Abstract
The genome of Borrelia burgdorferi is composed of one linear chromosome and approximately 20 linear and circular plasmids. Although some plasmids are required by B. burgdorferi in vivo, most plasmids are dispensable for growth in vitro. However, circular plasmid (cp) 26 is present in all natural isolates and has never been lost during in vitro growth. This plasmid carries ospC, which is critical for mammalian infection. We previously showed that cp26 encodes essential functions, including the telomere resolvase, ResT, and hence cannot be displaced. Here we identify two additional essential genes on cp26, bbb26 and bbb27, through a systematic attempt to inactivate each open reading frame (ORF). Furthermore, an incompatible plasmid carrying resT, bbb26 and bbb27 could displace cp26. Computational and experimental analyses suggested that both BBB26 and BBB27 are membrane-associated, periplasmic proteins. These data indicate that bbb26 and bbb27 encode essential but possibly redundant functions and that one or the other of these cp26 genes, in addition to resT, is required for bacterial viability. We conclude that the genetic linkage of critical physiological and virulence functions on cp26 is pertinent to its stable maintenance throughout the evolution of B. burgdorferi.
Introduction
Borrelia burgdorferi, the spirochetal agent of Lyme disease, is maintained in nature in a complex cycle between small mammals and a tick vector of the genus Ixodes. The genome of B. burgdorferi type strain B31 has been sequenced and consists of an approximately 900 kbp linear chromosome and more than 20 plasmids that together total ∼600 kbp in size (Stevenson et al., 1996; 1997; Fraser et al., 1997; Casjens et al., 2000; Miller et al., 2000). The 21 plasmids present in the sequenced B. burgdorferi B31 strain include 12 linear and nine circular replicons (Fraser et al., 1997; Casjens et al., 2000). Additional circular plasmids have been described in other stocks of strain B31, suggesting that at least 23 different plasmids were present in the original B31 isolate (Stevenson et al., 1996; 1997; Casjens et al., 2000; Miller et al., 2000). There is increasing evidence to suggest that plasmid-derived functions can be critical for B. burgdorferi viability at various stages in the natural mouse-tick infectious cycle (Purser and Norris, 2000; Labandeira-Rey and Skare, 2001; Purser et al., 2003; Grimm et al., 2004a; 2005; Pal et al., 2004; Yang et al., 2004; Revel et al., 2005; Strother et al., 2005; Jewett et al., 2007). For example, linear plasmids of 25 kbp (lp25), 28 kbp (lp28-1) and 36 kbp (lp36) are important for infectivity in the mouse (Purser and Norris, 2000; Labandeira-Rey and Skare, 2001; Purser et al., 2003; Jewett et al., 2007) and lp25 is also required for survival in the tick (Grimm et al., 2005; Revel et al., 2005; Strother et al., 2005). Although plasmid-encoded proteins may be required for B. burgdorferi survival in vivo, plasmid loss can be observed after limited in vitro propagation (Xu et al., 1996; Schwan et al., 1988; Grimm et al., 2003). In fact, serial passage coupled with immune selection against plasmid-encoded surface proteins yielded the B. burgdorferi clone B314 that lacks all linear and most circular plasmids (Sadziene et al., 1993). However, the loss of one circular plasmid of 26 kbp (cp26) has never been observed and this plasmid is present in all isolates that have been examined (Marconi et al., 1993; Tilly et al., 1998; Casjens et al., 2000; Byram et al., 2004; Terekhova et al., 2006). Furthermore, in contrast to other B. burgdorferi plasmids, cp26 cannot be displaced by an incompatible plasmid harbouring the cp26 replication machinery alone, suggesting that this plasmid carries essential genes (Byram et al., 2004).
One cp26 gene that appears to be critical for bacterial viability is resT, which is present as a single copy and encodes an enzyme required for resolution of the replicated telomeres of the linear DNA molecules, including the chromosome, into covalently closed hairpin ends (Kobryn and Chaconas, 2002; Byram et al., 2004; Chaconas, 2005). The resT gene could not be inactivated by allelic exchange unless a functional homologue was also present (Tourand et al., 2006) and is presumed to be required for spirochete growth (Byram et al., 2004). However, an incompatible plasmid that carried resT was unable to displace cp26, suggesting that telomere resolution is not the sole essential function encoded by cp26 (Byram et al., 2004).
Twenty-nine ORFs are present on cp26 and approximately half of these genes have known or putative functions (Fig. 1A) (Fraser et al., 1997; Casjens et al., 2000). Along with resT, cp26 carries genes that have been proposed or shown to be involved in purine biosynthesis (guaA and guaB) (Margolis et al., 1994; Zhou et al., 1997), peptide transport (oppAIV) (Bono et al., 1998), chitobiose transport (chbA, chbB and chbC) (Tilly et al., 2001), glucose transport (bbb29) (Fraser et al., 1997; Casjens et al., 2000; Byram et al., 2004) and purine transport (bbb22 and bbb23) (Byram et al., 2004). In addition, the cp26-encoded outer surface lipoprotein C (OspC) is required for spirochete survival in the mammalian host (Grimm et al., 2004a; Stewart et al., 2006; Tilly et al., 2006; 2007). Although some of the above-mentioned cp26 genes encode products that are known to be important to B. burgdorferi for growth and maintenance in its natural infectious cycle, ospC (Tilly et al., 1997), chbC (Tilly et al., 2001), oppAIV (Bono et al., 1998), guaB (Tilly et al., 1998) and bbb29 (Byram et al., 2004) have all been inactivated in previous studies without altering the spirochete's ability to propagate under normal in vitro growth conditions.
Fig. 1.
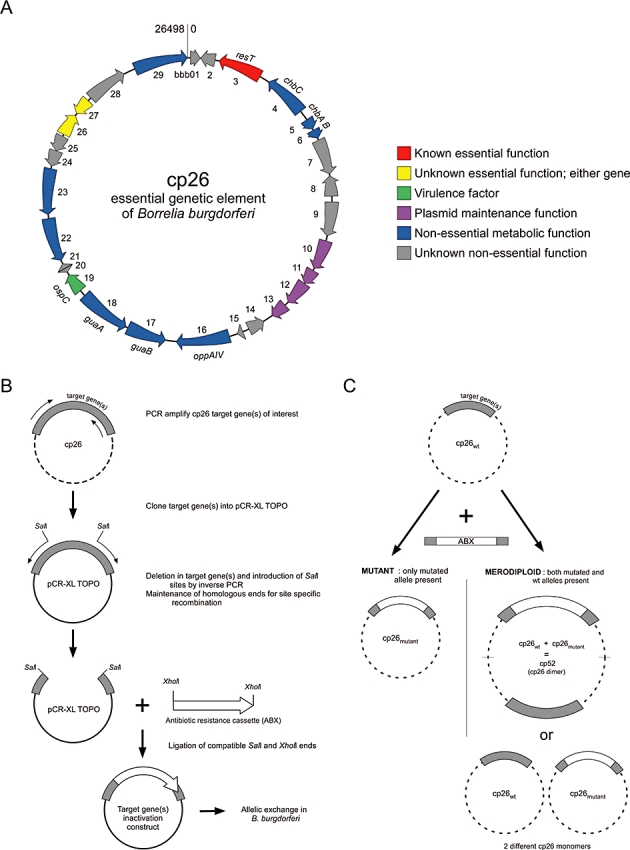
A. Graphical representation of cp26. The arbitrary position of bases 0/26498 is represented by a vertical line between bbb01 and bbb29 (Fraser et al., 1997). Genes are grouped according to functional class. Known essential function (red); unknown essential function (yellow); virulence factor (green); plasmid maintenance (purple); non-essential metabolic function (blue); unknown non-essential function (grey). B. Schematic diagram of the method used for targeted mutagenesis of most of the genes on cp26. Targeted mutagenesis was achieved by allelic exchange of an antibiotic resistance cassette with the gene(s) of interest. C. Graphical representation of the possible outcomes of allelic exchange with cp26 target genes. A single mutated copy of a gene(s) can be recovered if the gene(s) is non-essential for growth. Merodiploids are defined as clones that harbour both a wild-type and mutant copy of the target gene(s) within the same cell, in this case either on a single cp26 dimer (cp52) or on two separate, coexisting cp26 monomers.
In this study, we undertook to inactivate all remaining cp26 ORFs to determine which functions encoded by this plasmid, in addition to telomere resolution, are required for B. burgdorferi growth in vitro. We then attempted to selectively displace cp26 using an incompatible plasmid that carried, along with resT, those genes implicated to be important for bacterial survival. Our findings identify the complement of genes on cp26 that ensures stable retention of this replicon by B. burgdorferi throughout its life cycle.
Results
Inactivation of all cp26 ORFs except the essential resT gene and those required for plasmid maintenance
In a previous study we concluded that cp26 encodes functions essential to bacterial viability because of its presence in all natural isolates, its stability during in vitro growth, and its inability to be displaced by an incompatible plasmid harbouring only the cp26 replication machinery (Byram et al., 2004). We also discovered that telomere resolution, encoded by the cp26 gene resT, is a function vital for B. burgdorferi survival (Byram et al., 2004). However, transformation of B. burgdorferi with a cp26-incompatible plasmid carrying the essential resT gene did not result in displacement of cp26, suggesting that additional genes on cp26 encode critical functions (Byram et al., 2004). To examine which additional cp26 genes are important for in vitro growth, 17 previously uncharacterized cp26 ORFs were targeted for inactivation by allelic exchange (Fig. 1 and Tables 1 and 2). These included all remaining ORFs on cp26 that had not previously been inactivated, except for the four genes encompassing the plasmid replication region (bbb10–13) and two small ORFs (bbb20–21) of 110 and 95 base pairs respectively. The limited coding sequences of bbb20 and bbb21, and their lack of expression and conservation among other B. burgdorferi isolates (R. Byram, unpubl. data, Ojaimi et al., 2003; Glöckner et al., 2004), suggest that they are not protein-coding sequences. Gene inactivations were performed by independent transformation of B. burgdorferi strain B31 clone A (B31-A) with 12 different suicide plasmids designed for targeted allelic exchange with single genes or several adjacent genes (Table 1 and Fig. 1B). The allelic exchange event for each mutant was verified by polymerase chain reaction (PCR) with primers specific to the individual gene, followed by confirmation via Southern blot analysis (data not shown). Although the frequencies with which mutants were obtained varied for different constructs, all 17 remaining cp26 ORFs could be inactivated in some context (Table 2). Inactivation of each of these genes indicated that, unlike resT, they are not individually required for B. burgdorferi growth in vitro. This outcome was unanticipated because of our previous inability to displace cp26 with an incompatible plasmid carrying only resT (Byram et al., 2004). These conflicting results could be explained, however, if an essential function carried by cp26 were redundantly fulfilled by more than one cp26 gene, allowing deletion of each gene singly, whereas simultaneous deletion of all these genes would be lethal for the spirochete. Alternatively, distinct genes could encode important but unrelated functions, and the additive negative effect of their combined loss might render the spirochete non-viable.
Table 1.
Inactivation of cp26 ORFS in B31-A.
| 1 Gene designation and coordinates (1–26498) | 2 Primers used to amplify gene(s)a | 3 Deletion region | 4 Number of base pairs deleted | 5 Inactivation plasmid | 6 Primers used for inactivation | 7 Antibiotic resistance gene used | 8 Primers used to screen transformants |
|---|---|---|---|---|---|---|---|
| BBB01 (16–321) | 1 and 2 | 32–686 | 654 | XL-BBB01–02Δ | 3 and 4 | aacC1 | 1 and 2 |
| BBB02 (751–311) | 1 and 2 | 32–686 | 654 | XL-BBB01–02Δ | 3 and 4 | aacC1 | 1 and 2 |
| chbA (4428–4084) | 5 and 6 | 3951–4788 | 837 | XL-BBB05–07Δ | 7 and 8 | kan | 5 and 6 |
| chbB (4754–4440) | 5 and 6 | 3951–4788 | 837 | XL-BBB05–07Δ | 7 and 8 | kan | 5 and 6 |
| BBB07 (4769–5863) | 5 and 6 | 3951–4788 | 837 | XL-BBB05–07Δ | 7 and 8 | kan | 5 and 6 |
| BBB08 (6517–5891) | 9 and 10 | 6054–7096 | 1042 | XL-BBB08–09Δ | 11 and 12 | kan | 9 and 10 |
| BBB09 (6677–7711) | 9 and 10 | 6054–7096 | 1042 | XL-BBB08–09Δ | 11 and 12 | kan | 9 and 10 |
| BBB14 (11417–10923) | 13 and 14 | 11014–12011 | 997 | XL-BBB14–15Δ | 15 and 16 | aacC1 | 13 and 14 |
| BBB15 (11636–11737) | 13 and 14 | 11014–12011 | 997 | XL-BBB14–15Δ | 15 and 16 | aacC1 | 13 and 14 |
| guaA (16718–15135) | 41 and 42 | 15926–16159 | 233 | XL-guaAΔ | 17 and 18 | aacC1/kan | 41 and 42 |
| BBB22 (19321–17969) | 19 and 20 | 18565–19949 | 1384 | XL-BBB22–23Δ | 21 and 22 | aacC1 | 19 and 20 |
| BBB23 (20822–19434) | 19 and 20 | 18565–19949 | 1384 | XL-BBB22–23Δ | 21 and 22 | aacC1 | 19 and 20 |
| BBB24–27 (21364–22606) | 23 and 24 | 20897–23641 | 2744 | XL-BBB24–27Δ | NA | aacC1 | 31 and 32 |
| BBB24 (21364–20861) | 23 and 24 | 20897–21639 | 742 | XL-BBB24–25Δ | NA | aacC1 | 32 and 33 |
| BBB25 (21851–21342) | 23 and 24 | 20897–21639 | 742 | XL-BBB24–25Δ | NA | aacC1 | 32 and 33 |
| BBB26 (21898–22590) | 26 and 32 | 21923–22579 | 656 | XL-BBB26Δ | 37 and 39 | aacC1 | 35 and 36 |
| BBB27 (23154–22606) | 26 and 32 | 22653–23091 | 438 | XL-BBB27Δ | 36 and 40 | aacC1 | 33 and 34 |
| BBB26–27 (21898–23154) | 52 and 53 | 21923–23091 | 1168 | XL-BBB26–27Δ | 39 and 40 | aadA | 33 and 34 |
| BBB28 (23255–24496) | 24 and 25 | 23564–24037 | 473 | XL-BBB28Δ | 26 and 27 | aacC1/kan | 24 and 25 |
DNA fragment used in subsequent cloning of allelic exchange construct for targeted gene inactivation.
NA, not applicable.
Table 2.
Mutant alleles on cp26.
| Strains recovered | |||
|---|---|---|---|
| Gene(s) targeted for inactivationa | Merodiploid | Mutant | Reference |
| BBB01–2 | – | + | This study |
| BBB03 (resT) | + | – | Byram et al. (2004) |
| BBB04 (chbC) | – | + | Tilly et al. (2001) |
| BBB05–7 | – | + | This study |
| BBB08–9 | – | + | This study |
| BBB14–15 | – | + | This study |
| BBB16 (oppAIV) | – | + | Bono et al. (1998) |
| BBB17 (guaB) | – | + | Tilly et al. (1998) |
| BBB18 (guaA) | – | + | This study |
| BBB19 (ospC) | – | + | Tilly et al. (1997) |
| BBB22–23 | – | + | This study |
| BBB24–27 | + | – | This study |
| BBB24–25 | – | + | This study |
| BBB26–27 | + | – | This study |
| BBB26 | + | +b | This study |
| BBB27 | ±c | + | This study |
| BBB28 | – | + | This study |
| BBB29 | – | + | Byram et al. (2004) |
Merodiploid was resolved to a mutant following passage in liquid and solid media under antibiotic selection.
Mutants were readily obtained in the wild-type genetic background, whereas the merodiploid with pRB3 was resolved to a mutant only following passage in liquid and solid media under antibiotic selection.
Several pieces of data were consistent with the presence of critical but complementary genes on cp26. Mutations in such genes should be recoverable as single, but not double, mutants. Many genes on cp26 could be inactivated in conjunction with adjacent genes (Table 2). However, repeated attempts to delete the cp26 region encompassing bbb24–27 failed, yielding only merodiploid transformants in which both wild type and mutant alleles of these four genes were present in the same bacterium (as depicted in Fig. 1C), similar to the outcome of previous transformations in which the essential resT gene was targeted for inactivation (Table 2) (Byram et al., 2004). Subsequent attempts to inactivate just bbb24–25 from this region easily produced mutants lacking these genes (Table 2). We also obtained single mutants in both bbb26 and bbb27 when these genes were targeted individually (Table 2). In contrast, only merodiploid transformants were recovered following electroporation with a construct designed to delete both bbb26 and bbb27 (Table 2). PCR analyses of the targeted loci of different mutants in the bbb26–27 region illustrate the various outcomes of these transformations (Fig. 2). Additional analysis of the bbb26–27 merodiploid transformants using primers external to the allelic exchange construct (primers 24 and 30, Table S1) along with primers within the flaBp–aadA resistance cassette (primers 55 and 56, Table S1) demonstrated that recombination had occurred at the bbb26–27 locus on cp26 (data not shown). These data suggest that together bbb26 and bbb27 encode an essential function(s) and that at least one or the other of these cp26 genes, in addition to resT, is required for bacterial growth.
Fig. 2.
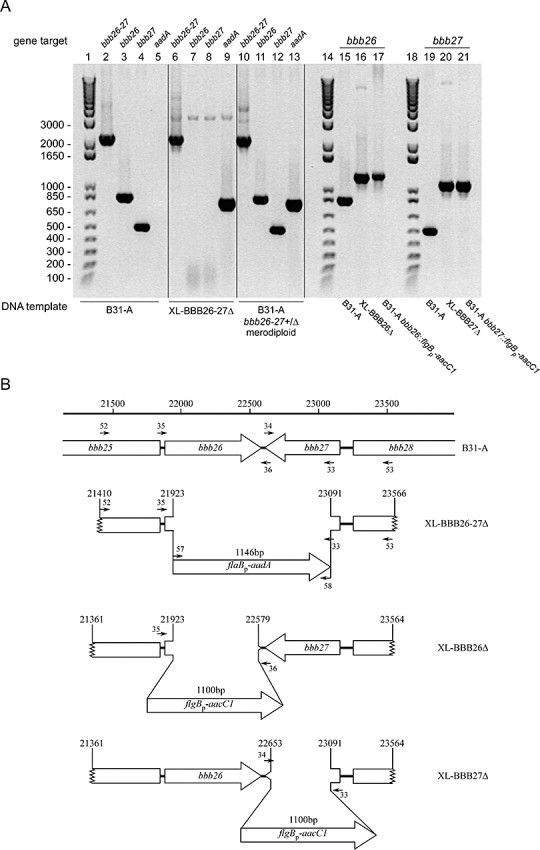
A. PCR analysis of genomic DNA from B. burgdorferi clones transformed with gene inactivation constructs targeting genes in the bbb26–27 region. Template DNAs from transformants are identified below the lanes and PCR amplification targets above the lanes. Template DNA from B31-A illustrates the PCR products from the wild-type alleles of bbb26–27, bbb26 and bbb27 (lanes 2–5, 15 and 19), whereas template DNAs from the gene inactivation constructs (XL-BBB26–27Δ, XL-BBB26Δ and XL-BBB27Δ) depict the PCR profiles of the mutated alleles (lanes 6–9, 16 and 20). The PCR products resulting from the clones transformed with the allelic exchange inactivation plasmids are illustrated in lanes 10–13, 17 and 21. The 1 kbp-plus size standards (Invitrogen) were run in lanes 1, 14 and 18 and sizes (base pairs) are indicated to the left of the panel. B. Graphical representation of the bbb26–27 region on cp26 (B31-A) and the cloned pieces of DNA used for the allelic exchange constructs (XL-BBB26–27Δ, XL-BBB26Δ and XL-BBB27Δ). The 1168 bp region of cp26 between nucleotides 21923 and 23091 was replaced with the 1146 bp flaBp–aadA resistance cassette to create XL-BBB26–27Δ for disruption of both bbb26 and bbb27. The 742 bp region of cp26 between nucleotides 21923 and 22579 was replaced with the 1100 bp flgBp–aacC1 resistance cassette to create XL-BBB26Δ for disruption of bbb26. The 438 bp region of cp26 between nucleotides 22653 and 23091 was replaced with the 1100 bp flgBp–aacC1 resistance cassette to create XL-BBB27Δ for disruption of bbb27. Locations of the primers used for analysis in (A) are indicated and the sequences are listed in Table S1.
Displacement of cp26 by an incompatible plasmid harbouring resT, bbb26 and bbb27
Previous experiments with shuttle vectors derived from non-essential plasmids of B. burgdorferi demonstrated specific displacement of the endogenous plasmid from which the replication/incompatibility region of a vector originated (Stewart et al., 2001; 2003; Eggers et al., 2002; Grimm et al., 2004b). We assumed therefore that we would likewise be able to displace cp26 with an incompatible vector, provided that it harboured all cp26 genes that encode essential functions. The results described earlier suggested that either the bbb26 or the bbb27 gene product is critical for B. burgdorferi viability, in addition to resT. To examine this hypothesis, we constructed the vector pRB3, a modified version of the cp26-incompatible plasmid pBSV26 (Byram et al., 2004), into which we cloned resT, bbb26 and bbb27 (Fig. 3A).
Fig. 3.

A. Graphical representation of pRB3, a cp26-incompatible shuttle vector that carries the resT, bbb26 and bbb27 genes of cp26. The bbb25 and bbb28 genes on pRB3 are truncated, lacking 3′ ends. Relevant restriction sites are shown. colE1, E. coli plasmid origin of replication; ZEO, zeocin-resistance marker; flgBP–kan, kanamycin resistance cassette; bbb10–13, ORFs from cp26 that confer autonomous plasmid replication and incompatibility with cp26 (Byram et al., 2004). B. Displacement of cp26 from B. burgdorferi by pRB3 as demonstrated by Southern blot analysis of transformants. Wild-type B. burgdorferi DNA (wt Bb), E. coli plasmid DNA of shuttle vector carrying resT, bbb26 and bbb27 (pRB3) and total genomic DNA from B. burgdorferi transformed with pRB3 (Bb/pRB3). Southern blot probed with the kanamycin resistance gene (left panel), which is present on pRB3, or the same blot stripped and reprobed with the resT gene (right panel), which is present on both cp26 and pRB3. An arrow indicates the supercoiled form of endogenous cp26; brackets denote supercoiled and nicked forms of pRB3. The mobilities of size standards (kbp) are indicated to the left of the panels.
We next attempted to displace cp26 by transforming B31-A with pRB3. Southern blot analysis of undigested genomic DNA demonstrated that cp26 was absent from pRB3 transformants (Fig. 3B). We conclude that the inclusion on pRB3 of essential functions encoded by resT, bbb26 and bbb27, in conjunction with the plasmid maintenance and incompatibility functions conferred by the bbb10–13 locus, permitted displacement of the endogenous cp26 plasmid of B. burgdorferi for the first time.
As described earlier, we found that either bbb26 or bbb27 could be inactivated in a strain carrying cp26, but that bbb26–27 double mutants were never recovered in this genetic background. We wondered if this restriction still applied to B. burgdorferi lacking cp26, or if both bbb26 and bbb27 were required when other cp26 genes were absent. To address this question, we attempted to individually and doubly inactivate both bbb26 and bbb27 in a strain in which cp26 had been displaced by pRB3. Single bbb26 or bbb27 mutants were each recovered in this background (data not shown and Table 3). However, no double bbb26–27 mutants were ever obtained in the strain lacking cp26, but harbouring pRB3. Likewise, attempts to displace pRB3 with pBSV26resTG, an incompatible vector carrying only resT (and a different selectable marker) were unsuccessful. These results further confirmed that bbb26 and bbb27 provide critical but complementary functions, and together with resT, determine the essential nature of cp26 in B. burgdorferi.
Table 3.
Attempted displacement of cp26 by incompatible plasmids carrying distinct cp26 gene complements.
| Strain | cp26 | cp26-incompatible shuttle vector | Reference |
|---|---|---|---|
| B31-A | + | None | Bono et al. (2000) |
| BbRB1a | coexisting | pBSV26 | Byram et al. (2004) |
| BbRB2b | coexisting | pBSV26resT | Byram et al. (2004) |
| BbRB3 | Displaced | pRB3 (pBSV26resTbbb26–27) | This study |
| BbRB4 | Displaced | pRB3Δbbb26c | This study |
| BbRB5 | Displaced | pRB3Δbbb27d | This study |
Strain previously denoted B31-A/pBSV26.
Strain previously denoted B31-A/pBSV26resT.
bbb26 on pRB3 inactivated by allelic exchange with a construct carrying flgBp–aacC1.
bbb27 on pRB3 inactivated by allelic exchange with a construct carrying flgBp–aacC1.
Altered growth of cp26 mutants
Although our data indicate that only the resT and bbb26 or bbb27 genes of cp26 are absolutely required for bacterial viability, mutants in which cp26 was displaced and bbb26 or bbb27 was inactivated were not easily recovered. These transformations initially yielded merodiploids, which only resolved into the ‘clean’ allelic exchange mutants described earlier after subsequent passage and replating under antibiotic selection. This suggested that mutants in which cp26 had been displaced and that carried only one of the bbb26–27 gene pair were at a selective growth disadvantage relative to wild-type organisms. To address this possibility, we compared the growth in liquid and solid media of several strains (Fig. 4).
Fig. 4.
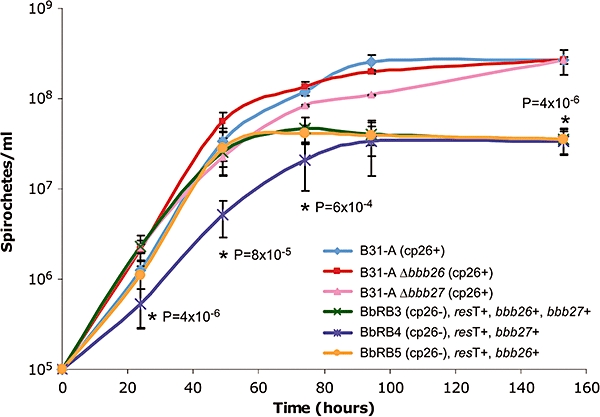
In vitro growth of B. burgdorferi strains with different cp26 gene complements. The presence and absence of cp26, resT, bbb26 and bbb27 from B31-A-derivative strains are as indicated. The densities of cultures from a starting dilution of 1 × 105 spirochetes ml−1 were determined every 24 h using a Petroff–Hausser counting chamber. Data are presented as averages of at least three separate experiments, with triplicate cultures per strain in each experiment, and error bars represent standard deviation from the mean. Statistical analyses (Student's t-test, two tailed) were used to compare pairs of data points. The exponential phase growth of BbRB4 was significantly slower than that of all other strains, as indicated by asterisks at 24, 49 and 72 h; the P-values represent comparisons with BbRB3. All three strains lacking cp26 reached significantly lower stationary phase densities relative to B31-A, as indicated by the asterisk and P-value at 153 h.
Consistent with the ability to recover single bbb26 and bbb27 mutants in the wild-type B31-A genetic background, no difference in growth rate or maximum stationary phase density in liquid medium was observed between B31-A and spirochetes lacking either bbb26 (B31-A, Δbbb26, cp26+) or bbb27 (B31-A, Δbbb27, cp26+) (Fig. 4). The BbRB3 strain (B31-A, cp26–, resT+, bbb26+, bbb27+), which lacks cp26 but contains resT, bbb26 and bbb27 (Table 3), displayed a doubling time similar to B31-A (6.1 h), but only reached an average stationary phase density of ∼4 × 107 spirochetes ml−1 (compared with ∼3 × 108 spirochetes ml−1 for B31-A), even after 120 h of growth (Fig. 4). BbRB3 and BbRB5 (B31-A, cp26–, resT+, bbb26+), the isogenic derivative lacking bbb27, demonstrated similar growth rates (Fig. 4). However, the isogenic strain lacking bbb26 (BbRB4) (B31-A, cp26–, resT+, bbb27+) grew at a slower rate than the other two mutant strains, with a doubling time of 9.5 h, and reached a comparable stationary phase density (3.5 × 107 spirochetes ml−1) approximately 20 h later (Fig. 4). Conversely, BbRB5 colony formation in solid medium was delayed up to 7 days relative to BbRB3 and BbRB4. The growth defects of BbRB4 and BbRB5 in liquid and solid media, respectively, are consistent with the difficulty with which these mutants were initially recovered.
Predicted cellular locations of the BBB26 and BBB27 proteins
Given the requirement for bbb26 and bbb27 by B. burgdorferi, we were interested in the predicted proteins that these genes encode. BBB26 and BBB27 are convergently transcribed ORFs (Fig. 1A) that encode unrelated basic proteins of 27.2 kDa (pI 9.2) and 21.5 kDa (pI 9.5), respectively. Both genes are described in the B. burgdorferi genome annotation (Fraser et al., 1997; Casjens et al., 2000) as hypothetical ORFs that lack sequence homology with genes in other organisms. The BBB26 and BBB27 ORFs are conserved, however, in the closely related Lyme disease spirochete Borrelia garinii, in which the predicted proteins share ∼90% amino acid identity with the B. burgdorferi homologues (Glöckner et al., 2004). No differences in protein profiles were detected when cellular lysates of bbb26 and bbb27 mutants were compared with wild-type B31-A on a silver-stained, single dimension SDS-polyacrylamide gel (data not shown), suggesting that, although critical, these proteins are not abundant components of in vitro grown organisms, a trait shared with ResT (R. Byram, unpubl. data).
Computational analyses of the BBB26 and BBB27 open reading frames suggested that both proteins could be membrane-associated. While BBB26 lacks a predicted amino-terminal signal sequence, Psort analysis (Nakai and Horton, 1999) identified hydrophobic residues 24–42 of BBB26 as a probable single transmembrane region, potentially anchoring the largely hydrophilic protein to the cytoplasmic membrane and localizing a majority of the protein to the periplasmic space. Psort analysis of BBB27 identified residues 1–22 as an amino-terminal signal sequence, but predicted that this leader peptide would be uncleaved. However, recent data suggest that Psort has failed to identify nearly half of the experimentally confirmed spirochetal lipoprotein sequences (Setubal et al., 2006). Because of the inaccuracy of Psort with regards to spirochete sequences, a novel spirochete-specific lipoprotein prediction algorithm, SpLip, has been developed from a confirmed spirochetal lipoprotein data set (Setubal et al., 2006). SpLip analysis identified BBB27 as a canonical lipoprotein with a characteristic spirochetal lipobox (LFYGC) (Setubal et al., 2006). Consistent with the findings of another lipoprotein signal peptide prediction algorithm LipoP (Juncker et al., 2003), SpLip predicted cleavage of the signal peptide between residues 15 and 16 (LLFYG/CSTIS), followed by amino-terminal acylation of BBB27 at the N-terminal Cys residue and association with either the spirochete inner or the outer membrane (Setubal et al., 2006).
In an initial attempt to experimentally address the computational predictions that both the BBB26 and BBB27 proteins are membrane-associated, polyclonal rabbit antisera were elicited against the recombinant proteins purified from insoluble inclusion bodies in Escherichia coli. However, the antibodies failed to recognize any proteins in B. burgdorferi lysates (data not shown). This suggested that either the epitopes of the denatured proteins were not exposed on the native proteins or that BBB26 and BBB27 were present in too low abundance in B. burgdorferi lysates to be detected by immunoblot analysis. To circumvent both of these potential limitations, bbb26 and bbb27 were cloned into the Borrelia expression vector pBSV2ex (Guyard et al., 2006) under the control of a strong Borrelia promoter (flgBp) and tagged with a C-terminal FLAG epitope (DYKDDDDK) (Hopp et al., 1988) to be used for immunoblot analysis. As expected, a protein of approximately 28 kDa was detected with monoclonal antiserum recognizing the FLAG epitope in total cell lysates of both E. coli and B. burgdorferi harbouring pBSV2ex bbb26-FLAG (Fig. 5A) and a protein of approximately 23 kDa was detected in total cell lysates of E. coli carrying pBSV2ex bbb27-FLAG (Fig. 5A). Consistent with the presence of an N-terminal signal peptidase II cleavage site in the BBB27 protein, a smaller protein of approximately 18 kDa was detected in total cell lysates of B. burgdorferi carrying the same construct (Fig. 5A), suggesting that BBB27 was processed when synthesized in B. burgdorferi but not when it was synthesized in E. coli. This may indicate a potential difference in the active site substrate specificity of the type II signal peptidase of spirochetes relative to other major bacterial groups, as suggested by Setubal et al. (2006) and further reflected in the inaccuracy of the Psort algorithm in predicting spirochetal lipoproteins (Setubal et al., 2006). The size difference between BBB27 synthesized in E. coli versus B. burgdorferi appears greater than predicted by cleavage of the signal peptide alone (∼21 kDa); this may reflect additional processing of BBB27 in B. burgdorferi. No proteins were detected in total cell lysates of B. burgdorferi carrying the pBSV2ex plasmid alone (data not shown), indicating that the FLAG antibody specifically recognized the engineered FLAG epitopes on the BBB26-FLAG and BBB27-FLAG proteins. Together these data demonstrated that the BBB26-FLAG and BBB27-FLAG proteins are synthesized in B. burgdorferi and can be detected using antiserum that recognizes the FLAG epitope.
Fig. 5.
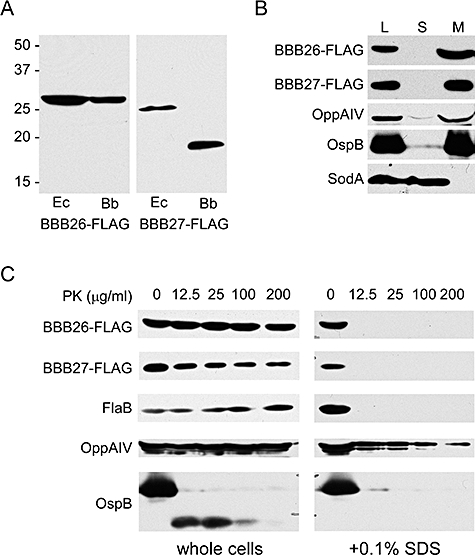
Expression and cellular localization of the BBB26-FLAG and BBB27-FLAG proteins. A. Proteins lysates from E. coli (Ec) or B. burgdorferi B31-A34 (Bb) harbouring either pBSV2ex bbb26-FLAG or pBSV2ex bbb27-FLAG were separated by SDS-PAGE and analysed by immunoblot with anti-FLAG antibodies. The mobilities of size standards (molecular weights in kDa) are indicated to the left of the figure. B. Protein lysates from B. burgdorferi clone A34 harbouring either pBSV2ex bbb26-FLAG or pBSV2ex bbb27-FLAG were harvested and separated into soluble and membrane fractions by ultracentrifugation. Protein fractions from equivalent numbers of spirochetes were subjected to SDS-PAGE and analysed by immunoblot with FLAG (BBB26 and BBB27), OppAIV (inner membrane), OspB (outer membrane) and SodA (cytoplasmic) antisera. Representative results for the localization of the OppAIV, OspB and SodA proteins from B. burgdorferi clone A34 harbouring either pBSV2ex bbb26-FLAG or pBSV2ex bbb27-FLAG are shown. L, total cell lysate; S, soluble protein fraction; M, membrane protein fraction. C. Equal numbers of whole or 0.1% SDS-treated (+0.1% SDS) cells of B. burgdorferi clone A34 harbouring either pBSV2ex bbb26-FLAG or pBSV2ex bbb27-FLAG were incubated with different concentrations (μg ml−1) of proteinase K (PK). Lysates of PK-treated bacteria were separated by SDS-PAGE and analysed by immunoblot with FLAG (BBB26 and BBB27), flagellin (FlaB, periplasmic marker), OppAIV (inner membrane marker) and OspB (surface exposed, outer membrane marker) antisera. Representative results for the proteinase K sensitivity of the FlaB, OppAIV and OspB proteins from B. burgdorferi clone A34 harbouring either pBSV2ex bbb26-FLAG or pBSV2ex bbb27-FLAG are shown.
To investigate the predicted membrane association of BBB26 and BBB27, B. burgdorferi harbouring either pBSV2ex bbb26-FLAG or pBSV2ex bbb27-FLAG were disrupted by French press and separated into total lysate, soluble and membrane fractions (see Experimental procedures). Cellular fractions were resolved by SDS-polyacrylamide gel electrophoresis (PAGE) and immunoblotted with FLAG, OppAIV, OspB and SodA antisera. Both the BBB26-FLAG and BBB27-FLAG proteins were detected in the total lysate and membrane fractions, but not the soluble fraction (Fig. 5B), as were the membrane proteins OppAIV and OspB. As anticipated, the cytoplasmic protein SodA (Fraser et al., 1997; Whitehouse et al., 1997) was limited to the soluble fraction (Fig. 5B). Together these data, along with the computational analysis, indicate that BBB26 and BBB27 are membrane-associated proteins.
To determine if either protein was surface exposed, intact borreliae harbouring either pBSV2ex bbb26-FLAG or pBSV2ex bbb27-FLAG were treated with varying amounts of proteinase K and bacterial lysates were immunoblotted with FLAG antiserum (Fig. 5C). Flagellin (FlaB), the major constituent of the flagella, was used as a periplasmic marker (Barbour et al., 1986; Bono et al., 1998; Bunikis and Barbour, 1999). The oligopeptide permease lipoprotein, OppAIV, served as a tightly associated inner membrane marker (Bono et al., 1998; Schulze and Zuckert, 2006), and the outer membrane lipoprotein OspB was used as a marker for surface exposed proteins (Bergström et al., 1989; Bono et al., 1998; Bunikis and Barbour, 1999). Little to no degradation of the BBB26-FLAG or the BBB27-FLAG protein was detected for intact cells across all concentrations of proteinase K, similar to the pattern observed for the internal proteins FlaB and OppAIV (Fig. 5C). In contrast, the surface exposed lipoprotein OspB was sensitive to degradation at the lowest concentration of proteinase K (12.5 µg ml−1, Fig. 5C), relative to untreated intact cells. Lysis of the borreliae with 0.1% SDS resulted in complete susceptibility of the BBB26-FLAG, BBB27-FLAG and FlaB proteins to protealysis (Fig. 5C), demonstrating that neither BBB26 nor BBB27 is intrinsically protease resistant. Conversely, only a slight increase in protease susceptibility was detected for OppAIV in SDS-treated lysates, relative to intact cells (Fig. 5C), suggesting that the proteinase K cleavage sites of this protein may have remained inaccessible despite the presence of detergent and/or this protein is resistant to proteolysis by proteinase K. OspB maintained sensitivity to proteolysis in SDS-treated lysates (Fig. 5C). Together these data suggest that neither BBB26-FLAG nor BBB27-FLAG is exposed on the B. burgdorferi outer surface and both are likely to be membrane-associated proteins localized to the periplasmic space.
Although a blast search of the current database did not identify any sequence homologues of BBB26 or BBB27, COG analysis revealed that both proteins share some amino acid similarity to peptide-cleaving enzymes (Altschul et al., 1990; Marchler-Bauer et al., 2005). However, no protease activity was detected when lysates of B. burgdorferi harbouring either pBSV2ex bbb26-FLAG or pBSV2ex bbb27-FLAG were analysed on casein zymogram gels or in spectrophotometric protease assays which used casein as a substrate, in contrast to lysates of B. burgdorferi producing a previously identified protease of Borrelia (Guyard et al., 2006) (data not shown).
Discussion
In this study we defined the full complement of genes carried by cp26 that are essential for spirochete survival in vitro. These include resT, a telomere resolvase (Kobryn and Chaconas, 2002; Byram et al., 2004), and either bbb26 or bbb27, adjacent genes unique to Borrelia. This study used two different genetic techniques to identify those genes encoded on cp26 that are crucial for B. burgdorferi growth: targeted gene inactivation by allelic exchange and plasmid displacement. Allelic exchange allows targeted deletion of a specific gene by replacement with a selectable marker, via homologous recombination with sequences flanking the targeted gene (Fig. 1B and C). Using this method, non-essential genes can be inactivated. Conversely, inactivation of essential genes would result in a lethal phenotype and mutants would not be recovered. An alternative outcome is merodiploid formation, in which both wild-type and mutated forms of the gene coexist in the same organism (Fig. 1C), suggesting that loss of the targeted gene is deleterious for the spirochete. Merodiploids may be resolved to allelic exchange mutants following repeated passage in the presence of selection if the target gene is not absolutely required. Therefore, failure to recover mutants using this method suggests the critical presence of the targeted gene for the in vitro survival of B. burgdorferi.
Plasmid displacement results in simultaneous loss of a number of genes by removal of an entire plasmid. This method asks whether the gene(s) present on an incompatible plasmid allow(s) displacement of the endogenous complement of genes carried on a particular replicon, suggesting that the reduced set of genes is sufficient for survival. Two plasmid species are considered incompatible when they contain identical replication and/or partitioning functions, resulting in the loss of one of the plasmids (Austin and Nordström, 1990). However, loss of one of the plasmids is not the only observed outcome of competition between incompatible plasmids. We previously demonstrated that the plasmid pBSV26 harbouring the cp26 replication machinery, as well as pBSV26resT, the same plasmid carrying the resT gene, when introduced into B. burgdorferi either coexisted with the endogenous cp26 or integrated into a cp26 replicon (Byram et al., 2004). Therefore, unless the minimal set of genes present on the introduced plasmid is sufficient for in vitro survival of B. burgdorferi, displacement will not be observed. In the current study, both targeted gene inactivation by allelic exchange and plasmid displacement suggest the central roles of bbb26 and bbb27, in addition to resT (Byram et al., 2004), for B. burgdorferi growth in vitro.
Strains lacking cp26, but harbouring only resT, bbb26 and bbb27, were impaired in growth compared with the parent B31-A strain (Fig. 4). These clones are lacking 22 of the 29 cp26 genes, including guaA, guaB, oppAIV, bbb22, bbb23 and bbb29, which likely play significant roles in the salvage of purines, peptides and sugars, functions that could be important for spirochete growth in vitro and/or in vivo (Margolis et al., 1994; Zhou et al., 1997; Bono et al., 1998; Botkin et al., 2006). Inactivation of bbb26 in the strain in which cp26 had been displaced by pRB3, carrying resT, bbb26 and bbb27, resulted in an even greater growth impairment in liquid medium compared with the parent (Fig. 4). In addition, deletion of bbb27 from pRB3 led to a delay in colony formation relative to the parent clone, reaffirming the physiological importance of the bbb26 and bbb27 gene products for B. burgdorferi growth.
The cp26 genes bbb26 and bbb27 appear to be required for B. burgdorferi survival in vitro, yet their functions remain unknown. Both genes are unique to Borrelia and neither shares convincing sequence similarity with genes of known function. Despite the lack of homology to genes from other bacteria, the BBB26 protein does contain a conserved domain of unknown function (DUF955) found among a number of bacterial and viral proteins. The BBB26 protein also demonstrates limited sequence similarity to members of COG 2856, a family of predicted zinc peptidases (Marchler-Bauer et al., 2005). Similar to BBB26, BBB27 displays limited sequence similarity to peptide-cleaving enzymes. Most notably, it shares some sequence similarity to protease-1 from the fungal pathogen Pneumocystis and to a probable peptidase from Rickettsia prowazekii (Altschul et al., 1990). If BBB26 or BBB27 does have peptidase activities, neither can use casein as a substrate, as no band of clearing was detected on a casein zymogram and no protease activity was detected by spectrophotometric assay (data not shown), suggesting that the putative peptidase activity(s), if any, of these proteins are not as general proteases and may be specific to peptides of a particular sequence or size.
Computational predictions as well as direct analysis of fractionated B. burgdorferi cells suggested that BBB26 and BBB27 are associated with a spirochetal membrane (Fig. 5B). Protease accessibility assays demonstrated that neither protein was exposed on the spirochete outer surface (Fig. 5C). BBB27 has a consensus spirochetal lipoprotein signal sequence (Setubal et al., 2006) that appears to be processed in B. burgdorferi (Fig. 5A) and therefore, similar to OppAIV (Bono et al., 1998; Schulze and Zuckert, 2006), may be a periplasmic protein tethered to the cytoplasmic membrane by an N-terminal acyl group. BBB26 is predicted to harbour a single transmembrane region that could allow association with the cytoplasmic membrane and potential periplasmic localization. Together these data suggest that both proteins may be present in the periplasmic space. Given its small size, C-terminal location and successful use in other systems (Molloy et al., 1994; Yamamoto et al., 2003; Mura et al., 2004), it is unlikely that the FLAG epitope tag used for these experiments interfered with the expression or misdirected the cellular localization of BBB26 and BBB27.
The uniquely segmented B. burgdorferi genome can contain as many as 23 genetic elements at one time (Stevenson et al., 1996; 1998; Fraser et al., 1997; Casjens et al., 2000; Miller et al., 2000). The reason that the genome of this bacterium is divided among such a large variety of replicons remains unclear. However, it appears that the distribution of genes across the numerous B. burgdorferi replicons, and on cp26 in particular, has not occurred at random. While cp26 carries genes important for bacterial growth, it also harbours a gene required for mammalian infectivity. Presumably, the linkage of the essential virulence gene ospC, which is only required at one specific stage of the B. burgdorferi life cycle (Grimm et al., 2004a; Stewart et al., 2006; Tilly et al., 2006; 2007), with constitutively required functions encoded by resT, bbb26 and bbb27, assures that this crucial virulence factor will not be lost during growth in environments in which ospC does not provide a selective advantage. Indeed, we found that when the ospC gene is carried by a non-essential plasmid, it was lost by spirochetes in infected mice subsequent to the development of the host acquired immune response (Tilly et al., 2006; 2007). Consistent with this observation, Xu and colleagues have also shown that persistent expression of ospC by B. burgdorferi in an infected mammal is deleterious to the spirochete (Xu et al., 2007). Although B. burgdorferi lacking ospC are efficiently acquired by feeding ticks, loss of OspC precludes infection of a new mammalian host during the next tick blood meal, thereby aborting the infectious cycle (Tilly et al., 2006; 2007). This demonstrates that ospC must be carried by an essential genetic element, such as cp26 or the chromosome, in order to assure its retention by the spirochete in the mammalian host. In sum, it appears that genetic linkage of critical physiological and virulence functions on cp26 has resulted in its stable maintenance throughout the evolution of B. burgdorferi.
In a previous study we showed that cp26 encodes functions essential to B. burgdorferi viability including, but not limited to, the telomere resolvase resT (Byram et al., 2004). Herein we conclude that in addition to resT, the membrane-associated proteins encoded by the cp26 genes bbb26 and bbb27 are important for spirochete viability, as the cp26 plasmid could only be displaced by an incompatible plasmid when it carried these three genes, and a mutant lacking both bbb26 and bbb27 was never obtained. Future studies are required to identify the contribution of bbb26 and bbb27 to basic spirochete physiology, both in vitro and in vivo. Together, the resT, bbb26 and bbb27 genes contribute to spirochete survival and thereby help to ensure the faithful maintenance of the cp26 replicon throughout the varying conditions encountered in the mouse-tick infectious cycle.
Experimental procedures
B. burgdorferi strains and growth conditions
B. burgdorferi strains were cultivated at 35°C either in liquid BSK-H complete medium (Sigma) or in Barbour-Stoenner-Kelly (BSKII) medium containing gelatin and supplemented with 6% rabbit serum (Pel-Freez Laboratories). B. burgdorferi cultures were plated in solid BSKII medium and grown at 35°C with 2.5% CO2 (Rosa et al., 1996). Kanamycin was used at a concentration of 200 μg ml−1, gentamicin at 40 μg ml−1 and streptomycin at 50 μg ml−1. B31 clone A (B31-A) (Bono et al., 2000) is a non-infectious derivative of type strain B31 (ATCC 35210), which was isolated from a tick collected on Shelter Island in New York (Burgdorfer et al., 1982). B31 clone A34 (A34) is a derivative of B31-A that also lacks lp56. For growth rate comparisons, strains were inoculated from freezer stocks into 5 ml of BSKII containing the appropriate antibiotic and grown to an approximate density of 1 × 107 spirochetes ml−1. Strains were subsequently diluted in triplicate to 1 × 105 spirochetes ml−1 in 5 ml of BSKII containing the appropriate antibiotic(s). Spirochete density was determined every 24 h using a Petroff–Hausser counting chamber. Time for colony formation was assessed by diluting various strains to 1 × 103 spirochetes ml−1 and plating 100 μl of culture in solid BSKII medium on duplicate plates. The plates were observed daily for about 18 days for appearance of colonies. Colony formation analysis was blinded and performed twice.
Constructs for gene inactivation by allelic exchange
The primer sequences used in this study were based on the previously described B31 genome sequence (Fraser et al., 1997; Casjens et al., 2000) and are shown in Table S1. The majority of the allelic exchange constructs for inactivation of cp26 genes were cloned by the following strategy, as diagramed in Fig. 1B. Details for each inactivation construct are provided in Table 1. A region of cp26 spanning the targeted gene(s) was amplified from B31-A genomic DNA (primers listed in column 2 of Table 1 and Table S1), using either Taq polymerase (fragments < 2.5 kb) (New England Biolabs) or the Expand Long Template PCR system (fragments > 2.5 kb) (Roche Molecular Biochemicals), and cloned into the vector pCR-XL-TOPO (Invitrogen). A deletion in each targeted gene(s) (as described in column 3 of Table 1) was constructed by inverse PCR performed with the primers listed in column 6 of Table 1, all of which introduce SalI sites, and the Expand Long Template PCR system. The flgBP–aacC1 gene cassette (Elias et al., 2003), which confers gentamicin resistance, was amplified using Taq polymerase and primers 43 and 44 and cloned into the pCR2.1-TOPO vector (Invitrogen). The flgBP–aacC1 gene cassette was removed from the pCR2.1-TOPO vector by XhoI digestion and ligated into inactivation constructs digested with SalI to create individual gene deletion plasmids. The flgBP–kan gene cassette (Bono et al., 2000), which confers kanamycin resistance, was amplified with Taq polymerase and primers 43 and 45 and cloned into the pCR2.1-TOPO vector. Inactivation constructs to confer kanamycin-resistance were created as above, except with a flgBP–kan fusion. The flaBP–aadA fusion, which confers spectinomycin/streptomycin resistance, was generated by PCR-amplifying the flaB promoter region from B31-A, using primers 50 and 54, and the aadA gene from pKFSS1 (Frank et al., 2003), using primers 55 and 56. Both the flaB and aadA DNA fragments were digested with NdeI, purified, ligated together and cloned into the pCR2.1-TOPO vector (Invitrogen). The flaBP–aadA cassette was subsequently PCR-amplified using primers 57 and 58, digested with XhoI and ligated into the appropriate SalI-digested inactivation construct. The structures of all plasmids were confirmed by DNA sequencing using the ABI Big Dye Terminator Cycle Sequencing Ready Reaction Kit (PE Applied Biosystems) on an ABI 3700 DNA sequencer.
Two constructs used for gene inactivation by allelic exchange were created by slightly different cloning strategies. To inactivate bbb24–27, a 4.9 kb region of cp26 including the full-length bbb24, bbb25, bbb26 and bbb27 genes was amplified from B31-A genomic DNA using the Expand Long Template PCR system and primers 23 and 24, and cloned into the pCR-XL-TOPO vector to create XL-bbb24–27. A 2.7 kb deletion in the bbb24–27 genes encompassing cp26 nucleotides 20897–23641 was constructed by XmnI restriction enzyme digestion. The flgBP–aacC1 gene cassette was amplified using Taq polymerase and primers 46 and 47, and cloned into the pCR2.1-TOPO vector. The flgBP–aacC1 gene cassette was removed from the pCR2.1-TOPO vector by PvuII digestion and ligated into XL-bbb24–27Δ20897-23641 that had been digested with XmnI. To inactivate just bbb24–25, a 742 bp deletion encompassing nucleotides 20897–21639 in the bbb24 and bbb25 genes carried by the plasmid XL-bbb24–27 was created by partial digestion of the plasmid with XmnI and SwaI restriction enzymes and then ligated with the flgBP–aacC1 gene cassette removed from the pCR2.1-TOPO vector by PvuII digestion.
Construction of shuttle vectors
To create pRB3 (pBSV26resTbbb26–27), a 3.4 kb region of cp26 encompassing the bbb26 and bbb27 genes was amplified using the Expand Long Template PCR system and primers 30 and 31, and cloned into the pCR-XL-TOPO vector. The 2.1 kb gene fragment including the bbb26 and bbb27 genes was subsequently removed from the pCR-XL-TOPO vector by KpnI digestion and ligated into the multiple cloning site of KpnI-digested pBSV26resT (Byram et al., 2004), to obtain pRB3.
To create pBSV26resTG, the flaBP–aacC1 resistance cassette was amplified using Taq DNA polymerase (New England Biolabs) and primers 50 and 51, and cloned into the pCR-XL-TOPO vector. The fragment was removed from the pCR-XL-TOPO vector by KpnI and BamHI digestion and ligated into the multiple cloning site of pBSV26resT digested with KpnI and BamHI to obtain the plasmid pBSV26resTG.
The bbb26 and bbb27 genes were FLAG-epitope tagged and individually cloned into the vector pBSV2ex (Guyard et al., 2006) under the control of the flaB promoter. The bbb26 and bbb27 genes were amplified from B31-A genomic DNA and C-terminally FLAG-epitope tagged using Vent DNA polymerase (New England Biolabs) and primer pairs 59 and 61, and 60 and 62, respectively, and cloned into the pCR-XL-TOPO vector. The DNA fragments were removed from the pCR-XL-TOPO vector by digestion with NdeI and KpnI and subcloned individually into pBSV2ex digested with NdeI and KpnI to obtain pBSV2ex-bbb26-FLAG and pBSV2ex-bbb27-FLAG. These plasmids were transformed into B. burgdorferi B31 clone A34. All plasmid constructs were verified by restriction digest and sequence analysis.
Transformation of B. burgdorferi
Transformation of B. burgdorferi by electroporation was performed as previously described (Elias et al., 2002). Briefly, 10–15 μg of plasmid DNA was resuspended in 8 μl of H2O and electroporated into B. burgdorferi. Following electroporation, the cells were resuspended in 5 ml of BSK-H complete medium (Sigma) or BSKII medium and allowed to recover for 18–24 h at 35°C. The spirochetes were then plated within solid BSKII medium supplemented with either 200 μg kanamycin ml−1, 40 μg gentamicin ml−1 or 50 μg spectinomycin ml−1.
Screening of B. burgdorferi transformants
B. burgdorferi colonies that arose in medium containing antibiotics were inoculated into 20 μl of PCR mixtures with sterile toothpicks. PCR amplification with primers specific for the kanamycin resistance cassette (primers 48 and 49) was used to identify shuttle vector transformants. Allelic exchange transformants were first identified by screening for either the presence of the kanamycin (primers 48 and 49), gentamicin (primers 46 and 47) or streptomycin (primers 55 and 56) resistance cassettes (Table S1). Transformants containing the appropriate resistance cassette were screened for inactivation of targeted genes by PCR with primers described in column 8 of Table 1. The PCR conditions were 94°C for 2 min, followed by 30 cycles of 94°C for 45 s, 55°C for 45 s and 68°C for 3 min in a GeneAmp PCR 9700 thermal cycler (Perkin Elmer) or a DNA engine tetrad 2 thermal cycler (MJ Research). PCR products were separated by agarose gel electrophoresis and visualized by ethidium bromide staining. Colonies of candidate transformants were aspirated with sterile Pasteur pipettes, placed in 5 ml of liquid BSK-H medium (Sigma), or BSKII medium, and allowed to grow to mid to late log phase. Total genomic DNA was then isolated from these cultures with a Wizard genomic DNA purification kit (Promega). PCR performed with total genomic DNA with primers specific for shuttle vectors or cp26 genes was used to further confirm the presence of foreign DNA or the structures of targeted loci in transformants (Table 1, column 8).
Southern hybridization analysis
Total genomic DNA of B. burgdorferi was isolated from 15 ml of cultures with a Wizard genomic DNA purification kit (Promega). Approximately 500 ng of uncut genomic DNA, or 500 ng of genomic DNA digested for 12–20 h with selected restriction enzymes, was separated by gel electrophoresis on a 0.3% agarose gel and visualized by ethidium bromide staining. Genomic DNA was depurinated, denatured and neutralized, and then blotted onto a Biotrans nylon membrane (ICN). A UV stratalinker 1800 (Stratagene) was used to cross-link the DNA to the membrane. The kan- and resT-specific probes were labeled with 32P and hybridizations were performed as previously described (Byram et al., 2004).
Cellular localization
All procedures were performed at 4°C. One litre cultures of B. burgdorferi at densities of 5 × 107 spirochetes ml−1 were harvested by centrifugation at 10 000 g for 10 min. Cell pellets were gently washed two times with 10 mM NaCl in 20 mM HEPES, pH 7.6. Cells were resuspended in 10 ml of 10 mM NaCl in 20 mM HEPES, pH 7.6 and lysed by three passes through a cold French press cell (14 000 lb/in2). Unlysed cells and cell debris were removed by centrifugation at 12 000 g for 10 min. The soluble and membrane fractions were separated by centrifugation in a Beckman 45Ti rotor (125 000 g, 3 h). The membrane pellets were washed in HEPES buffer and harvested by centrifugation in a Beckman 45Ti rotor (125 000 g, overnight). The membrane fractions were resuspended in 10 ml of 10 mM NaCl in 20 mM HEPES, pH 7.6. Fractions were normalized according to the total number of spirochetes for immunoblot analysis. Samples were subjected to PAGE with 12% acrylamide. Immunoblots were performed and proteins were detected using monoclonal anti-FLAG (Sigma) (1:500), monoclonal anti-OspB H6831 (Rosa et al., 1992) (1:50), monoclonal anti-FlaB H9724 (Barbour et al., 1986) (1:200), polyclonal anti-OppAIV (Bono et al., 1998) (1:2000) and polyclonal anti-SodA (NIH754) (1:1000) antibodies as previously described (Grimm et al., 2004a).
SodA purification and antibody production
The sodA ORF was amplified by PCR from B. burgdorferi strain B31-A3 using primers 63 and 64 (Table S1). The resulting DNA fragment was digested with XhoI/PstI and cloned into a similarly digested pBAD/HisA (Invitrogen) vector, generating pBADsodA. SodA-6xHis was overexpressed in E. coli Top10 (pLysE) cells (Invitrogen) and purified from the insoluble fraction under denaturing conditions (8 M urea) using a Ni-NTA agarose column (Qiagen) as per manufacturer's instructions. Purified SodA-6xHis was used to raise polyclonal antiserum in a female New Zealand White rabbit at Cocalico Biologicals (Reamstown, PA).
Proteinase K treatment of Borrelia cells
Intact B. burgdorferi or B. burgdorferi plus 0.1% SDS (Sigma) were treated with proteinase K (Invitrogen) as previously described (Bono et al., 1998; Bunikis and Barbour, 1999). Samples were subjected to PAGE with 12% acrylamide and transferred to nitrocellulose membranes. Immunoblots were performed and proteins were detected using monoclonal anti-FLAG, monoclonal anti-OspB, monoclonal anti-FlaB, polyclonal anti-OppAIV antibodies as previously described (Grimm et al., 2004a).
Acknowledgments
We are grateful to Philip Stewart, Michael Otto and Paul Brett for critical reading of the manuscript and helpful discussions. Thank you to Travis Jewett for technical expertise. We also thank Anita Mora, Gary Hettrick and Austin Athman for graphic design. This research was supported by the Intramural Research Program of the National Institute of Allergy and Infectious Diseases, National Institutes of Health.
Supplementary material
This material is available as part of the online article from:
http://www.blackwell-synergy.com/doi/abs/10.1111/j.1365-2958.2007.05969.x
(This link will take you to the article abstract).
Please note: Blackwell Publishing is not responsible for the content or functionality of any supplementary materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Austin S, Nordström K. Partition-mediated incompatibility of bacterial plasmids. Cell. 1990;60:351–354. doi: 10.1016/0092-8674(90)90584-2. [DOI] [PubMed] [Google Scholar]
- Barbour AG, Hayes SF, Heiland RA, Schrumpf ME, Tessier SL. A Borrelia-specific monoclonal antibody binds to a flagellar epitope. Infect Immun. 1986;52:549–554. doi: 10.1128/iai.52.2.549-554.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergström S, Bundoc VG, Barbour AG. Molecular analysis of linear plasmid-encoded major surface proteins, OspA and OspB, of the Lyme disease spirochete Borrelia burgdorferi. Mol Microbiol. 1989;3:479–486. doi: 10.1111/j.1365-2958.1989.tb00194.x. [DOI] [PubMed] [Google Scholar]
- Bono JL, Tilly K, Stevenson B, Hogan D, Rosa P. Oligopeptide permease in Borrelia burgdorferi: putative peptide-binding components encoded by both chromosomal and plasmid loci. Microbiol. 1998;144:1033–1044. doi: 10.1099/00221287-144-4-1033. [DOI] [PubMed] [Google Scholar]
- Bono JL, Elias AF, Kupko JJ, III, Stevenson B, Tilly K, Rosa P. Efficient targeted mutagenesis in Borrelia burgdorferi. J Bacteriol. 2000;182:2445–2452. doi: 10.1128/jb.182.9.2445-2452.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botkin DJ, Abbott A, Stewart PE, Rosa PA, Kawabata H, Watanabe H, et al. Identification of potential virulence determinants by HimarI transposition of infectious Borrelia burgdorferi B31. Infect Immun. 2006;74:6690–6699. doi: 10.1128/IAI.00993-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunikis J, Barbour AG. Access of antibody or trypsin to an integral outer membrane protein (P66) of Borrelia burgdorferi is hindered by Osp lipoproteins. Infect Immun. 1999;67:2874–2883. doi: 10.1128/iai.67.6.2874-2883.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgdorfer W, Barbour AG, Hayes SF, Benach JL, Grunwaldt E, Davis JP. Lyme disease – a tick-borne spirochetosis? Science. 1982;216:1317–1319. doi: 10.1126/science.7043737. [DOI] [PubMed] [Google Scholar]
- Byram R, Stewart PE, Rosa PA. The essential nature of the ubiquitous 26 kb circular replicon of Borrelia burgdorferi. J Bacteriol. 2004;186:3561–3569. doi: 10.1128/JB.186.11.3561-3569.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casjens S, Palmer N, van Vugt R, Huang WM, Stevenson B, Rosa P, et al. A bacterial genome in flux: the twelve linear and nine circular extrachromosomal DNAs in an infectious isolate of the Lyme disease spirochete Borrelia burgdorferi. Mol Microbiol. 2000;35:490–516. doi: 10.1046/j.1365-2958.2000.01698.x. [DOI] [PubMed] [Google Scholar]
- Chaconas G. Hairpin telomeres and genome plasticity in Borrelia: all mixed up in the end. Mol Microbiol. 2005;58:625–635. doi: 10.1111/j.1365-2958.2005.04872.x. [DOI] [PubMed] [Google Scholar]
- Eggers CH, Caimano MJ, Clawson ML, Miller WG, Samuels DS, Radolf JD. Identification of loci critical for replication and compatibility of a Borrelia burgdorferi cp32 plasmid and use of a cp32-based shuttle vector for expression of fluorescent reporters in the Lyme disease spirochaete. Mol Microbiol. 2002;43:281–295. doi: 10.1046/j.1365-2958.2002.02758.x. [DOI] [PubMed] [Google Scholar]
- Elias AF, Stewart PE, Grimm D, Caimano MJ, Eggers CH, Tilly K, et al. Clonal polymorphism of Borrelia burgdorferi strain B31 MI: implications for mutagenesis in an infectious strain background. Infect Immun. 2002;70:2139–2150. doi: 10.1128/IAI.70.4.2139-2150.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elias AF, Bono JL, Kupko JJ, Stewart PE, Krum JG, Rosa PA. New antibiotic resistance cassettes suitable for genetic studies in Borrelia burgdorferi. J Mol Microbiol Biotechnol. 2003;6:29–40. doi: 10.1159/000073406. [DOI] [PubMed] [Google Scholar]
- Frank KL, Bundle SF, Kresge ME, Eggers CH, Samuels DS. aadA confers streptomycin resistance in Borrelia burgdorferi. J Bacteriol. 2003;185:6723–6727. doi: 10.1128/JB.185.22.6723-6727.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser CM, Casjens S, Huang WM, Sutton GG, Clayton R, Lathigra R, et al. Genomic sequence of a Lyme disease spirochaete, Borrelia burgdorferi. Nature. 1997;390:580–586. doi: 10.1038/37551. [DOI] [PubMed] [Google Scholar]
- Glöckner G, Lehmann R, Romualdi A, Pradella S, Schulte-Spechtel U, Schilhabel M, et al. Comparative analysis of the Borrelia garinii genome. Nucleic Acids Res. 2004;32:6038–6046. doi: 10.1093/nar/gkh953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm D, Elias AF, Tilly K, Rosa PA. Plasmid stability during in vitro propagation of Borrelia burgdorferi assessed at a clonal level. Infect Immun. 2003;71:3138–3145. doi: 10.1128/IAI.71.6.3138-3145.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm D, Tilly K, Byram R, Stewart PE, Krum JG, Bueschel DM, et al. Outer-surface protein C of the Lyme disease spirochete: a protein induced in ticks for infection of mammals. Proc Natl Acad Sci USA. 2004a;101:3142–3147. doi: 10.1073/pnas.0306845101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm D, Eggers CH, Caimano MJ, Tilly K, Stewart PE, Elias AF, et al. Experimental assessment of the roles of linear plasmids lp25 and lp28-1 of Borrelia burgdorferi throughout the infectious cycle. Infect Immun. 2004b;72:5938–5946. doi: 10.1128/IAI.72.10.5938-5946.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm D, Tilly K, Bueschel DM, Fisher MA, Policastro PF, Gherardini FC, et al. Defining plasmids required by Borrelia burgdorferi for colonization of tick vector Ixodes scapularis (Acari: Ixodidae) J Med Entomol. 2005;42:676–684. doi: 10.1093/jmedent/42.4.676. [DOI] [PubMed] [Google Scholar]
- Guyard C, Battisti J, Raffel S, Schrumpf M, Whitney A, Krum JG, et al. Relapsing fever spirochetes produce a serine protease that provides resistance to oxidative stress and killing by neutrophils. Mol Microbiol. 2006;60:710–722. doi: 10.1111/j.1365-2958.2006.05122.x. [DOI] [PubMed] [Google Scholar]
- Hopp TP, Prickett KS, Price VL, Libby RT, March CJ, Cerretti DP, et al. A short polypeptide tag sequence useful for recombinant protein identification and purification. Biotechnol J. 1988;6:1204–1210. [Google Scholar]
- Jewett MW, Lawrence K, Bestor AC, Tilly K, Grimm D, Shaw P, et al. The critical role of the linear plasmid lp36 in the infectious cycle of Borrelia burgdorferi. Mol Microbiol. 2007;64:1358–1374. doi: 10.1111/j.1365-2958.2007.05746.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juncker AS, Willenbrock H, Von Heijne G, Burnak S, Nielsen H, Krogh A. Prediction of lipoprotein signal peptides in Gram-negative bacteria. Protein Sci. 2003;12:1652–1662. doi: 10.1110/ps.0303703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobryn K, Chaconas G. ResT, a telomere resolvase encoded by the Lyme disease spirochete. Mol Cell. 2002;9:195–201. doi: 10.1016/s1097-2765(01)00433-6. [DOI] [PubMed] [Google Scholar]
- Labandeira-Rey M, Skare JT. Decreased infectivity in Borrelia burgdorferi strain B31 is associated with loss of linear plasmid 25 or 28-1. Infect Immun. 2001;69:446–455. doi: 10.1128/IAI.69.1.446-455.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A, Anderson JB, Cherukuri PF, DeWeese-Scott C, Geer LY, Gwadz M, et al. CDD: a conserved domain database for protein classification. Nucleic Acids Res. 2005;33:D192–D196. doi: 10.1093/nar/gki069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marconi RT, Konkel ME, Garon CF. Variability of osp genes and gene products among species of Lyme disease spirochetes. Infect Immun. 1993;61:2611–2617. doi: 10.1128/iai.61.6.2611-2617.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolis N, Hogan D, Tilly K, Rosa PA. Plasmid location of Borrelia purine biosynthesis gene homologs. J Bacteriol. 1994;176:6427–6432. doi: 10.1128/jb.176.21.6427-6432.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JC, Bono JL, Babb K, El-Hage N, Casjens S, Stevenson B. A second allele of eppA. Borrelia burgdorferi strain B31 is located on the previously undetected circular plasmid cp9-2. J Bacteriol. 2000;182:6254–6258. doi: 10.1128/jb.182.21.6254-6258.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molloy SS, Thomas L, VanSlyke JK, Stenberg PE, Thomas G. Intracellular trafficking and activation of the furin proprotein convertase: localization to the TGN and recycling from the cell surface. EMBO J. 1994;13:18–33. doi: 10.1002/j.1460-2075.1994.tb06231.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mura CV, Cosmelli D, Munoz F, Delgado R. Orientation of Arabidopsis thaliana KAT1 channel in the plasma membrane. J Membr Biol. 2004;201:157–165. doi: 10.1007/s00232-004-0713-8. [DOI] [PubMed] [Google Scholar]
- Nakai K, Horton P. Psort: a program for detecting sorting signals in proteins and predicting their subcellular localization. Trends Biochem Sci. 1999;24:34–36. doi: 10.1016/s0968-0004(98)01336-x. [DOI] [PubMed] [Google Scholar]
- Ojaimi C, Brooks C, Casjens S, Rosa P, Elias A, Barbour AG, et al. Profiling temperature-induced changes in Borrelia burgdorferi gene expression using whole genome arrays. Infect Immun. 2003;71:1689–1705. doi: 10.1128/IAI.71.4.1689-1705.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal U, Yang X, Chen M, Bockenstedt LK, Anderson JF, Flavell RA, et al. OspC facilitates Borrelia burgdorferi invasion of Ixodes scapularis salivary glands. J Clin Invest. 2004;113:220–230. doi: 10.1172/JCI19894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purser JE, Norris SJ. Correlation between plasmid content and infectivity in Borrelia burgdorferi. Proc Natl Acad Sci USA. 2000;97:13865–13870. doi: 10.1073/pnas.97.25.13865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purser JE, Lawrenz MB, Caimano MJ, Radolf JD, Norris SJ. A plasmid-encoded nicotinamidase (PncA) is essential for infectivity of Borrelia burgdorferi in a mammalian host. Mol Microbiol. 2003;48:753–764. doi: 10.1046/j.1365-2958.2003.03452.x. [DOI] [PubMed] [Google Scholar]
- Revel AT, Blevins JS, Almazan C, Neil L, Kocan KM, de la Fuente J, et al. bptA (bbe16) is essential for the persistence of the Lyme disease spirochete, Borrelia burgdorferi, in its natural tick vector. Proc Natl Acad Sci USA. 2005;102:6972–6977. doi: 10.1073/pnas.0502565102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosa P, Samuels DS, Hogan D, Stevenson B, Casjens S, Tilly K. Directed insertion of a selectable marker into a circular plasmid of Borrelia burgdorferi. J Bacteriol. 1996;178:5946–5953. doi: 10.1128/jb.178.20.5946-5953.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosa PA, Schwan T, Hogan D. Recombination between genes encoding major outer surface proteins A and B of Borrelia burgdorferi. Mol Microbiol. 1992;6:3031–3040. doi: 10.1111/j.1365-2958.1992.tb01761.x. [DOI] [PubMed] [Google Scholar]
- Sadziene A, Wilske B, Ferdows MS, Barbour AG. The cryptic ospC gene of Borrelia burgdorferi B31 is located on a circular plasmid. Infect Immun. 1993;61:2192–2195. doi: 10.1128/iai.61.5.2192-2195.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze RJ, Zuckert WR. Borrelia burgdorferi lipoproteins are secreted to the outer surface by default. Mol Microbiol. 2006;59:1473–1484. doi: 10.1111/j.1365-2958.2006.05039.x. [DOI] [PubMed] [Google Scholar]
- Schwan TG, Burgdorfer W, Garon CF. Changes in infectivity and plasmid profile of the Lyme disease spirochete, Borrelia burgdorferi, as a result of in vitro cultivation. Infect Immun. 1988;56:1831–1836. doi: 10.1128/iai.56.8.1831-1836.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setubal JC, Reis M, Matsunaga J, Haake DA. Lipoprotein computational prediction in spirochaetal genomes. Microbiol. 2006;152:113–121. doi: 10.1099/mic.0.28317-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson B, Tilly K, Rosa PA. A family of genes located on four separate 32-kilobase circular plasmids in Borrelia burgdorferi B31. J Bacteriol. 1996;178:3508–3516. doi: 10.1128/jb.178.12.3508-3516.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson B, Casjens S, van Vugt R, Porcella SF, Tilly K, Bono JL, et al. Characterization of cp18, a naturally truncated member of the cp32 family of Borrelia burgdorferi plasmids. J Bacteriol. 1997;179:4285–4291. doi: 10.1128/jb.179.13.4285-4291.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson B, Casjens S, Rosa P. Evidence of past recombination events among the genes encoding the Erp antigens of Borrelia burgdorferi. Microbiol. 1998;144:1869–1879. doi: 10.1099/00221287-144-7-1869. [DOI] [PubMed] [Google Scholar]
- Stewart PE, Thalken R, Bono JL, Rosa P. Isolation of a circular plasmid region sufficient for autonomous replication and transformation of infectious Borrelia burgdorferi. Mol Microbiol. 2001;39:714–721. doi: 10.1046/j.1365-2958.2001.02256.x. [DOI] [PubMed] [Google Scholar]
- Stewart PE, Chaconas G, Rosa P. Conservation of plasmid maintenance functions between linear and circular plasmids in Borrelia burgdorferi. J Bacteriol. 2003;185:3202–3209. doi: 10.1128/JB.185.10.3202-3209.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart PE, Wang X, Bueschel DM, Clifton DR, Grimm D, Tilly K, et al. Delineating the requirement for the Borrelia burgdorferi virulence factor OspC in the mammalian host. Infect Immun. 2006;74:3547–3553. doi: 10.1128/IAI.00158-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strother KO, Broadwater A, De Silva A. Plasmid requirements for infection of ticks by Borrelia burgdorferi. Vector Borne Zoonotic Dis. 2005;5:237–245. doi: 10.1089/vbz.2005.5.237. [DOI] [PubMed] [Google Scholar]
- Terekhova D, Iyer R, Wormser GP, Schwartz I. Comparative genome hybridization reveals substantial variation among clinical isolates of Borrelia burgdorferi sensu stricto with different pathogenic properties. J Bacteriol. 2006;188:6124–6134. doi: 10.1128/JB.00459-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilly K, Casjens S, Stevenson B, Bono JL, Samuels DS, Hogan D, et al. The Borrelia burgdorferi circular plasmid cp26: conservation of plasmid structure and targeted inactivation of the ospC gene. Mol Microbiol. 1997;25:361–373. doi: 10.1046/j.1365-2958.1997.4711838.x. [DOI] [PubMed] [Google Scholar]
- Tilly K, Lubke L, Rosa P. Characterization of circular plasmid dimers in Borrelia burgdorferi. J Bacteriol. 1998;180:5676–5681. doi: 10.1128/jb.180.21.5676-5681.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilly K, Elias AF, Errett J, Fischer E, Iyer R, Schwartz I, et al. Genetics and regulation of chitobiose utilization in Borrelia burgdorferi. J Bacteriol. 2001;183:5544–5553. doi: 10.1128/JB.183.19.5544-5553.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilly K, Krum JG, Bestor A, Jewett MW, Grimm D, Bueschel D, et al. Borrelia burgdorferi OspC protein required exclusively in a crucial early stage of mammalian infection. Infect Immun. 2006;74:3554–3564. doi: 10.1128/IAI.01950-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilly K, Bestor A, Jewett MW, Rosa P. Rapid clearance of Lyme disease spirochetes lacking OspC from skin. Infect Immun. 2007;75:1517–1519. doi: 10.1128/IAI.01725-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tourand Y, Bankhead T, Wilson SL, Putteet-Driver AD, Barbour AG, Byram R, et al. Differential telomere processing by Borrelia telomere resolvases in vitro but not in vivo. J Bacteriol. 2006;188:7378–7386. doi: 10.1128/JB.00760-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehouse CA, Williams LR, Austin FE. Identification of superoxide dismutase activity in Borrelia burgdorferi. Infect Immun. 1997;65:4865–4868. doi: 10.1128/iai.65.11.4865-4868.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Q, McShan K, Liang FT. Identification of an ospC operator critical for immune evasion of Borrelia burgdorferi. Mol Microbiol. 2007;64:220–231. doi: 10.1111/j.1365-2958.2007.05636.x. [DOI] [PubMed] [Google Scholar]
- Xu Y, Kodner C, Coleman L, Johnson RC. Correlation of plasmids with infectivity of Borrelia burgdorferi sensu stricto type strain B31. Infect Immun. 1996;64:3870–3876. doi: 10.1128/iai.64.9.3870-3876.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto H, Kurosawa S, Sekiguchi J. Localization of the vegetative cell wall hydrolases LytC, LytE, and LytF on the Bacillus subtilis cell surface and stability of these enzymes to cell wall-bound or extracellular proteases. J Bacteriol. 2003;185:6666–6677. doi: 10.1128/JB.185.22.6666-6677.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang XF, Pal U, Alani SM, Fikrig E, Norgard MV. Essential role for OspA/B in the life cycle of the Lyme disease spirochete. J Exp Med. 2004;199:641–648. doi: 10.1084/jem.20031960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Cahoon M, Rosa P, Hedstrom L. Expression, purification, and characterization of inosine 5′-monophosphate dehydrogenase from Borrelia burgdorferi. J Biol Chem. 1997;272:21977–21981. doi: 10.1074/jbc.272.35.21977. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


