Abstract
Crosslinking of immunoreceptor tyrosine-based activation motif (ITAM)-containing receptor complexes on a variety of cells leads to their activation through the sequential triggering of protein tyrosine kinases. Recently, DAP12 has been identified as an ITAM-bearing signaling molecule that is noncovalently associated with activating isoforms of MHC class I receptors on natural killer cells. In addition to natural killer cells, DAP12 is expressed in peripheral blood monocytes, macrophages, and dendritic cells, suggesting association with other receptors present in these cell types. In the present study, we report the molecular cloning of the myeloid DAP12-associating lectin-1 (MDL-1), a DAP12-associating membrane receptor expressed exclusively in monocytes and macrophages. MDL-1 is a type II transmembrane protein belonging to the C type lectin superfamily and contains a charged residue in the transmembrane region that enables it to pair with DAP12. Crosslinking of MDL-1/DAP12 complexes in J774 mouse macrophage cells resulted in calcium mobilization. These findings suggest that signaling via MDL-1/DAP12 complexes may constitute a significant activation pathway in myeloid cells.
Crosslinking of immune receptors, such as the B or T cell receptor (BCR or TCR) or Fc receptors, on a variety of cells leads to their activation through the sequential activation of protein tyrosine kinases (PTKs) (1). These receptor complexes have in common that ligand binding and signal transduction occur in distinct receptor subunits. In their signal-transducing subunits (e.g., CD3 in the TCR), these complexes contain one or more immunoreceptor tyrosine-based activation motifs (ITAMs). Upon engagement of the ligand-binding subunit, the cytoplasmic ITAMs in the signaling subunits are tyrosine-phosphorylated by src-family PTKs. This leads to the recruitment of syk-family PTKs, which, in turn, triggers a cascade of intracellular phosphorylation that results in cellular activation.
A number of receptor complexes that recognize MHC class I molecules have been identified on natural killer (NK) cells (2–5). Although many of these receptors possess immunoreceptor tyrosine-based inhibition motifs (ITIMs) that recruit protein tyrosine phosphatases and inhibit NK cell activation, several of these MHC class I receptors lack intrinsic signaling sequences. These receptors express a charged residue in their transmembrane domain, suggesting the association with an adapter molecule that is capable of signaling. Recently, we identified DAP12, a type I glycoprotein containing an ITAM (6). DAP12 is expressed at the cell surface of NK cells noncovalently associated with the human KIR2DS receptor for HLA-C (6, 7), the mouse Ly49D and Ly49H receptors (8, 9), and the human heterodimer CD94/NKG2C receptor complex recognizing HLA-E (10).
In addition to NK cells, DAP12 is expressed in peripheral blood granulocytes, monocytes, and dendritic cells, suggesting association with other receptors present in these cell types. In the present study, we report the cloning of myeloid DAP12-associating lectin-1 (MDL-1), a DAP12-associating molecule expressed exclusively in monocytes and macrophages. Signaling via MDL-1/DAP12 complexes may constitute a significant activation pathway in myeloid cells.
MATERIALS AND METHODS
Transfectants.
A cDNA containing the CD8-leader segment, followed by the FLAG peptide epitope (DYKDDDDK) and joined to the extracellular, transmembrane and cytoplasmic regions of mouse DAP12, was subcloned into pREP10 (Invitrogen). This construct was used to transfect 293T cells. Using flow cytometry, 293T FLAG-DAP12 cells were selected that were devoid of cell surface DAP12 expression, but uniformly expressed FLAG-DAP12 in the cytoplasm. 293T cells expressing human FLAG-FcɛRIγ intracellularly were generated in the same manner.
A chimeric cDNA containing the intracellular, transmembrane, and stalk regions of human MDL-1 and the extracellular lectin domain of human CD69 (MDL-1/CD69) was constructed, sequenced, and subcloned into the pMX-neo retroviral vector (11). A control cDNA was generated that encoded an identical chimeric protein in which the charged lysine in the transmembrane region of MDL-1 was replaced for an isoleucine residue (MDL-1/CD69 TM K → I). J774 transfectants stably expressing MDL-1/CD69 at the cell surface were generated as described (11).
Expression Cloning.
A cDNA library from the mouse J774 macrophage cell line in the pJEF14 vector (12) was kindly provided by K. Moore (DNAX). cDNA cloning by transient expression in 293T FLAG-DAP12 cells was performed as described (13, 14), with minor modifications. We screened for cDNA-encoded proteins that were able to pair with DAP12, resulting in the translocation of DAP12 from the cytoplasm to the cell surface. Cell surface-expressed FLAG-DAP12 was visualized by using the anti-FLAG M2 mAb (Kodak). To avoid cloning Fc-receptor-encoding cDNAs, transiently transfected 293T FLAG-DAP12 cells were stained first with FITC-conjugated anti-CD16/32 2.4G2 mAb (PharMingen), subsequently with M2 mAb, followed by the biotinylated anti-mouse IgG1 A85–1 mAb (PharMingen) and streptavidin-phycoerythrin (PE) (Becton Dickinson). Single-positive transfected cells displaying bright PE fluorescence were collected for retrieval of plasmid DNA. After three rounds of selection, plasmid pools and, subsequently, single colonies were selected that were able to induce DAP12 cell surface expression.
RNA and DNA Analyses.
Northern and Southern blot analyses were performed as described (15). cDNA was prepared from 1 μg of total RNA by using AMV Reverse Transcriptase (GIBCO) and oligo(dT) (Boehringer Mannheim) primers in a total volume of 25 μl. PCRs were performed with 1 μl of cDNA by using AmpliTaq Gold polymerase (Perkin–Elmer) in the presence of 1.5 mM MgCl2 in a volume of 50 μl. For PCR analysis of human MDL-1 transcripts, the following primers were used: sense, 5′-CATCTGCAAGCAAGCCTATCC-3′; and antisense, 5′-TGTGGTAATGAAGCCGTTGG-3′. PCR was performed by using the program: 9 min at 94°, 30× (30 sec at 94°, 30 sec at 56°, 45 sec at 72°), 10 min at 72°. The integrity of the cDNA was assessed by PCR by using glyceraldehyde-3-phosphate dehydrogenase primers under identical conditions.
The chromosomal localization of the MDL-1 gene was determined by using a biotinylated MDL-1 probe for in situ hybridization to metaphases from two normal males as described (16).
Biochemistry.
MDL-1/CD69- and MDL-1/CD69 TM K → I-transfected J774 cells were solubilized in cold digitonin lysis buffer as described (17). Immunoprecipitation with anti-CD69 Leu-23 mAb (Becton Dickinson) or control Ig and Western blotting was performed as described (9). Immunoblots were probed with affinity-purified rabbit anti-mouse DAP12 antiserum (generated by standard methods against mouse DAP12-GST fusion proteins and DAP12 immunoprecipitates) or with rabbit anti-FcɛRIγ antiserum (generously provided by J.-P. Kinet, Harvard University, Cambridge, MA), followed by goat-anti-rabbit IgG-horseradish peroxidase (Jackson ImmunoResearch).
Calcium Mobilization.
J774 transfectants were loaded with Indo-1 AM (Sigma) as described (18). After loading, the cells were stained with saturating concentrations of the anti-CD16/32 2.4G2 mAb on ice for 10 min to block nonspecific Fc-mediated antibody binding and washed and analyzed on a fluorometer as described (18). After acquisition of a baseline for 75 sec, 10 μg of anti-H2-Dd 34–5-8S mAb (PharMingen) (used as a negative control) or anti-CD69 Leu-23 mAb followed by 10 μg of F(ab′)2 rabbit-anti-mouse IgG (Fc-specific) (Jackson ImmunoResearch) was added to the cells, and analysis was continued for up to 300 sec. After 250 sec, 2.5 μg of ionomycin (Sigma) was added to assess Indo-1 AM loading of the cells by depletion of intracellular calcium stores.
RESULTS
cDNA Cloning of MDL-1 by Using an Indirect Expression-Cloning Method.
To identify novel DAP12-associating molecules expressed in myeloid cells, we used an indirect expression-cloning strategy. When expressed in the absence of associating receptors, DAP12 largely remains located intracellularly. However, upon introduction of an associating partner, DAP12 complexes can be formed, resulting in the translocation of DAP12 to the cell surface (6). We exploited this phenomenon by generating a 293T cell line stably expressing N terminus FLAG-tagged mouse DAP12 in the cytoplasm, but not at the cell surface. To examine whether this cell line could be used for expression cloning of novel DAP12-associating molecules, we transfected it with a mouse NK cell cDNA library in an expression vector and sorted for cell surface FLAG-positive 293T cells. Plasmid DNA was retrieved from the sorted cells and was subjected to a second round of transfection. After three rounds of selection, we obtained a cDNA clone capable of rescuing DAP12 cell surface expression. This cDNA encoded Ly49H, a known partner of DAP12 in mouse NK cells (8, 9) (data not shown). No irrelevant cDNA clones were selected, demonstrating the selectivity of this indirect expression-cloning approach.
To identify DAP12-associating partners in myeloid cells, we transfected the 293T FLAG-DAP12 cell line with a J774 macrophage cell line-derived cDNA expression library and rescued plasmid DNA from transfected cells that expressed DAP12 at the cell surface. To avoid the cloning of Fc-receptor-encoding cDNAs, a mAb against CD16/32 was used to stain and exclude cells transfected with plasmids encoding these Ig receptors. After three rounds of selection, two cDNA clones were obtained of 733 and 856 bp that induced DAP12 cell surface expression upon transfection in 293T FLAG-DAP12 cells (Fig. 1A). Both clones were identical and differed only in the length of their 5′ untranslated regions. These cDNA had an ORF encoding a 166-aa type II transmembrane protein (Fig. 1B). Examination of the 5′ untranslated nucleotide sequence immediately upstream of the initiation codon revealed a termination codon in the same reading frame that would prevent translation from other potential upstream initiation codons. The predicted polypeptide has a short, intracellular region of only 2 aa, a 23-aa transmembrane region, and a 141-aa extracellular domain. The transmembrane region contains a positively charged amino acid [lysine (K)], which also is conserved in the activating MHC class I receptors, KIR2DS, Ly49D/H, and NKG2C. The presence of several characteristic structural motifs in the extracellular region indicates that this protein is a member of the C type lectin superfamily. Therefore, we named it MDL-1.
Figure 1.
(A) Rescue of DAP12 cell surface expression on 293T FLAG-DAP12 cells transfected with control plasmid or with pJEF14 vector containing mouse MDL-1 cDNA. (B) Nucleotide and predicted amino acid sequence of mouse MDL-1. The sequence of the cDNA encoding the short form of mouse MDL-1 is presented (GenBank accession no. AF139769). The predicted transmembrane region is underlined, with the charged lysine indicated in bold, and potential N-linked glycosylation sites are depicted with an asterisk. (C) Alignment of the predicted protein sequences of the long form of mouse MDL-1 (GenBank accession no. AA186015) and human MDL-1 (GenBank accession no. AF139768). The additional 25 aa in the stalk region of the long form of mouse MDL-1 are underlined.
Database searching revealed a GenBank expressed sequence tag (EST) encoding a mouse MDL-1 isoform containing an additional 25 aa in the extracellular region proximal to the transmembrane domain (Fig. 1C). As a result, this isoform has an extended stalk region positioning the lectin domain further from the cell membrane. In addition, it contains an extra, putative N-linked glycosylation site. These isoforms could be identified simultaneously in cDNA prepared from mouse macrophage cell lines (data not shown), indicating that they most likely result from alternative splicing. A similar splicing pattern resulting in different stalk regions of C type lectin proteins has been described for the NKG2A and NKG2B receptors (19). An EST database at our institute contained the full-length human homologue of MDL-1. Human MDL-1 has 69% identity at the amino acid level with the long isoform of mouse MDL-1 (Fig. 1C).
Expression Pattern and Genomic Localization of MDL-1.
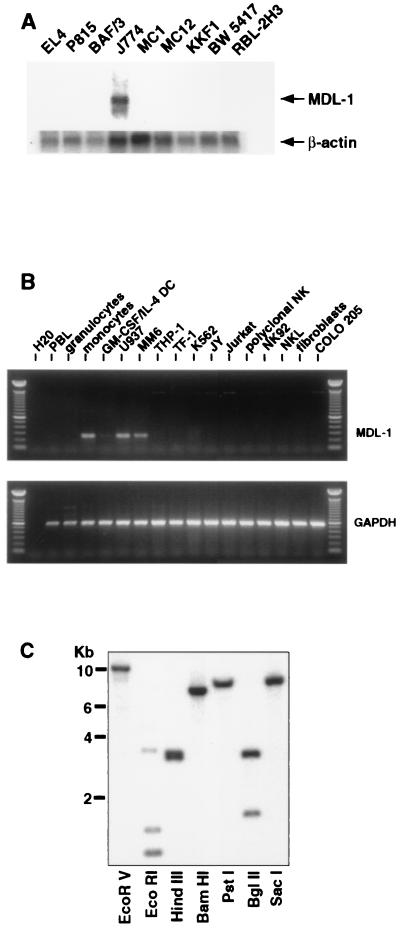
Northern blot analysis revealed MDL-1 transcripts of approximately 1.8 kb in the mouse macrophage cell line J774, but not in T cell lines, B cell lines, or mast cell lines (Fig. 2A). PCR analysis of a panel of human cDNAs revealed expression of MDL-1 in normal peripheral blood monocytes and in the monocyte/macrophage cell lines U937 and MonoMac6, but not in cell lines of other origins (Fig. 2B). Interestingly, when monocytes were differentiated toward dendritic cells by culturing them in the presence of granulocyte/macrophage colony-stimulating factor and IL-4, this resulted in a strong down-regulation of MDL-1 transcripts (Fig. 2B). These results indicate that MDL-1 is expressed predominantly in monocytes and macrophages.
Figure 2.
MDL-1 gene and expression. (A) Total RNA of mouse cell lines was analyzed for MDL-1 transcripts by using the short form of the mouse MDL-1 cDNA as a probe. EL4, KKF1, and BW 5417 are cell lines of T cell origin. P815 and BaF/3 are mastocytoma and pre-B cell lines, respectively. MC1 and MC12 are mast cell lines, J774 is a macrophage cell line, and RBL-2H3 is a rat basophilic leukemia. (Inset) Hybridization with a β-actin probe is shown. (B) PCR amplification of cDNA prepared by reverse transcription of RNA from healthy donor peripheral blood leukocyte subpopulations or primary cultures thereof or from human cell lines. U937, MM6, and THP-1 are cell lines of myeloid origin; TF-1 and K562 are of erythroid/myeloid and erythroid origins, respectively. JY is a B lymphoblastoid cell line, and Jurkat is a T cell leukemia cell line. NK92 and NKL are NK cell lines. Fibroblasts were derived from human foreskin samples. Colo 205 is a colon carcinoma cell line. (C) MDL-1 Southern blot analysis. Human genomic DNA was digested with the indicated enzymes and probed with the human MDL-1 cDNA.
Southern blot analysis of human genomic DNA using the human MDL-1 cDNA as a probe revealed a relatively simple banding pattern, suggesting that MDL-1 is encoded by a single gene (Fig. 2C). Using fluorescent in situ hybridization, we mapped the gene encoding human MDL-1 to chromosome 7q33 (data not shown). This region is distinct from loci encoding DAP12-associating receptors expressed in NK cells. Other known DAP12-associating molecules (KIR2DS, Ly49D/H, and NKG2C) are members of gene families consisting of activating as well as inhibitory receptors. These inhibitory NK receptors do not contain charged residues in their transmembrane regions. In contrast, they bear ITIMs in their intracellular region, which are responsible for the inhibition of cellular effector functions. Because MDL-1 does not appear to belong to a larger gene family, this receptor involved in the activation of myeloid cells may not have an inhibitory counterpart, although this cannot be excluded formally.
MDL-1 Selectively Associates with DAP12.
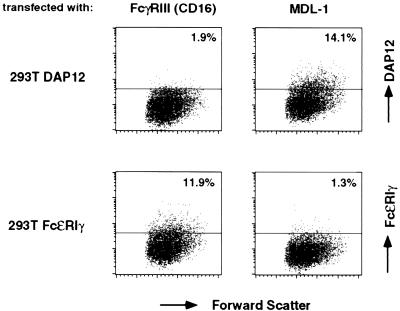
To determine whether MDL-1 associates with DAP12 and not with FcɛRIγ, which can serve as a signaling partner for FcγRIII (CD16) and FcαR (CD89) (17, 20–22), we transfected 293T cells expressing DAP12 or FcɛRIγ with cDNAs encoding either human MDL-1 or CD16. As shown in Fig. 3, 293T cells expressed DAP12 at significant levels at the cell surface only after transfection of MDL-1, whereas FcɛRIγ cell surface expression could be rescued only by CD16, indicating that MDL-1 selectively associates with DAP12.
Figure 3.
MDL-1 selectively rescues DAP12 cell surface expression. 293T cells stably expressing FLAG-DAP12 or FLAG-FcɛRIγ were transiently transfected with plasmids encoding human CD16 and human MDL-1, respectively. Cell surface expression of FLAG-DAP12 and FLAG-FcɛRIγ was analyzed by flow cytometry by using the anti-FLAG mAb M2.
To study the biochemical features of MDL-1, DAP12 complexes were immunoprecipitated from lysates of 293T FLAG-DAP12 cells transfected with the mouse and human MDL-1 cDNAs or from J774 cells by using an affinity-purified polyclonal anti-DAP12 rabbit antiserum. Although DAP12 clearly could be detected, no associating bands could be visualized in these immunoprecipitates from either iodinated or biotinylated cell lysates (data not shown). CD94, another member of the C type lectin superfamily, also cannot be detected via iodination or biotinylation (23). A DAP12-associating doublet of 30–32 kDa could be detected only in 35S-methionine-labeled cell lysates from human MDL-1-transfected 293T DAP12 cells, which most likely represent differentially glycosylated forms of the MDL-1 protein (data not shown). The generation of immune reagents against MDL-1 will allow future analysis of endogenously expressed MDL-1 proteins in myeloid cells.
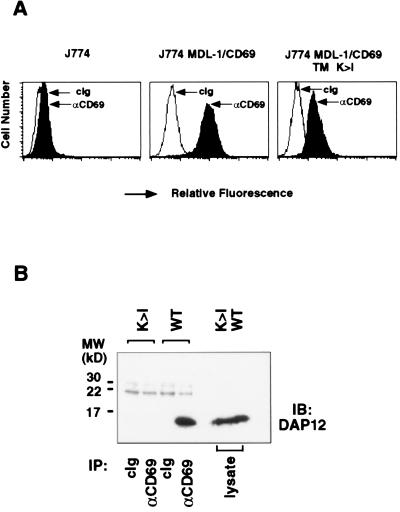
To examine whether MDL-1 noncovalently associates with DAP12, we generated J774 cells expressing a chimeric molecule containing the intracellular, transmembrane, and stalk regions of human MDL-1 and the lectin domain of human CD69 (Fig. 4A). Immunoprecipitation with anti-CD69 mAb from lysates of J774 MDL-1/CD69 cells followed by immunoblotting with anti-DAP12 antibody clearly showed the presence of associating DAP12 molecules (Fig. 4B). MDL-1/CD69 with its transmembrane Lys mutated to Ile (MDL-1/CD69 K → I) failed to associate with DAP12 in J774 cells (Fig. 4B), demonstrating the requirement of a charged residue in the transmembrane region of MDL-1 for its association with DAP12. This lack of DAP12 association also may account for the observed lower cell surface expression of the MDL-1/CD69 K → I mutant (Fig. 4A). Immunoblotting with anti-FcɛRIγ antibody showed no MDL-1-associated FcɛRIγ molecules, confirming the specific interaction of MDL-1 with DAP12 (data not shown).
Figure 4.
MDL-1 associates noncovalently with DAP12 on the cell surface. (A) Mouse J774 macrophage cells expressing MDL-1/CD69 or MDL-1/CD69 in which the transmembrane lysine was replaced by an isoleucine (TM K → I) were analyzed by flow cytometry by using isotype control or anti-CD69 mAb Leu-23. (B) J774 MDL-1/CD69 cells (WT) or J774 MDL-1/CD69 TM K → I cells (K → I) were lysed and were immunoprecipitated with control Ig (cIg) or anti-CD69 mAb. Immunoprecipitates were analyzed by SDS/PAGE under reducing conditions, blotted, and probed with affinity-purified rabbit anti-mouse DAP12 antiserum.
Crosslinking of MDL-1/DAP12 Complexes Results in Calcium Mobilization.
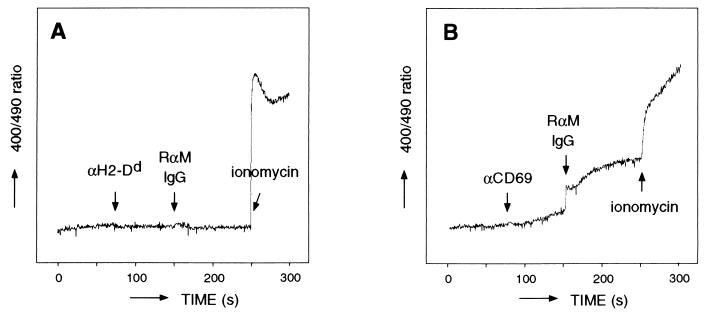
To analyze the stimulatory function of MDL-1/DAP12 complexes, we tested whether MDL-1 receptors can trigger Ca2+ mobilization in CD69/MDL-1-transfected J774 cells. As shown in Fig. 5B, crosslinking of CD69/MDL-1 by using anti-CD69 mAb induced significant Ca2+ mobilization in J774 cells, whereas crosslinking of MHC class I (as a control) left the cells unaffected (Fig. 5A). Vector control-transfected J774 cells or J774 cells transfected with the MDL-1/CD69 K → I transmembrane mutant cDNA showed no Ca2+ mobilization upon crosslinking with anti-CD69 mAb (data not shown). These results demonstrate that the MDL-1 receptor induces cell activation in myeloid cells via DAP12-mediated signaling.
Figure 5.
MDL-1 receptor induces intracellular Ca2+ mobilization. Indo-1 AM-loaded MDL-1/CD69-transfected J774 cells were incubated with anti-H2-Dd mAb (used as a negative control) (A) or anti-CD69 mAb (B), followed by a crosslinking Ab. Intracellular calcium levels were measured as described in Materials and Methods.
DISCUSSION
MDL-1 is a novel member of the C type lectin family that is the first DAP12-associating receptor described in myeloid cells. A physiological role for the MDL-1/DAP12 receptor complex has not yet been determined. Most likely, the ligand for MDL-1 will be a cell-bound glycoprotein or, perhaps, a glycolipid. The high degree of homology between mouse and human MDL-1 suggests that its counterstructure may have been conserved during evolution. Engagement of MDL-1/DAP12 receptor complexes on macrophages with its ligand may cause cellular activation, resulting in the onset of an oxidative burst, the production of cytokines, or the induction of costimulatory molecules.
Other likely candidates associating with DAP12 in myeloid cells are members of the Ig-like transcripts/myeloid Ig-like receptors (ILT-1) and paired Ig-like receptors (PIR-A1) families expressed by human monocytes and murine myeloid cells and B cells, respectively (24–26). However, we could not demonstrate association of these molecules with DAP12 (data not shown), which is consistent with the recent finding that these molecules associate with FcɛRIγ (27–29). We could detect only very low levels of MDL-1 transcripts in cDNA from dendritic cells that had been generated by culturing human monocytes in the presence of granulocyte/macrophage colony-stimulating factor and IL-4. However, these cells do express a significant amount of DAP12, suggesting the presence of other receptors. Candidate dendritic cell receptors that associate with DAP12 currently are under investigation. Our results suggest that MDL-1 receptors may be involved in the proinflammatory activation of macrophages, whereas DAP12 complexes involving other novel receptors may activate dendritic cells.
Acknowledgments
We thank Kevin Moore for providing the J774 expression library and Ken Davis for his gift of the Leu-23 mAb; Chitra Saraiya, Yaoli Song, Holly Cherwinski, and Mike Bigler for excellent technical assistance; Jim Cupp, Eleni Callas, Dixie Polakoff, Melinda Cook, and Jennifer Maskrey for cell sorting; Debbie Liggett for synthesis of oligonucleotides; Dan Gorman for sequencing; Maribel Andonian and Gary Burgett for graphics; JoAnn Katheiser for secretarial assistance; and Jun Wu and Stuart Tangye for stimulating discussions. DNAX Research Institute of Molecular and Cellular Biology, Inc., is supported by Schering-Plough Corporation.
ABBREVIATIONS
- ITAM
immunoreceptor tyrosine-based activation motif
- ITIM
immunoreceptor tyrosine-based inhibition motif
- MDL-1
myeloid DAP12-associating lectin-1
- NK
natural killer
Footnotes
References
- 1.Weiss A, Littman D R. Cell. 1994;76:263–274. doi: 10.1016/0092-8674(94)90334-4. [DOI] [PubMed] [Google Scholar]
- 2.Lanier L L. Annu Rev Immunol. 1998;16:359–393. doi: 10.1146/annurev.immunol.16.1.359. [DOI] [PubMed] [Google Scholar]
- 3.Biassoni R, Cantoni C, Falco M, Verdiani S, Bottino C, Vitale M, Conte R, Poggi A, Moretta A, Moretta L. J Exp Med. 1996;183:645–650. doi: 10.1084/jem.183.2.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mason L H, Anderson S K, Yokoyama W M, Smith H R C, Winkler-Pickett R, Ortaldo J R. J Exp Med. 1996;184:2119–2128. doi: 10.1084/jem.184.6.2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Houchins J P, Lanier L L, Niemi E, Phillips J H, Ryan J C. J Immunol. 1997;158:3603–3609. [PubMed] [Google Scholar]
- 6.Lanier L L, Corliss B C, Wu J, Leong C, Phillips J H. Nature (London) 1998;391:703–707. doi: 10.1038/35642. [DOI] [PubMed] [Google Scholar]
- 7.Olcese L, Cambiaggi A, Semenzato G, Bottino C, Moretta A, Vivier E. J Immunol. 1997;158:5083–5086. [PubMed] [Google Scholar]
- 8.Mason L H, Willette-Brown J, Anderson S K, Gosselin P, Shores E W, Love P E, Ortaldo J R, McVicar D W. J Immunol. 1998;160:4148–4152. [PubMed] [Google Scholar]
- 9.Smith K M, Wu J, Bakker A B H, Phillips J H, Lanier L L. J Immunol. 1998;161:7–10. [PubMed] [Google Scholar]
- 10.Lanier L L, Corliss B, Wu J, Phillips J H. Immunity. 1998;8:693–701. doi: 10.1016/s1074-7613(00)80574-9. [DOI] [PubMed] [Google Scholar]
- 11.Onishi M, Kinoshita S, Morikawa Y, Shibuya A, Phillips J, Lanier L L, Gorman D M, Nolan G P, Miyajima A, Kitamura T. Exp Hematol. 1996;24:324–329. [PubMed] [Google Scholar]
- 12.Ho A S, Liu Y, Khan T A, Hsu D H, Bazan J F, Moore K W. Proc Natl Acad Sci USA. 1993;90:11267–11271. doi: 10.1073/pnas.90.23.11267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aruffo A, Seed B. Proc Natl Acad Sci USA. 1987;84:8573–8577. doi: 10.1073/pnas.84.23.8573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lanier L L, Chang C, Phillips J H. J Immunol. 1994;153:2417–2428. [PubMed] [Google Scholar]
- 15.Chang C, Rodriguez A, Carretero M, Lopez-Botet M, Phillips J H, Lanier L L. Eur J Immunol. 1995;25:2433–2437. doi: 10.1002/eji.1830250904. [DOI] [PubMed] [Google Scholar]
- 16.Callen D F, Baker E, Eyre H J, Chernos J E, Bell J A, Sutherland G R. Ann Genet. 1990;33:219–221. [PubMed] [Google Scholar]
- 17.Lanier L L, Yu G, Phillips J H. J Immunol. 1991;146:1571–1576. [PubMed] [Google Scholar]
- 18.Bacon K B, Premack B A, Gardner P, Schall T J. Science. 1995;269:1727–1730. doi: 10.1126/science.7569902. [DOI] [PubMed] [Google Scholar]
- 19.Plougastel B, Jones T, Trowsdale J. Immunogenetics. 1996;44:286–291. doi: 10.1007/BF02602558. [DOI] [PubMed] [Google Scholar]
- 20.Ra C, Jouvin M-H E, Blank U, Kinet J-P. Nature (London) 1989;341:752–754. doi: 10.1038/341752a0. [DOI] [PubMed] [Google Scholar]
- 21.Pfefferkorn L C, Yeaman G R. J Immunol. 1994;153:3228–3236. [PubMed] [Google Scholar]
- 22.Morton H C, van den Herik-Oudijk I E, Vossebeld P, Snijders A, Verhoeven A J, Capel P J, van de Winkel J G. J Biol Chem. 1995;270:29781–29787. doi: 10.1074/jbc.270.50.29781. [DOI] [PubMed] [Google Scholar]
- 23.Phillips J H, Chang C, Mattson J, Gumperz J E, Parham P, Lanier L L. Immunity. 1996;5:163–172. doi: 10.1016/s1074-7613(00)80492-6. [DOI] [PubMed] [Google Scholar]
- 24.Wagtmann N, Rojo S, Eichler E, Mohrenweiser H, Long E O. Curr Biol. 1997;7:615–618. doi: 10.1016/s0960-9822(06)00263-6. [DOI] [PubMed] [Google Scholar]
- 25.Samaridis J, Colonna M. Eur J Immunol. 1997;27:660–665. doi: 10.1002/eji.1830270313. [DOI] [PubMed] [Google Scholar]
- 26.Hayami K, Fukuta D, Nishikawa Y, Yamashita Y, Inui M, Ohyama Y, Hikida M, Ohmori H, Takai T. J Biol Chem. 1997;272:7320–7327. doi: 10.1074/jbc.272.11.7320. [DOI] [PubMed] [Google Scholar]
- 27.Maeda A, Kurosaki M, Kurosaki T. J Exp Med. 1998;188:991–995. doi: 10.1084/jem.188.5.991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yamashita Y, Ono M, Takai T. J Immunol. 1998;161:4042–4047. [PubMed] [Google Scholar]
- 29.Nakajima H, Samaridis J, Angman L, Colonna M. J Immunol. 1999;162:5–8. [PubMed] [Google Scholar]