Abstract
The sequence-tagged connector (STC) strategy proposes to generate sequence tags densely scattered (every 3.3 kilobases) across the human genome by arraying 450,000 bacterial artificial chromosomes (BACs) with randomly cleaved inserts, sequencing both ends of each, and preparing a restriction enzyme fingerprint of each. The STC resource, containing end sequences, fingerprints, and arrayed BACs, creates a map where the interrelationships of the individual BAC clones are resolved through their STCs as overlapping BAC clones are sequenced. Once a seed or initiation BAC clone is sequenced, the minimum overlapping 5′ and 3′ BAC clones can be identified computationally and sequenced. By reiterating this “sequence-then-map by computer analysis against the STC database” strategy, a minimum tiling path of clones can be sequenced at a rate that is primarily limited by the sequencing throughput of individual genome centers. As of February 1999, we had deposited, together with The Institute for Genomic Research (TIGR), into GenBank 314,000 STCs (≈135 megabases), or 4.5% of human genomic DNA. This genome survey reveals numerous genes, genome-wide repeats, simple sequence repeats (potential genetic markers), and CpG islands (potential gene initiation sites). It also illustrates the power of the STC strategy for creating minimum tiling paths of BAC clones for large-scale genomic sequencing. Because the STC resource permits the easy integration of genetic, physical, gene, and sequence maps for chromosomes, it will be a powerful tool for the initial analysis of the human genome and other complex genomes.
The Human Genome Project is committed to generating four types of maps: genetic, physical, gene, and sequence. In the final analysis, each of these maps is being created by the localization of unique DNA sequence markers by PCR analysis or hybridization to individual chromosomes or fragments thereof (1–4). The genetic map positions genetic markers (simple sequence repeats or single-nucleotide polymorphisms) by following their segregation in families (5–7). The physical map positions sequence-tagged sequences (STSs) by binning to radiation hybrid chromosomal fragments (8, 9) or determining ordered STS positions across large insert clones such as yeast artificial chromosomes or bacterial artificial chromosomes (BACs) (1, 2). The gene sequences, expressed sequence tags (ESTs) or cDNA clones, are mapped either to chromosomes by in situ hybridization (10) or to yeast artificial chromosome or BAC clones by hybridizations (1, 2). Finally, the sequence map can be determined by identifying a minimum overlapping (tiling) path of large-insert clones across each chromosome (generally BAC clones) and then sequencing the individual BAC clones on the minimum tiling path. The commonly used shotgun sequencing approach randomly cleaves each 100- to 200-kilobase (kb) BAC insert into 1- to 4-kb fragments, subclones them into plasmid or phage vectors, and sequences one or both ends of these small inserts to ≈10-fold sequence coverage of the original insert. These fragments are computationally assembled into a consensus sequence, and directed sequencing of incomplete or low-quality areas then permits finishing the sequence to an accuracy of less than 1 error in 10,000.
Human genome sequencing has moved into a new phase with the announcements by Celera, Inc. that it will carry out a shotgun sequence analysis of the entire human genome in 3 years (11) and by the National Institutes of Health and the Department of Energy that the human genome will be sequenced in “first draft” (5- to 10-fold shotgun sequence coverage without finishing) by spring of 2000, and in a highly accurate final draft by 2003 (12, 13). The National Institutes of Health/Department of Energy efforts face two challenging problems: (i) how can seed or initiation BAC clones spaced as evenly as possible across the genome [ideally every 1–2 megabases (Mb)] be identified and (ii) how can a minimum tiling path of BAC clones be generated efficiently connecting all of the sequenced seed BAC clones?
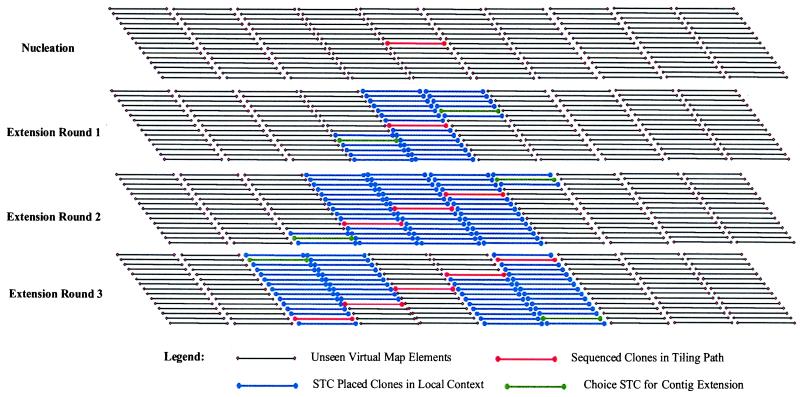
In 1996, we proposed a new strategy called the sequence-tagged connector (STC) approach that will solve both of these problems (14). Since then, we have proposed a human STC resource with three components: (i) an array of 450,000 BAC clones (>20-fold clone coverage of the genome) averaging 150 kb or greater in insert length in 384-well microtiter plates; (ii) the sequence of both ends of each arrayed BAC clone generating 900,000 STC sequences; and (iii) a single restriction enzyme analysis of each BAC clone generating 450,000 fingerprints. The BAC clones would be widely available and distributed to major genome centers. The 900,000 STCs would, on average, be scattered every 3.3 kb across the genome. Thus, the STCs create a virtual map joining together all 450,000 BAC clones (Fig. 1). These virtual relationships can be made explicit by a process termed sequence extension. In this approach, the minimum tiling paths are identified by sequencing a seed BAC clone and analyzing it against the STC database. On average, 45 STC matches (1 every 3.3 kb) would align their corresponding BACs across the seed BAC sequence. The BAC clones minimally overlapping the 5′ and 3′ ends could be verified for genomic fidelity (e.g., identifying possible deletions, insertions, or rearrangements) by matching their fingerprints against those of the BAC clones overlapping them and these, in turn, could be sequenced in a reiterative process (Fig. 1). In this way, large genome centers could sequence from many seed BAC clones and small genome centers from a few.
Figure 1.
A model for the virtual genome map generated by the STC resource. The sequence of a nucleation (seed) BAC clone reveals overlapping BAC clones through a search of the STC database with this sequence. The minimal overlapping 5′ and 3′ BAC clones can then be chosen for sequence extension. As this process is reiterated, the virtual genome map becomes explicit.
Over the past year, the University of Washington and The Institute for Genomic Research (TIGR) have sequenced 314,000 STCs to generate a 135-Mb scan of the human genome. Computational searches of these data suggest that the STC approach will readily permit the construction of a minimum tiling path of BAC clones in the analyzed genomic regions. Moreover, by virtue of STC hits against mapped sequence markers (simple sequence repeats, STSs, ESTs, cDNAs), thousands of BAC clones have been placed on chromosomal maps and now represent potential positioned seed BAC clones. This analysis has also allowed the assembly of a deep, genome-wide repeat library (J.C.W., T.H., L.H., and G.G.M., unpublished data). Finally, the STC resource will be a very powerful tool for integrating the four types of chromosomal maps: genetic, physical, gene, and sequence.
METHODS
Collaborations.
As of February 1999, the University of Washington and TIGR have collaborated in sequencing ≈160,000 and 150,000 STCs, respectively. The Internal Review Board-approved BAC clone libraries are the California Institute of Technology library D (with one aliquot of HindIII partial digest and a second EcoRI partial digest) supplied by Hiroaki Shizuya and Mel Simon (California Institute of Technology, Pasadena, CA), and the RPCI-11 library (a partial EcoRI digest) supplied by Pieter de Jong (Roswell Park Cancer Institute, Buffalo, NY).
High-Throughput STC Mapping and Sequencing Facility at the University of Washington.
The BAC DNA is purified by using an Autogen 740, a DNA extraction robot capable of purifying 288 samples every 24 hours. Packard Multiprobe 204 and 208 robots were used to transfer sample liquids 96 at a time. BAC-end sequencing was carried out with BigDye terminators (Applied Biosystems Division of Perkin–Elmer) by using about 0.75–1.0 μg of DNA and ABD 377 sequencers (3–5 96-lane runs per day). BAC DNA was digested with the restriction enzyme HindIII and separated by size by using 0.8% agarose gels and electrophoresis for 18 hours (384 single enzyme fingerprints per day per gel box). The agarose gels were stained with Vistra Green dye and imaged with a Molecular Dynamics 595 Flourimager. Band sizes and total lengths were determined by frag, a custom software program (A.K., S.S., C. Abajian, and G.M., unpublished data). Thus, this program permits an estimate of BAC-insert lengths. Plates containing these BAC clones were initially bar-coded, and the data were automatically tracked by a laboratory information management system. All data were automatically transferred to a central server for processing and analysis.
RESULTS AND DISCUSSION
General Features of STC Sequences.
An analysis of 314,000 human STCs exhibited an average STC sequence length of 432 bp. These data resulted in a scan of 135 Mb of human DNA sequence [(3.14 × 105 STCs) × (4.32 × 102 bp per STC)]. STCs generated by the the University of Washington High-Throughput Sequencing Center (HTSC) averaged >200 bp of nucleotides with a Phred quality of 20 (a Phred 20 quality base has a 99% probability that it is correct) (15–17). About 85% of the STCs have 100 bp or more of contiguous unique (not matching known repeats) sequence, representing unique genome addresses. The average STC has 281 bp of unique (nonrepetitive) sequence. Sixty-six percent of the STCs are matching pairs on individual BAC clones.
STC Fingerprints.
As of February 1999, we have generated >75,000 DNA fingerprints (from 77% of the BAC clones with HTSC end sequences). We estimate that the HindIII component of Caltech Library D has an average insert size of 121 kb (40,000 fingerprints), the EcoRI component of Caltech Library D has an average insert size of 128 kb (40,000 fingerprints), and segments 3 and 4 of the Roswell Park BAC library (20,000 fingerprints) indicate an average insert size of 168 kb.
A comparison of insert sizes estimated from fingerprints by using frag and actual insert sizes from frag clones that have both ends aligning on large contiguous genomic sequences establish that frag typically underestimates the BAC insert sizes by ≈10%, presumably because of the failure to detect fragments smaller than 2 kb (A.K., S.S., C. Abajian, and G.M., unpublished data).
The STC Resource Facilitates Genomic Sequencing.
We have analyzed the distribution of STC hits across ≈1.8 Mb of human chromosomal sequence [the BRCA2 (0.7 Mb) and the α/δ T cell receptor (1.1 Mb) loci (18). In Table 1, we present an analysis from a database of 509,000 STCs on the mean distance between STCs and the number of STC spacings that fall into 5-kb bins. This analysis is done for all STCs (in either orientation) and STCs in the same 3′ or 5′ orientations (a given minimally overlapping STC has a 50% chance of being oriented in the correct direction for sequence extension). Two points are obvious. First, the mean spacing distribution for 509,000 STCs for either orientation is about half that for spacings in the same orientation. This observation means that in these examples, there are similar numbers of STCs oriented in the 3′ and 5′ directions. Moreover, the mean STC spacing across these two loci is about what is expected (for 314,000 STCs, 10 kb theoretically vs. 11.2 kb observed, and for 509,000 STCs, 6 kb theoretically vs. 6.6 kb observed). Second, with an increased number of STCs, the numbers of STCs separated by larger spaces decreases markedly. Together with TIGR, we will generate about 900,000 STCs (1 STC per 3.3 kb) by the end of August 1999, thus reducing the minimal overlap among BAC clones by approximately one third [ranging from 3.3 to 6.6 kb (one and both orientations, respectively; see Table 1) and averaging 5 kb (3.3 kb + 6.6 kb/2) from the 10-kb map].
Table 1.
Spacing of STC markers across the BRCA2 (700 kb) and the T cell receptor α/δ loci (1.1 Mb)
| Distance, kb |
n = 509,000 STCs
|
n = 314,000 STCs
|
||
|---|---|---|---|---|
| Same orientation, BRCA2 | Either orientation, TCR α/δ | Same orientation, BRCA2 | Either orientation, TCR α/δ | |
| 0 | 17 | 20 | 6 | 7 |
| 0–5 | 85 | 138 | 35 | 59 |
| 5–10 | 49 | 52 | 17 | 30 |
| 10–15 | 34 | 31 | 20 | 19 |
| 15–20 | 31 | 15 | 18 | 18 |
| 20–25 | 12 | 10 | 9 | 9 |
| 25–30 | 13 | 5 | 11 | 11 |
| 30–35 | 12 | 4 | 8 | 3 |
| 35–40 | 4 | 1 | 5 | 1 |
| 40–45 | 6 | 1 | 8 | 2 |
| 45–50 | 2 | 0 | 5 | 0 |
| >50 | 10 | 0 | 18 | 3 |
| Total inter-end distances | 275 | 277 | 160 | 162 |
| >35 kb | 8% | 0.72% | 22.5% | 3.70% |
| Mean distance | 13.1 kb | 6.6 kb | 22.4 kb | 11.2 kb |
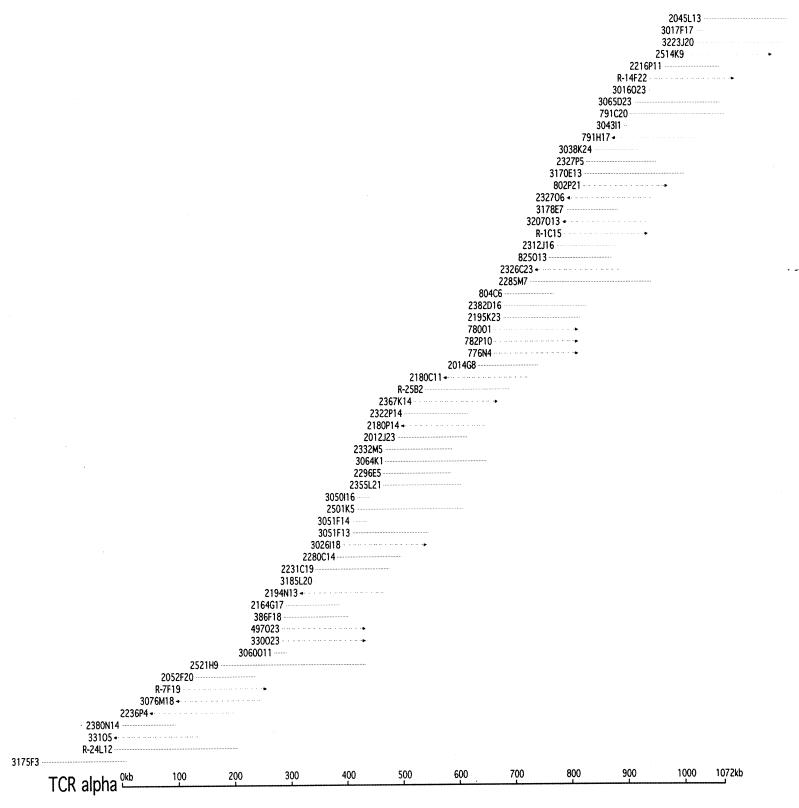
If 20,000 150-kb BAC clones (1-fold BAC coverage) must be sequenced to cover the human genome (150 kb per BAC × 20,000 BACs = 3,000 Mb), then the 5-kb map requires sequencing an extra 100,000 kb (5 kb per BAC × 20,000 BACs), whereas the 17-kb map (22.4 kb + 11.2 kb/2) requires sequencing an extra 340,000 kb of overlapping DNA (17 kb per BAC × 20,000 BACs). If genomic sequencing costs $0.35 per bp, the higher-resolution map would require the sequence analysis of 1,600 fewer BAC clones (240,000 kb/150 kb per BAC), resulting in $100 million in savings (19). Thus, sequencing by extension with a deeper STC resource allows for both high-throughput sequencing and a very significant cost savings, given that the total cost for the STC resource will be less than $14 million. Moreover, it will not require any experimental mapping for closure, an additional significant savings. Finally, the STC resource provides efficient minimal tiling paths for sequencing, as can be seen by the STC distribution across the 1.1-Mb α/δ T cell receptor locus (Fig. 2).
Figure 2.
STC map across the 1.1-Mb human α/δ T cell receptor locus. This diagram depicts a BAC contig constructed by using a 1,071-kb sequence from the T cell receptor α locus to query the STC database. The sequence was masked for repeats (http://repeatmasker.genome.washington.edu/) and used to search the TIGR STC database (http://www.tigr.org/tdb/humgen/bac_end_search/bac_end_search.html) which, at the time, contained around 310,000 sequences. BAC clones shown as solid lines had both end sequences available in the STC database; dotted lines represent BAC clones where only one end sequence was available The arrow-head points in the direction of the unsequenced end, and the insert size is arbitrarily set at 150 kb. The bottom scale depicts the length in kb of this locus.
The approach now being used to identify seed BAC clones for sequencing the human genome is to sequence randomly selected clones or contigs up to a coverage of 90% (12, 13). In contrast, the use of the STC resource to select more evenly distributed clones (ideally every 1–2 Mb) ensures that the contig closure process will sequence minimal tiling paths of BAC clones without excessive overlaps. Therefore, problems relating to both the determination of a minimal tiling path and the long-range selection of seed BAC clones are solved efficiently by the STC resource.
STC fingerprints provide a powerful aid to genomic sequencing. (i) Fingerprints permit an accurate estimation of insert size, thus permitting an estimate of the number of small insert clones that should be sequenced in the shotgun process (e.g., a 240-kb insert requires sequencing three times as many small insert clones as would an 80-kb BAC insert). (ii) The ability to identify the 5% or so of BAC clones containing sequence artifacts (e.g., deletions, insertions, rearrangements, chimeras) by comparing overlapping fingerprints is important. A minimally overlapping clone contains an artifact detectable by fingerprint analysis; one may then sequence an adjacent BAC clone containing the second-best overlap. (iii) In some cases, multiple STCs cluster at a single site. Fingerprints frequently reveal that the corresponding BAC clones are all unrelated, thus suggesting that the clustered STCs contain a heretofore unidentified similar repeat sequence.
Random Distribution of STCs Across the Genome.
There has been concern about the randomness of the inserts in BAC libraries. Several lines of evidence suggest the STC sequences are relatively evenly distributed throughout the genome: (i) The mean distribution of 275 STCs across more than 1.8 Mb of genomic sequence is 1 STC per 6.6 kb (Table 1). As noted earlier, the expected distribution of these STCs across the genome is 1 STC/6 kb. (ii) The distribution of STCs across all human chromosomes is about what one would expect from the amount of cDNA, EST, and genomic sequence available for each chromosome (see below). (iii) The G/C content of the STCs is 40.2%, accurately reflecting that of the entire genome (20, 21). (iv) The STC sequences after removing common repetitive elements are rarely identical (or even very similar). Thus, the same chromosomal site is not represented at the ends of multiple BAC clones. Most instances of identical STCs arise in clones derived from adjacent wells in the same 384-well plate and lead us to conclude that these identical clones are artifacts of clone picking. Taken together, these observations suggest that the distribution of BAC clones (STCs) across the human genome is relatively random.
The STC Resource Provides a Powerful Means to Scan the Human Genome.
Together with TIGR, we have sequenced more than 135 Mb (4.5%) of the human genome in STCs. A blast (21) analysis of 314,000 STCs (Table 2) showed that 15,174 (4.9%) were complementary to sequences in the nonredundant nucleotide database, and these represented 7,377 (2.4%) unique sequences; 3,977 (1.3%) were complimentary to the nonredundant protein database and 2,939 (0.9%) of these represented unique proteins; 9,869 (3.2%) were complementary to sequences in the nonredundant EST database, of which 8,259 (2.7%) were unique; and 5,178 (1.65%) were complementary to sequences in the Unigene database, and 4,289 (1.36%) of these were unique (see Table 2). This blast analysis required a >90% match with at least 90 identities over 100 nt (90% match over 30% aa residues). Two percent (6, 280) of the STCs contain CpG islands by criteria given by Cross and Bird (23) and Cross et al. (24), suggesting that some of these STCs mark the regions upstream of genes.
Table 2.
Repeat analysis of 314,000 human STCs generating 135 Mb of human genomic sequence
| Repeat type | % Repeat |
|---|---|
| Composite | 0.046 |
| DNA | 2.153 |
| LINE | 14.722 |
| LTR | 5.625 |
| Low complexity | 0.606 |
| RNA | 0.002 |
| SINE | 7.267 |
| Satellite | 2.667 |
| Simple repeat | 0.829 |
| Unknown | 0.259 |
| rRNA | 0.002 |
| scRNA | 0.005 |
| snRNA | 0.008 |
| srpRNA | 0.007 |
| tRNA | 0.002 |
| Total Repeat | 35.3% |
About 35% of the 314,000 STC sequences (≈47 Mb) represent genome-wide repeats (25): 14.7% long interspersed nuclear elements, 5.6% long terminal repeats, 7.2% short interspersed nuclear elements, 2.7% microsatellites, and 2.15% DNA transposons (Table 3). Thus, it is possible from STC data to construct a relatively complete repeat library, which will be useful for the characterizations of complex genomes, because known repeat sequences can be masked or removed before sequence assembly and similarity searches against the STC database (J.C.W., T.H., L.H., and G.M., unpublished data).
Table 3.
The distribution of simple sequence repeats identified from 314,219 STSs
| Length | Number | Simple sequence repeats, % | STCs, % |
|---|---|---|---|
| Dinucleotide | 4,217 | 56 | 1.34 |
| Trinucleotide | 680 | 9 | 0.22 |
| Tetranucleotide | 1,163 | 15 | 0.37 |
| Pentanucleotide | 1,487 | 20 | 0.47 |
| Total | 7,547 | 2.40 |
Data were generated by the criteria of ref. 7.
The important point is that in the case of each of these features, the STC sequence corresponds to a particular sequence-ready BAC clone; hence, interesting STC features may immediately be sequenced at the genomic level. Moreover, by sequence extension chromosomal regions extending 5′ and 3′ from these seed BAC clones can readily be sequenced.
Accuracy of the STC Sequences.
The quality of the STC sequences (length of phred 20 scores, base calling and quality assessment program developed by Phil Green) is lower than the typical genomic sequences obtained from plasmid or M13 inserts. However, the quality of STC sequence is more than sufficient to find BAC clones for walking by sequence extension (minimum tiling path), to find genes (ESTs, cDNAs), to find chromosomal markers (STSs), to localize BAC clones to previously mapped chromosomal sequences, and to identify genome-wide repeats for a masking library (see below). These findings indicate that (i) even 100 bp of unique sequence constitutes a precise genome address and (ii) sequence matches set at 90% similarity are capable of readily identifying genes, chromosomal markers, and BACs for sequence extension. The fact that we found nearly the number of STC hits expected across two fully sequenced loci 1.8 Mb in length argues that our search criteria are effectively utilizing most STCs. Indeed, the HTSC and the TIGR STCs have been equally successful at hitting ESTs, cDNAs, STSs, and chromosomal sequences. Because the STCs are mapping markers and do not require the same accuracy as assembled sequence, it would seem important to minimize their cost and increase their throughput, rather than spending unnecessary resources for greater quality.
STC Resource Provides the Means for Generating Integrated Physical, Genetic, Gene, and Sequence Maps.
The STC resource provides an effective (and inexpensive) approach to creating integrated physical and gene maps. For example, the STCs may hit mapped chromosomal markers (e.g., STSs, ESTs, Unigene clusters, chromosomal sequence, etc.) and, thus, map the corresponding BAC clones to precise chromosomal locations. As noted earlier, of 314,000 human STCs, 8,256 (2.7%) hit unique ESTs and 4,289 (1.4%) hit Unigene clusters, many of which are now chromosomally mapped, and soon, all will be. An analysis of STCs against the genemap ’98 EST (26) and mRNA database revealed 2,520 complimentary sequences, of which 2,177 were unique. These genemap ’98 hits were distributed on all chromosomes more or less in proportion to available genomic sequence. Because the numbers of mapped chromosomal markers are increasing so rapidly, we eventually expect to have a mapped BAC clone, on average, every 200 kb across the genome. Some of these mapped BACs would be ideal seed BACs. These mapped BACs constitute a dense high-resolution physical map.
Genetic markers can be readily identified through the STC sequences. Weissenbach et al. (7) defined criteria for microsatellite or simple sequence repeat markers based on the probability of polymorphism among Centre d’Études du Polymorphism Humaine pedigrees. These criteria are a minimum of 21 nt per repeat, 11 repeat units per dinucleotide repeat, and 7 repeat units for trinucleotide repeats. By using these criteria, simple sequence repeat analysis of 314,219 STCs revealed 7,547 simple sequence repeats (2.4%), of which 4,217 were dinucleotide, 680 trinucleotide, 1,163 tetranucleotide, and 1,487 pentanucleotide repeats (Table 3). These are potential genetic markers, many of which may represent human polymorphisms. Moreover, the STC sequences could be used to generate PCR primers and amplified sequence from different individuals to identify single-nucleotide polymorphisms across the human genome. If the STC polymorphisms are determined from STC BAC clones that have been physically mapped (see above), then the genetic and physical maps will be integrated.
Because the STC BAC clones mapped by genetic, coding, and physical criteria are used to initiate chromosomal sequencing, the integration of genetic, physical, gene, and sequence maps will be complete. In addition, mapped chromosomal markers (STSs, mapped Unigene clusters, polymorphic STSs, etc.) as well as ESTs can readily be assigned to STC resource BACs by hybridization. Indeed, 10,000 ESTs are being mapped to the Caltech BAC library (W. Kim and M. Simon, personal communication), and 8,000 ESTs are being mapped to the Roswell Park BAC library (P. de Jong, personal communication). In addition, high-resolution radiation hybrid maps are now being developed using STS assays defined from the STCs (D. Cox, personal communication). Thus, the STC resource affords a powerful opportunity for the ongoing integration of the four types of genomic maps as well as other interesting unmapped chromosomal sequence markers.
The STC Mapping Approach Has Several Advantages over Other Mapping Procedures for Large-Scale Sequencing.
The creation of minimum tiling paths of clones using chromosomal PCR (STS) or hybridization (EST or cDNA) markers requires time-consuming experimental procedures with significant levels of false-positives. In contrast, the STC approach maps by sequencing BACs and analyzing their sequences computationally against the STC and fingerprint data. Moreover, STC sequence data are far more precise than PCR or hybridization analyses because false-positive and -negative results can more easily be controlled. Even more significant is that the STC sequences each are associated with a specific sequence-ready clone. The density of the STC probes is more than 20 times as great as the existing STS probes and soon will be 60 times as great, thus minimizing the cost of the sequencing process itself by reducing the amount of overlapping sequence analyzed. By simultaneous sequence walking with multiple seed BACs, large genome centers can readily generate the numbers of BAC clones they need for large-scale sequencing. Importantly, with the human STC resource, the proposed strategy of sequencing significant numbers of random BACs for the first draft sequence is no longer necessary. Rather, genome centers can assume responsibility for regional chromosomal sequencing and at the same time assume the responsibility for finishing these discrete regions to a highly accurate final draft.
The STC Approach Provides a Relatively Inexpensive and Efficient Approach to the Sequencing of Other Animal, Plant, and Even Some Microbial Genomes.
The STC approach will be very effective for complex genomes for which little or no genomic information is available (e.g., mouse, other mammals, vertebrates, and invertebrates). A variety of plant genomes including rice, corn, sorghum, and tomato could be rapidly characterized by using the STC approach. Three points are important with regard to the STC characterization of uncharacterized genomes. First, the STC genome scan samples genes without respect to their relative levels of expression, identifying both highly and infrequently expressed genes. Second, the simple sequence repeats identified in STCs offer the possibility of easily generating a multiplicity of genetic markers (or single-nucleotide polymorphisms). Finally, the STCs provide the sequence to generate a table of genome-wide repeats that need to be masked from the genomic sequence before undertaking detailed similarity analyses against the genome itself (e.g., to find gene families) and against the databases of known sequence. The investment in the STC resource is modest compared with the enormous benefits arising from these genome-wide sequence scans.
Acknowledgments
This research was supported by a grant from the Department of Energy. We would also like to acknowledge the staff of the High-Throughput Sequencing Center for their outstanding sequencing efforts.
ABBREVIATIONS
- STC
sequence-tagged connector
- BAC
bacterial artificial chromosome
- STS
sequence-tagged sequence
- EST
expressed sequence tag
- kb
kilobase
- Mb
megabase
Note Added in Proof
A recent updata of the STC data and analysis is provided at www.htsc.washington.edu/human/info/index.cfm.
References
- 1.Green E D, Olson M V. Science. 1990;250:94–98. doi: 10.1126/science.2218515. [DOI] [PubMed] [Google Scholar]
- 2.Green E D, Mohr R M, Idol J R, Jones M, Buckingham J M, Deaven L L, Moyzis R K, Olson M V. Genomics. 1991;11:548–564. doi: 10.1016/0888-7543(91)90062-j. [DOI] [PubMed] [Google Scholar]
- 3.Kere J, Nagaraja R, Mumm S, Ciccodicola A, D’Urso M, Schlessinger D. Genomics. 1992;14:241–248. doi: 10.1016/s0888-7543(05)80212-5. [DOI] [PubMed] [Google Scholar]
- 4.Green E D, Green P. PCR Methods Appl. 1991;1:77–90. doi: 10.1101/gr.1.2.77. [DOI] [PubMed] [Google Scholar]
- 5.Boysen C, Carlson C, Hood E, Hood L, Nickerson D A. Immunogentics. 1996;44:121–127. [PubMed] [Google Scholar]
- 6.Nickerson D A, Kaiser R, Lappin S, Stewart J, Hood L, Landegren U. Proc Natl Acad Sci USA. 1990;87:8923–8927. doi: 10.1073/pnas.87.22.8923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weissenbach J, Gyapay G, Dib C, Vignal A, Morissette J, Millasseau P, Vaysseisx G, Lathrop M. Nature (London) 1992;359:794–801. doi: 10.1038/359794a0. [DOI] [PubMed] [Google Scholar]
- 8.Stewart E A, McKusick K B, Aggarwal A, Bajorek E, Brady S, Chu A, Fang N, Hadley D, Harris M, Hussain S, et al. Genome Res. 1997;7:422–433. doi: 10.1101/gr.7.5.422. [DOI] [PubMed] [Google Scholar]
- 9.Cox D R, Burmeister M, Price E R, Kim S, Myers R M. Science. 1990;250:245–250. doi: 10.1126/science.2218528. [DOI] [PubMed] [Google Scholar]
- 10.Hildmann T, Kong X, O’Brien J, Riesselman L, Christensen H M, Dagand E, Lehrach H, Yaspo M L. Genome Res. 1999;9:360–372. [PMC free article] [PubMed] [Google Scholar]
- 11.Venter J C, Adams M D, Sutton G G, Kerlavage A R, Smith H O, Hunkapillar M. Science. 1998;280:1540–1542. doi: 10.1126/science.280.5369.1540. [DOI] [PubMed] [Google Scholar]
- 12.Marshall E. Science. 1998;281:1774–1775. doi: 10.1126/science.281.5384.1774. [DOI] [PubMed] [Google Scholar]
- 13.Pennisi E. Science. 1999;283:1822–1823. doi: 10.1126/science.283.5409.1822. [DOI] [PubMed] [Google Scholar]
- 14.Venter J C, Smith H O, Hood L. Nature (London) 1996;381:364–366. doi: 10.1038/381364a0. [DOI] [PubMed] [Google Scholar]
- 15.Ewing B, Green P. Genome Res. 1998;8:186–194. [PubMed] [Google Scholar]
- 16.Ewing B, Hillier L, Wendl M C, Green P. Genome Res. 1998;8:175–185. doi: 10.1101/gr.8.3.175. [DOI] [PubMed] [Google Scholar]
- 17.Richterich P. Genome Res. 1998;8:251–259. doi: 10.1101/gr.8.3.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boysen C, Simon M I, Hood L. Genome Res. 1997;7:330–338. doi: 10.1101/gr.7.4.330. [DOI] [PubMed] [Google Scholar]
- 19.Siegel A F, Trask B, Roach J C, Mahairas G G, Hood L, van den Engh G. Genome Res. 1999;9:297–307. [PMC free article] [PubMed] [Google Scholar]
- 20.Saccone S, De Sario A, Wiegant J, Raap A K, Della Valle G, Bernardi G. Proc Natl Acad Sci USA. 1993;90:11929–11933. doi: 10.1073/pnas.90.24.11929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bernardi G. Mol Biol Evol. 1993;10:186–204. doi: 10.1093/oxfordjournals.molbev.a039994. [DOI] [PubMed] [Google Scholar]
- 22.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cross S H, Bird A P. Curr Opin Genet Dev. 1995;5:309–314. doi: 10.1016/0959-437x(95)80044-1. [DOI] [PubMed] [Google Scholar]
- 24.Cross S H, Clark V H, Bird A P. Nucleic Acids Res. 1999;27:2099–2107. doi: 10.1093/nar/27.10.2099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smit A F. Curr Opin Genet Dev. 1996;6:743–748. doi: 10.1016/s0959-437x(96)80030-x. [DOI] [PubMed] [Google Scholar]
- 26.Deloukas P, Schuler G D, Gyapay G, Beasley E M, Soderlund C, Rodriguez-Tomé P, Hui L, Matise T C, McKusik K B, Beckmann J S, et al. Science. 1998;282:744–746. doi: 10.1126/science.282.5389.744. [DOI] [PubMed] [Google Scholar]