinc finger protein, Y-linked (ZFY), mRNA |
| GE58359 | 0.35 | GCHFR | NM_005258 | GTP cyclohydrolase I feedback regulatory protein (GCHFR), mRNA |
| GE87376 | 0.36 | BAIAP2L1 | NM_018842 | insulin receptor tyrosine kinase substrate (LOC55971), mRNA |
| GE561268 | 0.39 | FOXD4L2 | NM_199135 | FOXD4-like 2 (FOXD4L2), mRNA |
| GE54486 | 0.4 | DAPK2 | NM_014326 | death-associated protein kinase 2 (DAPK2), mRNA |
| GE79999 | 0.4 | CAMK1 | NM_003656 | calcium/calmodulin-dependent protein kinase I (CAMK1), mRNA |
| GE58178 | 0.4 | KAL1 | NM_000216 | Kallmann syndrome (KAL) mRNA, complete cds |
| GE82446 | 0.4 | BAIAP2L1 | NM_018842 | insulin receptor tyrosine kinase substrate (LOC55971), mRNA |
| GE58327 | 0.41 | CD83 | NM_004233 | CD83 antigen (activated B lymphocytes, immunoglobulin superfamily) (CD83), mRNA |
| GE567535 | 0.42 | NGFRAP1 | AF187064 | p75NTR-associated cell death executor (NADE) mRNA |
| GE62339 | 0.42 | CSE-C | NM_018978 | sialic acid-specific 9-O-acetylesterase I mRNA, complete cds |
| GE82815 | 0.42 | FA2H | NM_024306 | fatty acid 2-hydroxylase (FA2H), mRNA |
| GE82367 | 0.44 | LAPTM4B | NM_018407 | lysosomal associated protein transmembrane 4 beta (LAPTM4B), mRNA phosphodiesterase 4A, cAMP-specific (phosphodiesterase E2 dunce homolog, Drosophila) |
| GE81609 | 0.45 | PDE4A | NM_006202 | (PDE4A), mRNA |
| GE61123 | 0.45 | ZNF239 | NM_005674 | zinc finger protein 239 (ZNF239), mRNA |
| GE53189 | 0.46 | AUTS2 | NM_015570 | autism susceptibility candidate 2 (AUTS2), mRNA |
| GE81426 | 0.46 | USP9Y | NM_004654 | ubiquitin specific protease 9, Y chromosome (fat facets-like Drosophila) (USP9Y), mRNA |
| GE63274 | 0.48 | CENTG3 | AL537305 | FETAL BRAIN cDNA clone CS0DF025YC08 5-PRIME |
| GE83170 | 0.48 | BCL2L14 | NM_030766 | apoptosis regulator BCL-G (BCLG), transcript variant 2, mRNA |
| GE58554 | 0.49 | PRO1914 | NM_014106 | PRO1914 protein (PRO1914), mRNA |
| GE86889 | 0.49 | SLC26A7 | NM_052832 | solute carrier family 26, member 7 (SLC26A7), transcript variant 1, mRNA |
| GE81520 | 0.49 | BCAT1 | NM_005504 | branched chain aminotransferase 1, cytosolic (BCAT1), mRNA |
Fold change = diseased/normal
Hierarchical clustering
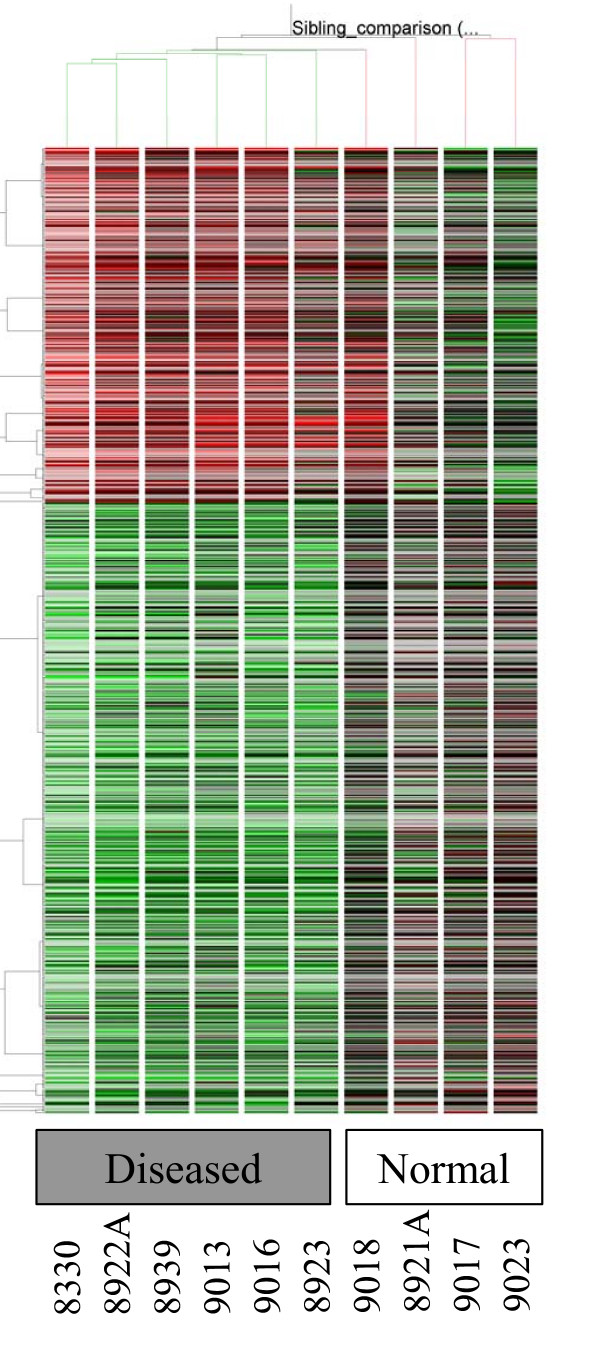
This gene list was subjected to a hierarchical clustering routine in GeneSpring using a Pearson correlation similarity measure across conditions and a standard correlation similarity measure across genes. The final tree is represented in Figure 1.
Figure 1.
Hierarchical clustering of all statistically significant genes that show differential expression between diseased and normal samples. Upregulated are colored red and downregulated genes are colored green. Clustering of genes is shown on the left while condition based tree is shown on the top.
Pathway Analysis
The 1.5 fold list was further analyzed using Onto-Express and Pathway-Express, part of the Onto-Tools package available from the Draghici laboratory. This software allows for identification of pathways that show enrichment in the microarray data and is a novel tool that translates lists of differentially regulated genes into functional profiles characterizing the impact of the condition studied.
Quantitative PCR validation
To verify expression data obtained by the Codelink analysis, Taqman gene expression assays were obtained (Applied Biosystems) to evaluate the expression of selected genes by RT-qPCR. The assays utilized were as follows: TCF2 (Applied Biosystems assay Hs01001609_m1), CXCL10 (Hs00171042_m1), FUT4 (Hs00275643_s1), SERPINB1 (Hs00229084_m1), CD9 (Hs01124026_m1), LYPLA3 (Hs00202399_m1), SERPINB9 (Hs00394497_m1), CD83 (Hs00188486_m1), CDC5L (Hs00904622_m1), TNFRSF19 (Hs00218634_m1), HLA-DRA (Hs00219578_m1). All assays are designed to detect the appropriate transcripts at an exon-exon junction to ensure the detection of mRNA and not genomic DNA.
The expression levels of these 11 genes were evaluated by two-step Taqman-based RT-qPCR. First strand cDNA was synthesized with M-MLV reverse transcriptase (Ambion, Inc., Austin, TX) and primed with oligo-dT per the manufacturer's specifications. The cDNA equivalent of 20 ng starting RNA was then included in reactions containing 1× Taqman Master Mix (Applied Biosystems, Foster City, CA), and 1× gene-specific assay reagents as recommended by the manufacturer (Applied Biosystems). All reactions were run in triplicate. Reactions that did not contain template cDNA were included as negative controls.
Reaction plates were processed on an Applied Biosystems 7900 HT Sequence Detection System. The AmpliTaq Gold polymerase was activated at 95°C for 10 min followed by 40 cycles consisting of denaturation for 15 seconds at 95°C and annealing and extension for 60 seconds at 60°C.
Amplification data was analyzed with the ABI Prism SDS 2.1 software (Applied Biosystems). Although run individually, samples GM8921A, 9017, 9018, and 9023 were pooled for analysis as the control sample, and samples GM8830, 8922A, 8923, 8839, 9013, and 9016 were pooled for analysis as the MS sample, unless otherwise noted. Relative quantification of gene expression was performed by the ΔΔCt method [18]. GAPDH expression was used as an endogenous control to normalize expression within each sample.
Transcription network analysis
We employed a software program that one of our authors continues to develop (G.A.) to recast the matrix of regulatory interactions found in a prior step as a directed graph overlaying color information for gene responses. Our regulatory network was constructed by integrating transcription factor binding site information (from TRANSFAC) with gene expression data using graph TFs (Bioconductor functions for transcription network analysis). The standard microarray experiments cannot measure the transcription factor activities (TFAs) directly, since TFAs are subject to post-translational modifications or ligand binding in order to exert their function.
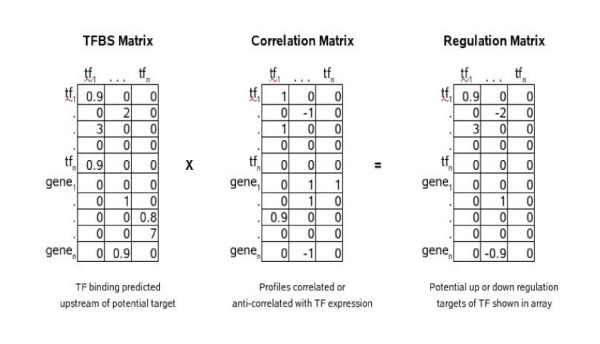
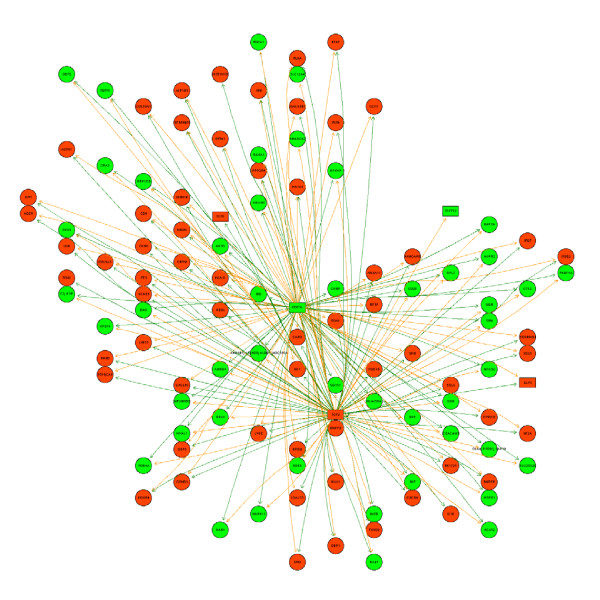
We hypothesized that decoding some of the important TFAs involved in B cells of MS patients might provide important clues to disease pathogenesis or uncover potential molecular targets for therapy. We started with an initial list of all genes changing at the 1.5-fold level. In this dual sample study, we assumed all pairs of genes moving in the same direction to be concordant and all pairs moving in opposite directions to be discordant. Combining this with the potential for regulatory relationships inferred from a TRANSFAC binding site dataset, we were able to extrapolate the potential transcription regulatory networks driving expression changes. We found that TCF2 and CDC5L bind very similar promoter motifs and seem to control overlapping sets of up and down regulated genes in this study. The transcripts we uncovered in this dataset are most notably, CCR1 and CXCR4, molecules which are associated with MS. The multiplication of corresponding cells in the TFBS and correlation matrices yields a regulation matrix where the sign of the interaction score reflects positive or negative regulation and the magnitude of the score reflects the amount of supporting evidence, as shown in the grid below (Figure 2). We examined these networks for various thresholds of fold-change and TRANSFAC evidence scores to obtain the picture of regulation of downstream molecules by tcf2 and cdcl5 shown in Figure 3.
Figure 2.
Analysis of transcription networks from microarray data.
Figure 3.
Transcription factor network deciphered from genes differentially expressed at a 1.5 fold cutoff in microarray data. The color 'red" shows upregulation while "green" shows downregulated expression. Genes are depicted by circles while transcription factors are represented as rectangles. Green arrows indicate that the nodes at each end of the arrow are regulated in the same direction (up or down) while the red arrows connect the nodes that are anticorrelated.
Results
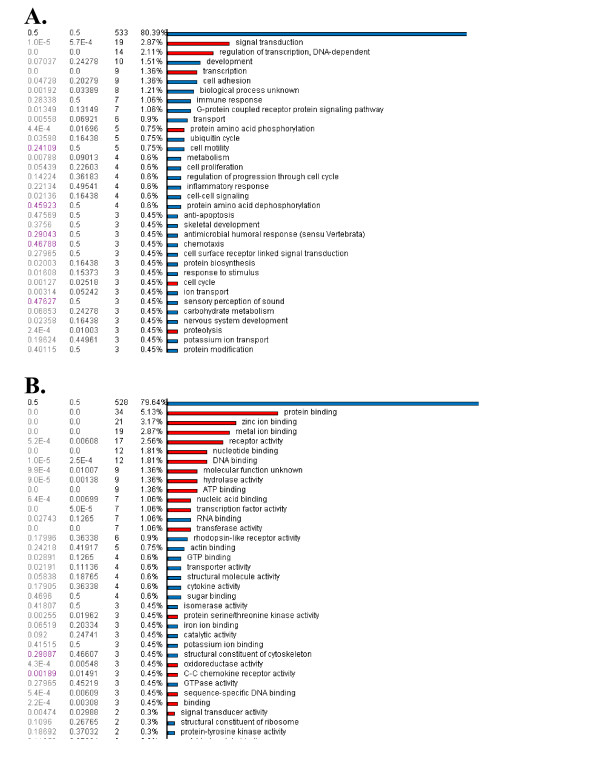
As shown in Figure 1, the rows represent hierarchical clustering of all differentially expressed genes between the diseased (MS) and control (normal siblings) samples. The MS samples were 8923, 8922 A, 8830, 8839, 9013 and 9016, respectively. The unaffected siblings were 9017, 9018, 9023 and 8921A, respectively. Onto-Express analyses depicted in Figure 4 shows that transcription factor binding is a highly significant component in our microarray data. Results from Pathway-Express analysis are listed in Table 4, and these show that immune function and neurospecific pathways are also highly implicated in this data set.
Figure 4.
Results from functional profiling of data using Onto-Express. (A) The figure shows the significant (red) biological processes represented in a set of genes differentially expressed between two clinical groups. (B) The figure shows the significant (red) molecular function represented in a set of genes differentially expressed between two clinical groups.
Table 4.
Results of pathway profiling using Pathway Express.
| Rank | Database Name | Pathway Name | Impact Factor | #Genes in Pathway | #Input Genes in Pathway | #Pathway Genes on Chip | %Input Genes in Pathway | %Pathway Genes in Input | p-value |
| 1 | KEGG | Cell adhesion molecules (CAMs) | 6.94 | 132 | 11 | 126 | 2.099 | 8.333 | 9.69E-04 |
| 2 | KEGG | Axon guidance | 6.94 | 130 | 11 | 126 | 2.099 | 8.462 | 9.69E-04 |
| 3 | KEGG | Leukocyte transendothelial migration | 5.288 | 117 | 9 | 113 | 1.718 | 7.692 | 0.005054 |
| 4 | KEGG | Type I diabetes mellitus | 5.232 | 44 | 5 | 40 | 0.954 | 11.364 | 0.005344 |
| 5 | KEGG | Regulation of actin cytoskeleton | 4.461 | 206 | 12 | 198 | 2.29 | 5.825 | 0.011556 |
| 6 | KEGG | Natural killer cell mediated cytotoxicity | 4.11 | 128 | 8 | 114 | 1.527 | 6.25 | 0.016401 |
| 7 | KEGG | Focal adhesion | 3.949 | 194 | 11 | 188 | 2.099 | 5.67 | 0.019276 |
| 8 | KEGG | Tight junction | 3.187 | 119 | 7 | 112 | 1.336 | 5.882 | 0.041314 |
| 9 | KEGG | Antigen processing and presentation | 3.112 | 86 | 5 | 68 | 0.954 | 5.814 | 0.044499 |
| 10 | KEGG | Adherens junction | 2.721 | 77 | 5 | 76 | 0.954 | 6.494 | 0.065823 |
| 11 | KEGG | Colorectal cancer | 2.676 | 77 | 5 | 77 | 0.954 | 6.494 | 0.068828 |
| 12 | KEGG | Circadian rhythm | 2.386 | 18 | 2 | 18 | 0.382 | 11.111 | 0.091979 |
| 13 | KEGG | Maturity onset diabetes of the young | 1.975 | 25 | 2 | 23 | 0.382 | 8 | 0.138816 |
| 14 | KEGG | VEGF signaling pathway | 1.972 | 72 | 4 | 70 | 0.763 | 5.556 | 0.139208 |
| 15 | KEGG | Fc epsilon RI signaling pathway | 1.83 | 75 | 4 | 74 | 0.763 | 5.333 | 0.160493 |
| 16 | KEGG | mTOR signaling pathway | 1.811 | 49 | 3 | 49 | 0.573 | 6.122 | 0.16353 |
| 17 | KEGG | Huntington's disease | 1.558 | 30 | 2 | 30 | 0.382 | 6.667 | 0.210455 |
| 18 | KEGG | Wnt signaling pathway | 1.474 | 147 | 6 | 144 | 1.145 | 4.082 | 0.229071 |
| 19 | KEGG | MAPK signaling pathway | 1.441 | 273 | 10 | 268 | 1.908 | 3.663 | 0.236637 |
| 20 | KEGG | Toll-like receptor signaling pathway | 1.416 | 91 | 4 | 88 | 0.763 | 4.396 | 0.242667 |
| 21 | KEGG | B cell receptor signaling pathway | 1.318 | 63 | 3 | 63 | 0.573 | 4.762 | 0.267565 |
| 22 | KEGG | T cell receptor signaling pathway | 1.294 | 93 | 4 | 93 | 0.763 | 4.301 | 0.274064 |
| 23 | KEGG | Long-term potentiation | 1.235 | 67 | 3 | 66 | 0.573 | 4.478 | 0.290928 |
| 24 | KEGG | Insulin signaling pathway | 1.143 | 135 | 5 | 131 | 0.954 | 3.704 | 0.318761 |
| 25 | KEGG | Type II diabetes mellitus | 1.117 | 44 | 2 | 41 | 0.382 | 4.545 | 0.327167 |
| 26 | KEGG | Dentatorubropallidoluysian atrophy (DRPLA) | 1.044 | 15 | 1 | 15 | 0.191 | 6.667 | 0.352194 |
| 27 | KEGG | Epithelial cell signaling in Helicobacter pylori infection | 0.997 | 46 | 2 | 45 | 0.382 | 4.348 | 0.368822 |
| 28 | KEGG | Parkinson's disease | 0.992 | 16 | 1 | 16 | 0.191 | 6.25 | 0.370684 |
| 29 | KEGG | Cytokine-cytokine receptor interaction | 0.963 | 256 | 8 | 241 | 1.527 | 3.125 | 0.381922 |
| 30 | KEGG | Jak-STAT signaling pathway | 0.936 | 153 | 5 | 144 | 0.954 | 3.268 | 0.392344 |
| 31 | KEGG | Apoptosis | 0.86 | 84 | 3 | 83 | 0.573 | 3.571 | 0.423249 |
| 32 | KEGG | Complement and coagulation cascades | 0.6 | 69 | 2 | 64 | 0.382 | 2.899 | 0.548544 |
| 33 | KEGG | Olfactory transduction | 0.523 | 31 | 1 | 31 | 0.191 | 3.226 | 0.592482 |
| 34 | KEGG | SNARE interactions in vesicular transport | 0.468 | 36 | 1 | 34 | 0.191 | 2.778 | 0.62642 |
| 35 | KEGG | Phosphatidylinositol signaling system | 0.421 | 79 | 2 | 78 | 0.382 | 2.532 | 0.656227 |
| 36 | KEGG | Taste transduction | 0.317 | 53 | 1 | 45 | 0.191 | 1.887 | 0.728441 |
| 37 | KEGG | GnRH signaling pathway | 0.277 | 97 | 2 | 95 | 0.382 | 2.062 | 0.758254 |
| 38 | KEGG | Gap junction | 0.27 | 99 | 2 | 96 | 0.382 | 2.02 | 0.763352 |
| 39 | KEGG | Adipocytokine signaling pathway | 0.145 | 69 | 1 | 69 | 0.191 | 1.449 | 0.864684 |
| 40 | KEGG | Long-term depression | 0.124 | 75 | 1 | 74 | 0.191 | 1.333 | 0.882976 |
| 41 | KEGG | Neuroactive ligand-receptor interaction | 0.102 | 302 | 5 | 279 | 0.954 | 1.656 | 0.902917 |
| 42 | KEGG | ECM-receptor interaction | 0.086 | 87 | 1 | 86 | 0.191 | 1.149 | 0.91743 |
| 43 | KEGG | Cell cycle | 0.042 | 112 | 1 | 110 | 0.191 | 0.893 | 0.958921 |
| 44 | KEGG | Calcium signaling pathway | 0.041 | 176 | 2 | 173 | 0.382 | 1.136 | 0.959904 |
We selected a subset of genes that were up and down regulated in the MS samples for further validation by RT-qPCR. Our results generally confirmed the results obtained with the Codelink arrays. TCF2, CXCL10, and FUT4 were all up regulated in the MS samples whereas CDC5L, TNFRSF19 and HLA-DR were down regulated (Table 5).
Table 5.
Relative Expression of Selected Genes by RT-qPCR.
| Control | MS | |||
| RE | +/- SD | RE | SD | |
| TCF2 | 1.00 | 0.68 | 7.95 | 4.25 |
| CXCL10 | 1.00 | 0.47 | 5.93 | 3.85 |
| FUT4 | 1.00 | 0.43 | 3.93 | 2.23 |
| SERPIN B1 | 1.00 | 0.57 | 1.71 | 1.07 |
| CDC5L | 1.00 | 0.62 | 0.54 | 0.26 |
| TNFRSF19 | 1.00 | 0.40 | 0.52 | 0.28 |
| HLA-DRA | 1.00 | 0.49 | 0.33 | 0.16 |
RE; relative expression.
+/-; calculated by determining relative expression for the value obtained by adding the ΔΔCt and ΔCt standard deviation (RE +/-), and then subtracting the RE +/- value from the RE.
For three other genes, we obtained disparate results for expression when the control and MS samples were pooled for analysis. CD83 was up regulated by RT-qPCR while down regulated by array analysis, SERPINB9 was down regulated by RT-qPCR while up regulated on the arrays, and CD9 was only minimally up regulated by RT-qPCR while clearly up regulated on the arrays (Table 6). For each of these genes, some samples were negative for expression by RT-qPCR (control sample 8921A for SERPINB9 and CD9, MS sample 8922A for CD83), and the influence of the negative values is not fully accounted for when pooling the samples prior to analysis by the ΔΔCt method. Thus, for these genes we further evaluated the RT-qPCR data by analyzing the control and MS samples individually and then averaging, rather than pooling the samples prior to the ΔΔCt analysis. After evaluating individual samples, we also found that some control samples were significant outliers for the CD83 and SERPINB9 genes. Control sample 8921A had a 26-fold increase in CD83 expression relative to the next highest control sample, and control sample 9018 had a 255-fold increase in SERPINB9 expression and a 156-fold increase in CD9 expression relative to the next highest samples for those genes (data not shown). To compensate for these outliers, we eliminated the highest expression samples from both the control and MS samples from the analysis and also included the negative samples. When analyzed in this way, CD83 expression in the MS samples was down regulated by 0.58-fold, SERPINB9 expression was up regulated by 2.47-fold, and CD9 expression was up regulated 3.5-fold relative to the Control samples (Table 6) and these results are in concordance with array data for these genes.
Table 6.
Analysis of SERPINB9, CD83, and CD9 Expression by RT-qPCR.
| SERPINB9 | CD83 | CD9 | ||||
| Pool | Indiv | Pool | Indiv | Pool | Indiv | |
| Control | 1 | 1 | 1 | 1 | 1 | 1 |
| MS | 0.5 | 2.47 | 1.1 | 0.58 | 1.23 | 3.5 |
1. Pool; after RT-qPCR, Control and MS samples pooled prior to calculation of RE by ΔΔCt method.
2. Indiv; RE of samples first determined by ΔΔCtCt method, then Control and MS sample REs averaged (see text for details).
Although we included a Taqman assay for LYPLA3 in our validation studies, we were not able evaluate expression of this gene by RT-qPCR. None of the four Control samples were consistently positive for expression of this gene by RT-qPCR, and only one of the six MS samples was consistently positive. The probe for this gene on the Codelink array is located near the 3' end of the coding sequence whereas the Taqman assay detects sequences at the 5' end of the coding sequence, at the junction between exons 1 and 2 (6 total exons). Thus, full length cDNA would be required to detect LYPLA3 expression with the Taqman assay we employed. Our inability to consistently detect this gene suggests that we did not generate full length LYPLA3 cDNA during the RT step - either due to mRNA instability or inefficient reverse transcription.
Additional data
A list of genes at 1.5 fold differential expression from microarray data can be found in Additional file 1.
Discussion and Conclusion
The genes involved in our analyses encode proteins involved in apoptosis, cytokine pathways and inflammation. Although up or downregulation of gene transcription is not reflected in a 1:1 translation of protein expression, the gene product generally follows gene regulation dynamics [19]. A growing number of expression profiling studies provide experimental evidence indicating the presence of a transcriptionally distinct gene pattern in MS. Much of our current knowledge of MS stems from the analysis of a mouse model, experimental autoimmune encephalomyelitis (EAE) that is thought to be similar to MS. While EAE has striking clinical and histopathological similarities to MS, it has failed in predicting the efficacy of new therapeutics [20]. Our discussion is largely restricted to those genes that had > 2.5 fold up-regulation and the two transcription factors that had the most effect on other genes. Among the total list of differentially expressed genes, the proportion of downregulated genes was higher than that of upregulated ones (262 vs. 81), an observation that suggests an interplay involving complex inflammatory cascades in MS.
Study limitations
The findings in our study are limited by a) sample size and b) the cellular pathways that Epstein-Barr virus (EBV) regulates remain poorly characterized and there are few data on how EBV may influence gene pathways in B cells derived from MS patients vs. controls. The ideal starting material would be to obtain native B cells from controls and MS patients; however, collection and processing bias are hard to eliminate in any microarray-based study.
Upregulated genes
The gene for CXCL10 (interferon-γ-inducible protein-10) encodes for an interferon (gamma)-induced, secreted protein of 10 kDa, a chemokine of the CXC subfamily that is one of the ligands for the receptor CXCR3. The binding of this protein to CXCR3 causes pleiotropic effects, including stimulation of monocytes, natural killer and T-cell migration, and modulation of adhesion molecule expression (UCSC web browser). Its levels are increased in cerebrospinal fluid of MS patients with symptomatic attacks of demyelination, suggesting a role for this molecule in MS [21]. Following mouse hepatitis virus (MHV) infection in mice [22], CXCL10 (IFN inducible protein 10 kDa) protein was expressed during both acute and chronic stages of disease suggesting a role for this protein in disease exacerbation and chronicity. Previous studies have shown that during the acute phase of infection, T lymphocytes are recruited into the CNS by CXCL10. Our finding that CXCL10 upregulation (5.8-fold) in B cells from MS siblings is supportive of prior observations.
Serpin B1 is a leukocyte elastase inhibitor and regulates the activity of the neutrophil proteases elastase, cathepsin G and proteinase-3. In humans, serpins constitute 10% of the plasma proteins and are known as critical regulators of both thrombotic and fibrinolytic systems. Serpins participate in the regulation of the complement cascade, angiogenesis, apoptosis and innate immunity. Most of the human clade B serpins inhibit serine and/or papain-like cysteine proteinases and protect cells from exogenous and endogenous proteinase-mediated injury. As some serpins also guard cells against the deleterious effects of promiscuous proteolytic activity [23], it is possible that the cytoprotective function is a common feature of intracellular serpin clades and we hypothesize that up-regulation of serpin B1 gene may be protective in MS. While our findings need to be validated, serpin B1 gene may represent a novel therapeutic target to ameliorate MS, considering the importance of these molecules in regulating proteolytic cascades.
We found that FUT4 gene was upregulated by 2.5 fold. It is a member of the interleukin 1 cytokine family and the protein encoded by the gene is produced by activated macrophages as a pro-protein and processed to its active form by caspase 1 (CASP1/ICE). This cytokine is an important mediator of the inflammatory response, cell proliferation, differentiation, and apoptosis. The induction of cyclooxygenase-2 (PTGS2/COX2) by this cytokine in the central nervous system (CNS) is found to contribute to inflammatory pain hypersensitivity. FUT4 mRNA is increased in Jurkat cells undergoing apoptosis.
LYPLA3 (lysosomal phospholipase A2): Lysophospholipases are enzymes that act on biological membranes to regulate the multifunctional lysophospholipids. The protein encoded by this gene hydrolyzes lysophosphatidylcholine to glycerophosphorylcholine and a free fatty acid. LYPLA3 is present in the plasma and thought to be associated with high-density lipoprotein. Cellular phospholipases are key participants in cellular transduction [24] and are thought to be involved in the pathogenesis of local and systemic inflammatory disorders [25]. Interestingly, cytosolic phospholipase A2 levels were found to be low, possibly reflecting proteolysis or inactivation of enzyme activity in brain MS lesions [26].
The gene for XCL1 was upregulated; it encodes for chemokines, a group of small (8-14 kD) molecules that regulate cell trafficking of leukocytes through interactions with a subset of 7-transmembrane, G protein-coupled receptors. Chemokines also play fundamental roles in the development, homeostasis, and function of the immune system, and they have effects on cells of the central nervous system as well as on endothelial cells involved in angiogenesis or angiostasis. The protein product of XCL1 has chemotactic activity for neutrophils may play a role in inflammation and exerts its effects on endothelial cells in an autocrine fashion. We also found PSTPIP1 (proline-serine-threonine phosphatase interacting protein1) upregulated; its role in MS is unclear.
Downregulated genes
An important finding in our study is an 11.7-fold downregulation of eukaryotic translation initiation factor 1A, Y-linked (EIF1AY) gene. It is thought that EIF1AY encodes for minor histocompatibility antigen (mHA) and B-cell mediated antibody response to Y-chromosome encoded histocompatibility antigens (H-Y antigens) is associated with maintenance of disease remission in graft-vs-host disease [27]. The B cells may provide a new target for immune intervention in chronic graft vs. host disease and whether such a correlation can be translated to chronic MS requires further study.
We found that the tumor necrosis factor receptor (TNFR) superfamily 19 gene (TNFRSF 19) was downregulated by 5.3-fold. In a recent study by Achiron et al., TNF superfamily members 14, 1B, 4, and 6 were downregulated. The TNFR superfamily of cell surface receptors share approximately 80 amino acids within their cytoplasmic region called the death domain (DD), critical for recruiting the death machinery [28]. Individual TNF receptors are expressed in different cell types and have a range of affinities for various intracellular molecules that provide signaling and biological specificities. However, much is unknown about how individual signal transduction pathways are regulated upon activation by any particular TNF receptor, under physiological conditions [29] and while TNFR superfamily 19 genes may mediate important functions in immunity, inflammation, differentiation, cell proliferation control, and apoptosis, their specific effects in MS are unknown.
The gene for CD83 was downregulated in MS siblings. CD83 molecules are expressed at a high level on immune-competent, activated and mature dendritic cells. In MS, CD83-positive dendritic cells are present in occasional perivascular cuffs in lesions, suggesting that mature dendritic cells migrate to the CNS in response to chemokine signals [30].
The downregulation of insulin receptor tyrosine kinase substrate (BAIAP2L1) gene in B cells of MS siblings is significant. The insulin-like growth factor I receptor (IGF-IR), is a member of the receptor tyrosine kinase family of growth factor receptors. In rheumatoid arthritis (RA) it has been shown that IgG antibodies from the sera of patients can stimulate synovial fibroblasts through interaction with insulin-like growth factor receptor 1 (IGF-R1), provoking trafficking of T cells [31]. The data demonstrate, for the first time, a bridging link between B-cell activity and T-cell trafficking. In addition, they are of potential importance for the development of innovative therapeutic strategies, in which interrupting the IGF-1/IGF-1R axis could result in sustained disease modification by affecting both the growth-factor triggered activation of fibroblasts and the accumulation of T lymphocytes. A similar link in MS has not been demonstrated but is plausible. Other genes downregulated include ADAMTS16, BID, MIF and DAPK2, involved in the apoptosis pathway.
TCF2, diabetes and MS
Our transcription and microarray analysis showed that TCF2 was upregulated. TCF2 encodes transcription factor 2, a liver-specific factor of the homeobox-containing basic helix-turn-helix family. The TCF2 protein forms heterodimers with another liver-specific member of this transcription factor family, TCF1; depending on the TCF2 isoform, activation or inhibition of transcription of target genes occurs. We found that TCF2 upregulates the CC chemokine receptor 1 (CCR1) a molecule expressed on macrophages and T cells; in MS plaques, numerous CCR1-positive infiltrating macrophages and microglial cells, associated with CCL3, were found [2] and CCR1 has been associated with newly infiltrating monocytes in MS lesions [32]. CCR1-deficient mice show an attenuated EAE course [33]. Additionally, CXCR1, a pro-inflammatory molecule is upregulated by TCF2. Furthermore, TCF2 downregulated CD28, a co-stimulatory molecule involved in T-cell stimulation and IL6R, a pro-inflammatory molecule. Taken together, it is plausible that TCF2 plays a central role in the pathogenesis of MS.
Interestingly, a mutation in TCF2 has been linked to the etiology of MODY5 (Maturity-Onset of Diabetes, Type 5) and in a recent Danish nationwide cohort study, intra-individual and, to a lesser degree, intra-familial co-occurrence was evident in MS and type 1 diabetes. The underlying mechanisms may involve both genetic and environmental factors [34]. Individuals with type 1 diabetes are more than three times more likely to develop MS than controls; in addition, the two diseases appear to be linked, albeit to a weak extent, within families. The exact mechanism of how mutations in these transcription factors cause diabetes remains unknown. To reiterate, a link between TCF2 mutation and MODY5 has been established, while type 1 diabetics have an increased risk of MS development. Our study points to TCF2 having a prominent role in regulation of other transcription factors. In summary, the role of TCF2 in MS needs to be further explored since MODY5 and TCF2 are linked and type 1 diabetes confers increased risk of MS development.
Another transcription factor that we found in our transcription analysis, CDC5L, was downregulated. It is a well-characterized pre-mRNA splicing factor and involved in cell cycle kinetics in yeast, S. pombe; its role in MS, if any, is not characterized.
Competing interests
The author(s) declare that they have no competing interests.
Authors' contributions
JRA (Jagannadha R Avasarala) designed, conceptualized the study format and wrote the manuscript; SVC (Sridar V Chittur) did the microarray studies, analyzed the data and authored the materials and methods section; JAT (John A Tine) did quantitative RT-PCR while ADG (Ajish D George) performed the transcription factor network analysis. All authors have read and approved the final version of the manuscript.
Pre-publication history
The pre-publication history for this paper can be accessed here:
Supplementary Material
List of genes at 1.5 fold differential expression from microarray data. The data provided represents that complete list of statistically significant genes from the microarray analysis, which show differential expression at a 1.5 fold cut-off.
Acknowledgments
Acknowledgements
No funding was obtained to complete this study.
Contributor Information
Jagannadha R Avasarala, Email: javasarala@yahoo.com.
Sridar V Chittur, Email: schittur@albany.edu.
Ajish D George, Email: ajishg@gmail.com.
John A Tine, Email: jtine@csc.albany.edu.
References
- Hafler DA. Multiple sclerosis. J Clin Invest. 2004;113:788–94. doi: 10.1172/JCI200421357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balashov KE, Rottman JB, Weiner HL, Hancock WW. CCR5(+) and CXCR3(+) T cells are increased in multiple sclerosis and their ligands MIP-1alpha and IP-10 are expressed in demyelinating brain lesions. Proc Natl Acad Sci USA. 1999;96:6873–8. doi: 10.1073/pnas.96.12.6873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsey JW. Familial recurrence rates and genetic models of multiple sclerosis. Am J Med Genet A. 2005;135:53–8. doi: 10.1002/ajmg.a.30702. [DOI] [PubMed] [Google Scholar]
- Ebers GC, Bulman DE, Sadovnick AD, Paty DW, Warren S, Hader W, Murray TJ, Seland TP, Duquette P, Grey T, et al. A population-based study of multiple sclerosis in twins. N Engl J Med. 1986;315:1638–42. doi: 10.1056/NEJM198612253152603. [DOI] [PubMed] [Google Scholar]
- Mackay RP, Myrianthopoulos NC. Multiple sclerosis in twins and their relatives. Arch Neurol. 1966;15:449–62. doi: 10.1001/archneur.1966.00470170003001. [DOI] [PubMed] [Google Scholar]
- Robertson NP, O'Riordan JI, Chataway J, Kingsley DP, Miller DH, Clayton D, Compston DA. Offspring recurrence rates and clinical characteristics of conjugal multiple sclerosis. Lancet. 1997;349:1587–90. doi: 10.1016/S0140-6736(96)07317-5. [DOI] [PubMed] [Google Scholar]
- Sospedra M, Muraro PA, Stefanová I, Zhao Y, Chung K, Li Y, Giulianotti M, Simon R, Mariuzza R, Pinilla C, Martin R. Redundancy in antigen-presenting function of the HLA-DR and -DQ molecules in the multiple sclerosis-associated HLA-DR2 haplotype. J Immunol. 2006;176:1951–61. doi: 10.4049/jimmunol.176.3.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luomala M, Lehtimäki T, Elovaara I, Wang X, Ukkonen M, Mattila K, Laippala P, Koivula T, Hurme M. A study of interleukin-1 cluster genes in susceptibility to and severity of multiple sclerosis. J Neurol Sci. 2001;185:123–7. doi: 10.1016/S0022-510X(01)00482-8. [DOI] [PubMed] [Google Scholar]
- Luomala M, Elovaara I, Ukkonen M, Koivula T, Lehtimaki T. Plasminogen activator inhibitor 1 gene and risk of MS in women. Neurology. 2000;54:1862–4. doi: 10.1212/wnl.54.9.1862. [DOI] [PubMed] [Google Scholar]
- Mattila KM, Luomala M, Lehtimaki T, Laippala P, Koivula T, Elovaara I. Interaction between ESR1 and HLA-DR2 may contribute to the development of MS in women. Neurology. 2001;56:1246–7. doi: 10.1212/wnl.56.9.1246. [DOI] [PubMed] [Google Scholar]
- Tourtellotte WW, Baumhefner RW, Syndulko K, Shapshak P, Osborne M, Rubinshtein G, Newton L, Ellison G, Myers L, Rosario I. The long march of the cerebrospinal fluid profile indicative of clinical definite multiple sclerosis; and still marching. J Neuroimmunol. 1988;20:217–27. doi: 10.1016/0165-5728(88)90163-4. [DOI] [PubMed] [Google Scholar]
- Qin Y, Duquette P, Zhang Y, Talbot P, Poole R, Antel J. Clonal expansion and somatic hypermutation of V(H) genes of B cells from cerebrospinal fluid in multiple sclerosis. J Clin Invest. 1998;102:1045–50. doi: 10.1172/JCI3568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genain CP, Cannella B, Hauser SL, Raine CS. Identification of autoantibodies associated with myelin damage in multiple sclerosis. Nat Med. 1999;5:170–5. doi: 10.1038/5532. [DOI] [PubMed] [Google Scholar]
- Cepok S, Zhou D, Srivastava R, Nessler S, Stei S, Büssow K, Sommer N, Hemmer B. Identification of Epstein-Barr virus proteins as putative targets of the immune response in multiple sclerosis. J Clin Invest. 2005;115:1352–60. doi: 10.1172/JCI200523661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lock C, Hermans G, Pedotti R, Brendolan A, Schadt E, Garren H, Langer-Gould A, Strober S, Cannella B, Allard J, Klonowski P, Austin A, Lad N, Kaminski N, Galli SJ, Oksenberg JR, Raine CS, Heller R, Steinman L. Gene-microarray analysis of multiple sclerosis lesions yields new targets validated in autoimmune encephalomyelitis. Nat Med. 2002;8:500–8. doi: 10.1038/nm0502-500. [DOI] [PubMed] [Google Scholar]
- Bittner M, Meltzer P, Chen Y, Jiang Y, Seftor E, Hendrix M, Radmacher M, Simon R, Yakhini Z, Ben-Dor A, Sampas N, Dougherty E, Wang E, Marincola F, Gooden C, Lueders J, Glatfelter A, Pollock P, Carpten J, Gillanders E, Leja D, Dietrich K, Beaudry C, Berens M, Alberts D, Sondak V. Molecular classification of cutaneous malignant melanoma by gene expression profiling. Nature. 2000;406:536–40. doi: 10.1038/35020115. [DOI] [PubMed] [Google Scholar]
- Gene Expression Omnibus Viewer http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=pnsxvqskiumgevk&acc=GSE10064
- Winer J, Jung CK, Shackel I, Williams PM. Development and validation of real-time quantitative reverse transcriptase-polymerase chain reaction for monitoring gene expression in cardiac myocytes in vitro. Anal Biochem. 1999;270:41–9. doi: 10.1006/abio.1999.4085. [DOI] [PubMed] [Google Scholar]
- Gebicke-Haerter PJ. Microarrays and expression profiling in microglia research and in inflammatory brain disorders. J Neurosci Res. 2005;81:327–41. doi: 10.1002/jnr.20479. [DOI] [PubMed] [Google Scholar]
- Steinman L. Immunotherapy of multiple sclerosis: the end of the beginning. Curr Opin Immunol. 2001;13:597–600. doi: 10.1016/S0952-7915(00)00266-1. [DOI] [PubMed] [Google Scholar]
- Galimberti D, Scalabrini D, Fenoglio C, Comi C, De Riz M, Venturelli E, Lovati C, Mariani C, Monaco F, Bresolin N, Scarpini E. CXCL10 haplotypes and multiple sclerosis: association and correlation with clinical course. Eur J Neurol. 2007;14:162–7. doi: 10.1111/j.1468-1331.2006.01629.x. [DOI] [PubMed] [Google Scholar]
- Liu MT, Keirstead HS, Lane TE. Neutralization of the chemokine CXCL10 reduces inflammatory cell invasion and demyelination and improves neurological function in a viral model of multiple sclerosis. J Immunol. 2001;167:4091–7. doi: 10.4049/jimmunol.167.7.4091. [DOI] [PubMed] [Google Scholar]
- Silverman GA, Whisstock JC, Askew DJ, Pak SC, Luke CJ, Cataltepe S, Irving JA, Bird PI. Human clade B serpins (ov-serpins) belong to a cohort of evolutionarily dispersed intracellular proteinase inhibitor clades that protect cells from promiscuous proteolysis. Cell Mol Life Sci. 2004;61:301–25. doi: 10.1007/s00018-003-3240-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis EA, Rhee SG, Billah MM, Hannun YA. Role of phospholipase in generating lipid second messengers in signal transduction. Faseb J. 1991;5:2068–77. doi: 10.1096/fasebj.5.7.1901288. [DOI] [PubMed] [Google Scholar]
- Pruzanski W, Vadas P. Phospholipase A2 - a mediator between proximal and distal effectors of inflammation. Immunol Today. 1991;12:143–6. doi: 10.1016/S0167-5699(05)80042-8. [DOI] [PubMed] [Google Scholar]
- Huterer SJ, Tourtellotte WW, Wherrett JR. Alterations in the activity of phospholipases A2 in postmortem white matter from patients with multiple sclerosis. Neurochem Res. 1995;20:1335–43. doi: 10.1007/BF00992509. [DOI] [PubMed] [Google Scholar]
- Miklos DB, Kim HT, Miller KH, Guo L, Zorn E, Lee SJ, Hochberg EP, Wu CJ, Alyea EP, Cutler C, Ho V, Soiffer RJ, Antin JH, Ritz J. Antibody responses to H-Y minor histocompatibility antigens correlate with chronic graft-versus-host disease and disease remission. Blood. 2005;105:2973–8. doi: 10.1182/blood-2004-09-3660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen HM, Pervaiz S. TNF receptor superfamily-induced cell death: redox-dependent execution. Faseb J. 2006;20:1589–98. doi: 10.1096/fj.05-5603rev. [DOI] [PubMed] [Google Scholar]
- Dempsey PW, Doyle SE, He JQ, Cheng G. The signaling adaptors and pathways activated by TNF superfamily. Cytokine Growth Factor Rev. 2003;14:193–209. doi: 10.1016/S1359-6101(03)00021-2. [DOI] [PubMed] [Google Scholar]
- Plumb J, Armstrong MA, Duddy M, Mirakhur M, McQuaid S. CD83-positive dendritic cells are present in occasional perivascular cuffs in multiple sclerosis lesions. Mult Scler. 2003;9:142–7. doi: 10.1191/1352458503ms890oa. [DOI] [PubMed] [Google Scholar]
- Pritchard J, Tsui S, Horst N, Cruikshank WW, Smith TJ. Synovial fibroblasts from patients with rheumatoid arthritis, like fibroblasts from Graves' disease, express high levels of IL-16 when treated with Igs against insulin-like growth factor-1 receptor. J Immunol. 2004;173:3564–9. doi: 10.4049/jimmunol.173.5.3564. [DOI] [PubMed] [Google Scholar]
- Trebst C, Sørensen TL, Kivisäkk P, Cathcart MK, Hesselgesser J, Horuk R, Sellebjerg F, Lassmann H, Ransohoff RM. CCR1+/CCR5+ mononuclear phagocytes accumulate in the central nervous system of patients with multiple sclerosis. Am J Pathol. 2001;159:1701–10. doi: 10.1016/s0002-9440(10)63017-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rottman JB, Slavin AJ, Silva R, Weiner HL, Gerard CG, Hancock WW. Leukocyte recruitment during onset of experimental allergic encephalomyelitis is CCR1 dependent. Eur J Immunol. 2000;30:2372–7. doi: 10.1002/1521-4141(2000)30:8<2372::AID-IMMU2372>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- Nielsen NM, Westergaard T, Frisch M, Rostgaard K, Wohlfahrt J, Koch-Henriksen N, Melbye M, Hjalgrim H. Type 1 diabetes and multiple sclerosis: A Danish population-based cohort study. Arch Neurol. 2006;63:1001–4. doi: 10.1001/archneur.63.7.1001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
List of genes at 1.5 fold differential expression from microarray data. The data provided represents that complete list of statistically significant genes from the microarray analysis, which show differential expression at a 1.5 fold cut-off.