Abstract
Background
To identify thermophile-specific proteins, we performed phylogenetic patterns searches of 66 completely sequenced microbial genomes. This analysis revealed a cluster of orthologous groups (COG1618) which contains a protein from every thermophile and no sequence from 52 out of 53 mesophilic genomes. Thus, COG1618 proteins belong to the group of thermophile-specific proteins (THEPs) and therefore we here designate COG1618 proteins as THEP1s. Since no THEP1 had been analyzed biochemically thus far, we characterized the gene product of aq_1292 which is THEP1 from the hyperthermophilic bacterium Aquifex aeolicus (aaTHEP1).
Results
aaTHEP1 was cloned in E. coli, expressed and purified to homogeneity. At a temperature optimum between 70 and 80°C, aaTHEP1 shows enzymatic activity in hydrolyzing ATP to ADP + Pi with kcat = 5 × 10-3 s-1 and Km = 5.5 × 10-6 M. In addition, the enzyme exhibits GTPase activity (kcat = 9 × 10-3 s-1 and Km= 45 × 10-6 M). aaTHEP1 is inhibited competitively by CTP, UTP, dATP, dGTP, dCTP, and dTTP. As shown by gel filtration, aaTHEP1 in its purified state appears as a monomer. The enzyme is resistant to limited proteolysis suggesting that it consists of a single domain. Although THEP1s are annotated as "predicted nucleotide kinases" we could not confirm such an activity experimentally.
Conclusion
Since aaTHEP1 is the first member of COG1618 that is characterized biochemically and functional information about one member of a COG may be transferred to the entire COG, we conclude that COG1618 proteins are a family of thermophilic NTPases.
Background
The comparison of protein coding sequences from complete genomes led to a classification, based on sequence similarities, that assigns proteins to clusters of orthologous groups (COGs) [1,2]. Phylogenetic patterns search [3] is a tool to retrieve COGs that contain protein sequences that match a certain predefined pattern of organisms. Recently, Forterre [4] used this tool to identify reverse gyrase as the only hyperthermophile-specific protein. Another survey identified 30 COGs enriched in hyperthermophilic procaryotes [5]. In contrast, we here extended a similar search with respect to a group consisting of both thermophiles and hyperthermophiles i. e. the currently 3 organisms with optimal growth temperatures between 55 and 80°C were included. This search retrieved COG1618 as one of the high ranking clusters containing a protein from every of the 13 thermophilic and, by including only one representative, the fewest sequences from mesophilic genomes of the current COG database. Thus, amongst others, the COG1618 proteins belong to a group of thermophile-specific proteins (THEPs) and in this report we designate them as THEP1s.
Orthologs typically have the same function, allowing the transfer of functional information from one member of a COG to the entire COG. To the best of our knowledge, no function could be established for any of the THEP1s thus far. On the other hand, Cort et al. predicted THEP1s being ATPases [6]. This paper confirms their hypothesis experimentally by characterizing the gene product of aq_1292 from Aquifex aeolicus (aaTHEP1) as a first example representative for this protein family. Aquifex aeolicus is a hyperthermophilic bacterium growing optimally at 95°C [7].
Results
Identification of COG1618
Among the 4,873 COGs of the current COG database, phylogenetic patterns search revealed 167 COGs that include a sequence from each of the 13 thermophilic genomes (thermophiles and hyperthermophiles). A further analysis showed that 63 COGs contain a sequence from every genome, indicating that those conserved proteins are probably essential for microbial life. Consequently, the remaining 104 COGs are more or less thermophile-specific proteins. To retrieve absolute thermophile-specific COGs the search parameters were chosen in the way that the output COGs must contain a sequence from every thermophilic and must not contain any sequence from a mesophilic genome. Whereas no COG exactly met this search condition, less thermophile-specific COGs with only one or a few exceptions from the exact search condition were detected (table 1). Among those, COG1618 is the only one that contains a sequence from each thermophilic and only one further sequence from a mesophilic genome (MA3402).
Table 1.
COGs containing the THEPs with the highest thermophile-specificity.
| Number of mesophilic genomes in COG | |||||||
| 0 | 1 | 2 | 3 | 4 | 5 | ||
| Number of thermophilic genomes in COG | 13 | -/- | COG1618 | COG3635 | -/- | COG1355 COG1371 |
COG2078 |
| 12 | COG1980 | -/- | -/- | COG1144 COG1820 |
COG1730 | COG1503 COG1867 |
|
| 11 | COG1581 COG1888 |
COG1350 COG1909 |
|||||
| 10 | COG1110 | ||||||
| 9 | COG1318 COG1630 COG2250 |
||||||
Given are the COGs that contain sequences from the most among the 13 thermophilic and the fewest among the 53 mesophilic genomes of the current COG database.
Sequence analysis of THEP1s
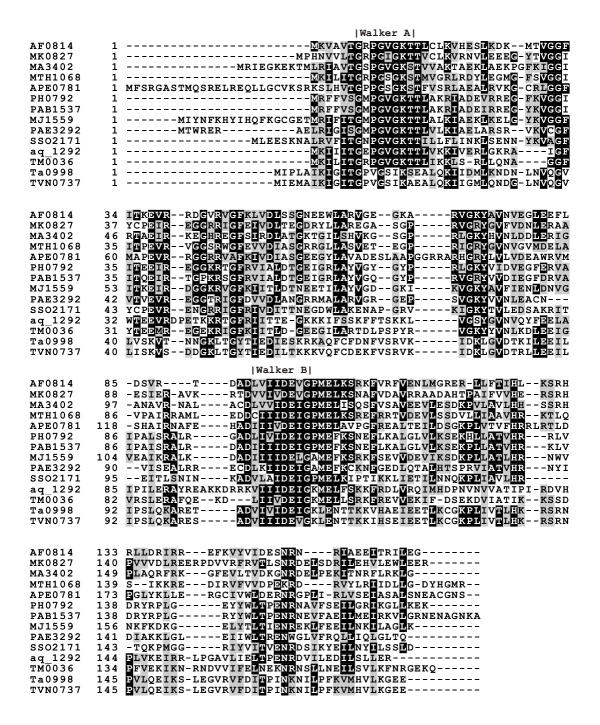
CLUSTALW analysis of all THEP1s led to the design of suitable experiments to assay catalytic in vitro function of aaTHEP1. All proteins contain both the Walker A (GxxxxGK [ST]) and Walker B motif (4 × hydrophobic [DE]xxG) indicating that aaTHEP1 is a P-loop NTPase (figure 1). Since the distal [NT]KxD motif responsible for guanine specificity [8] is absent, aaTHEP1 probably do not belong to the GTPase superclass of the P-loop NTPases. Cort et al. stated that THEP1s "probably represent a novel family of ATPases in both sequence and structural terms" [6]. The authors determined the structure of MTH538 from Methanobacterium thermoautotrophicum by NMR and based on extensive sequence and structure comparisons proposed that the relationship between MTH583 and THEP1s might be similar to that between CheY and ATPase/phosphatase members of the HAD family as suggested by Ridder and Dijkstra [9].
Figure 1.
THEP1s contain Walker A and Walker B motifs. CLUSTALW analysis of all THEP1s. The positions of the Walker A and Walker B motifs are indicated. The shown sequences are from Archaeoglobus fulgidus (AF0814), Methanopyrus kandleri AV19 (MK0827), Methanosarcina acetivorans (MA3402), Methanothermobacter thermautotrophicus (MTH1068), Aeropyrum pernix (APE0781), Pyrococcus horikoshii (PH0792), Pyrococcus abyssi (PAB1537), Methanococcus jannaschii (MJ1559), Pyrobaculum aerophilum (PAE3292), Sulfolobus solfataricus (SSO2171), Aquifex aeolicus (aq_1292), Thermotoga maritima ( TM0036), Thermoplasma acidophilum (Ta0998), and Thermoplasma volcanium (TVN0737).
Purification of recombinant aaTHEP1
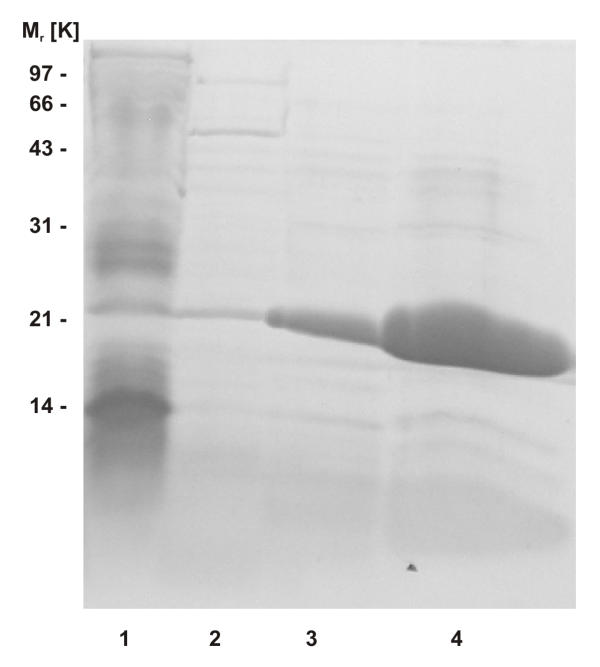
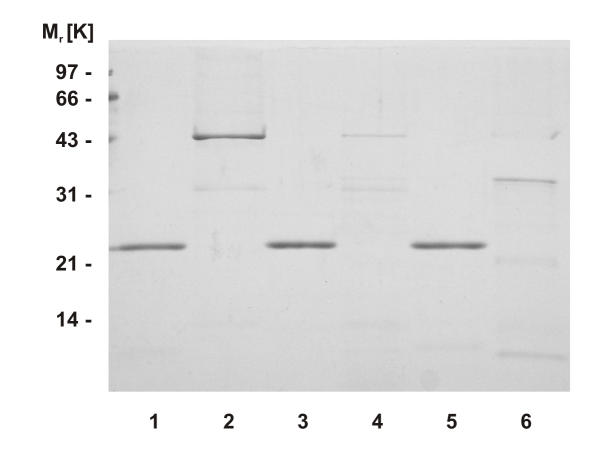
The bulk of E. coli proteins could be removed by heating the crude cell extract for 10 min at 75°C. As expected from the sequence-based theoretical pI of 9.88, aaTHEP1 binds to the matrix of a cation exchanger. Ion exchange followed by hydrophobic interaction chromatography finally resulted in a homogenous aaTHEP1 preparation that migrates slightly higher than the calculated molecular weight of 20,555 Da (figure 2).
Figure 2.
Recombinant aaTHEP1 can be expressed and purified. SDS-PAGE after different steps during purification of recombinant aaTHEP1. Lane 1: crude cell extract, lane 2: supernatant after heat treatment, lane 3: eluate after cation exchange chromatography, lane 4: eluate after hydrophobic interaction chromatography. To demonstrate the purity of the final preparation, lane 4 intentionally was overloaded.
Functional activity of aaTHEP1
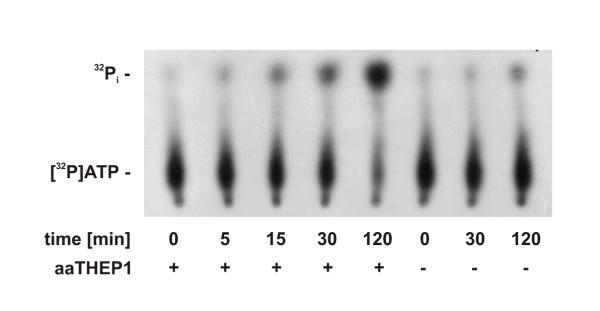
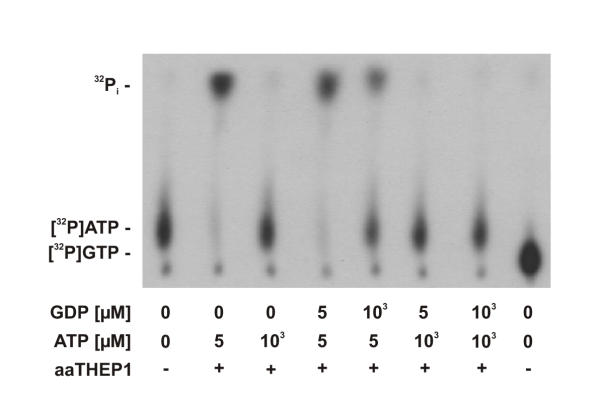
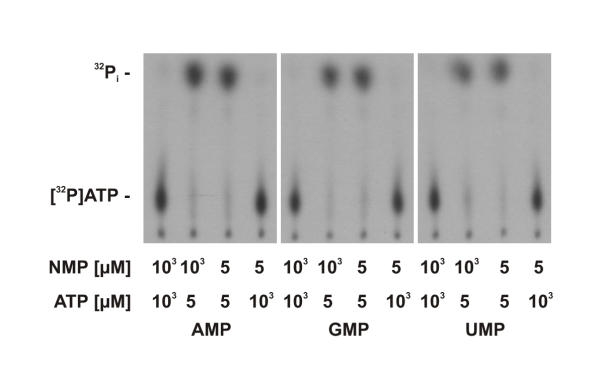
As shown in figure 3, purified aaTHEP1 clearly catalyzes the hydrolysis of ATP to ADP and Pi. As can be seen, the longer the reaction mixture was incubated the more 32Pi was released from [γ-32P]ATP. Since THEP1s are annotated as "predicted nucleotide kinases", we assayed aaTHEP1 for nucleoside diphosphate kinase and nucleoside monophosphate kinase activities. Using ATP as the phosphate donor and GDP (figure 4), GMP, AMP, and UMP (figure 5) as acceptors we could not detect the predicted phosphoryl transfer.
Figure 3.
aaTHEP1 is an ATPase. Autoradiography of thin-layer chromatograms showing samples containing [γ-32P]ATP after different times of incubation at 70°C. Measurements were performed at 50 μM ATP. 0.5 μg of purified aaTHEP1 was used for each assay in 25 μl buffer.
Figure 4.
aaTHEP1 is no NDP kinase. Autoradiography of thin-layer chromatograms showing samples containing [γ-32P]ATP after five minutes of incubation at 70°C and at different concentrations of ATP and GDP. 1 μg of purified aaTHEP1 was used for each assay in 25 μl buffer.
Figure 5.
aaTHEP1 is no NMP kinase. Autoradiography of thin-layer chromatograms showing samples containing [γ-32P]ATP after five minutes of incubation at 70°C and at different concentrations of ATP, AMP, GMP, and UMP, respectively. 1 μg of purified aaTHEP1 was used for each assay in 25 μl buffer.
Temperature dependence
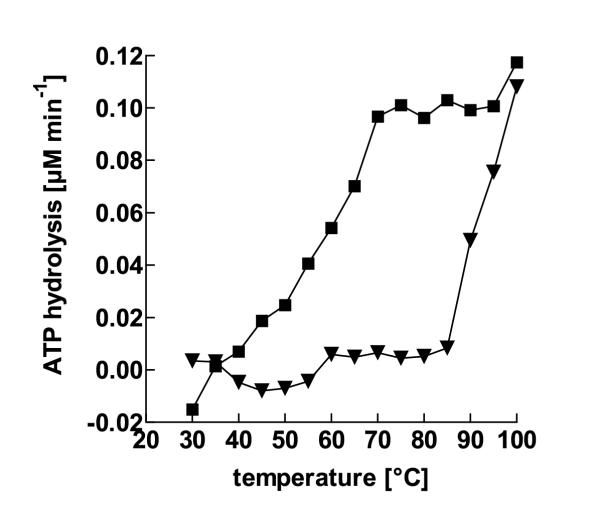
To show that aaTHEP1 is a thermophilic enzyme, thermal activity was determined by measuring ATP hydrolysis as catalyzed by the purified enzyme at different temperatures. Since spontaneous ATP hydrolysis also occurs at higher temperatures, those rates were measured and shown as well. As can be seen in figure 6, aaTHEP1 is still active at 90°C. As an optimum temperature with respect to the signal to noise ratio, 70°C was chosen for all further kinetic measurements.
Figure 6.
aaTHEP1 is a thermophilic enzyme. Temperature dependence of aaTHEP1 catalyzed ATP hydrolysis. Measurements were performed at 5 μM ATP in buffer A. ATP-hydrolysis was measured in the presence of aaTHEP1 (squares) and spontaneous ATP-degradation was determined in the absence of aaTHEP1 (triangles). 0.5 μg of purified aaTHEP1 was used for each assay in 25 μl buffer.
Steady-state kinetics
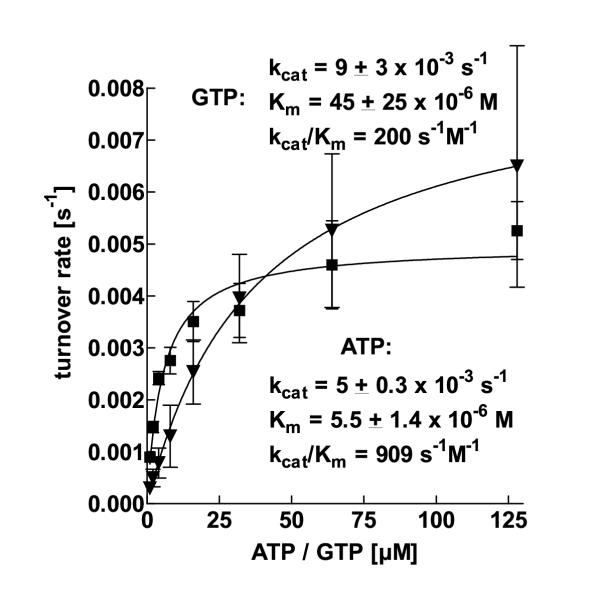
aaTHEP1 catalyzes both ATP and GTP hydrolysis. Measuring the turnover rates at different substrate concentrations under steady-state conditions resulted in hyperbolic curves if presented in a double linear plot i. e. no cooperativity could be observed (figure 7). For that reason, steady-state kinetics could be further analyzed by fitting the data points to curves obeying the Michaelis-Menten equation retrieving kcat- and Km-values for each substrate. Compared to the hydrolysis of ATP, the maximum turnover rate kcat for GTP is faster by a factor of 2. On the other hand, the enzyme's substrate affinity to ATP as represented by Km exceeds that to GTP by one order of magnitude. The catalytic efficiency of an enzyme is defined as kcat/Km. Thus, the catalytic efficiency of ATP hydrolysis as catalyzed by aaTHEP1 significantly exceeds that of GTP hydrolysis. The specific activities for ATP and GTP hydrolysis corresponding to the kcat-values given in figure 7 are 14.6 and 26.3 nmol min-1 mg-1, respectively.
Figure 7.
aaTHEP1 hydrolyzes ATP and GTP obeying the Michaelis-Menten-equation. Steady-state kinetics of ATP (squares) and GTP hydrolysis (triangles) as catalyzed by aaTHEP1 at 70°C. Each data point for ATP hydrolysis is the mean ± SD of 4, each data point for GTP-hydrolysis the mean ± SD of 3 independent measurements. Since the signal to noise ratio of the activity measurement dramatically increases at higher substrate concentrations, 128 μM was the highest concentration under investigation. 0.5 μg of purified aaTHEP1 was used for each assay in 25 μl buffer.
Inhibition of ATP-hydrolysis by other nucleotides
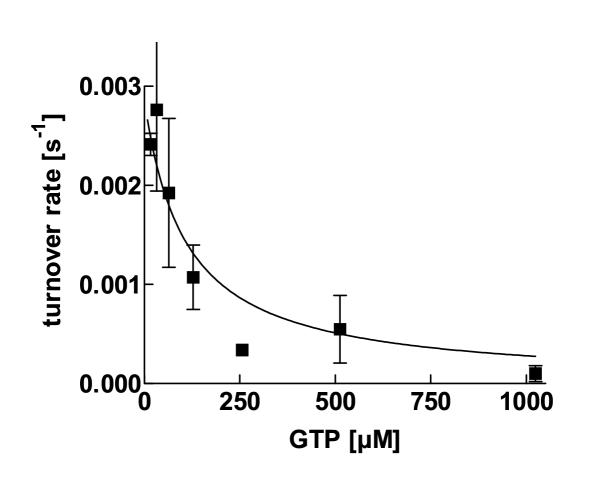
Competition experiments demonstrated that all nucleosidetriphosphates inhibit aaTHEP1 in hydrolyzing ATP. The data points fit to a model for competitive inhibition. As an example, inhibition by GTP is shown in figure 8. With the exception of GTP and dGTP, the Ki-values of all nucleotides are approximately in a similar range as the Km for ATP (table 3). Whereas GDP also inhibits ATP-hydrolysis, GMP, AMP, and UMP show no inhibition (figures 4 and 5).
Figure 8.
ATP hydrolysis is inhibited by GTP. Inhibition of aaTHEP1 catalyzed ATP hydrolysis by GTP. Each data point represents the mean ± SD of two independent measurements. 0.5 μg of purified aaTHEP1 were used for each assay in 25 μl buffer.
Table 3.
Inhibition constants of ATP hydrolysis by nucleoside-3-phosphates.
| GTP | CTP | UTP | dATP | dGTP | dCTP | dTTP | |
| Ki [μM] | 57 ± 20 | 6 ± 2 | 5 ± 2 | 3 ± 0.2 | 18 ± 2 | 9 ± 3 | 6 ± 2 |
The Ki-values of inhibition of ATP hydrolysis as determined at 5 μM ATP at 70°C are summarized. 0.5 μg of purified aaTHEP1 was used for each assay in 25 μl buffer.
Possible homologous non-covalent interactions
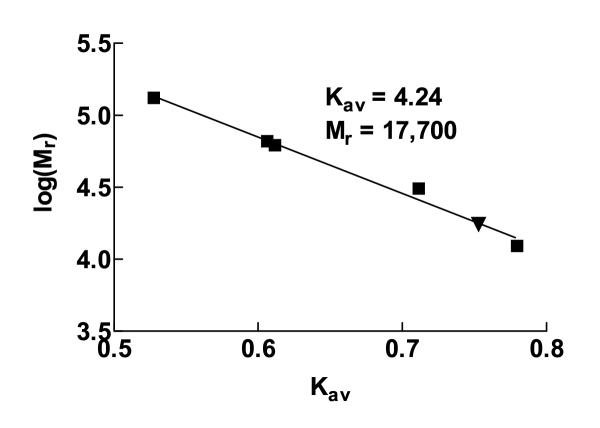
Gel filtration on Superose 6 prep grade was performed under native conditions to detect possible aaTHEP1 oligomers. We observed a single peak at Kav = 4.24 corresponding to a relative molecular weight of 17,700 indicating that purified aaTHEP1 exists as a highly compact folded monomer (figure 9). In addition, several attempts to crosslink aaTHEP1 by glutaraldehyde resulted in a non-crosslinked protein whereas under the same experimental conditions β-tryptophan synthase resulted in a crosslinked dimer (data not shown).
Figure 9.
Isolated aaTHEP1 appears a monomer. Gel filtration on calibrated Superose™ 6 prep grade. Native marker protein peaks are represented as squares and aaTHEP1 is shown as a triangle in a linear regression of Kav versus log(Mr).
Domain structure
A possible multidomain structure of aaTHEP1 was probed by limited proteolysis. As a control, the β-subunit of E. coli tryptophan synthase as a typical protein composed of distinct domains connected by a hinge region [10] was also proteolyzed. In contrast to β-tryptophan synthase, aaTHEP1 is resistant to proteolytic cleavage by both trypsin and endoproteinase Glu-C (figure 10). Compared to published experiments on the proteolytic cleavage of β-tryptophan synthase [10,11], the experimental conditions were chosen in a way that fragmented β-tryptophan synthase (~30 and ~10 kDa fragments) already were further degraded. Even under these conditions, aaTHEP1 remains stable although it contains 36 (20 × K + 16 × R) possible trypsin and 17 (17 × E) possible endoproteinase Glu-C cleavage sites as predicted from the sequence.
Figure 10.
aaTHEP1 consists of a single domain. SDS-PAGE after limited proteolysis by trypsin and endoproteinase Glu-C. aaTHEP1 is shown on lanes 1, 3 and 5. As a control, the β-subunit of tryptophan synthase from E. coli is run on lanes 2, 4, and 6. Lanes 1 and 2 show the native proteins without the addition of proteases. Proteolysis by trypsin is seen on lanes 3 and 4 whereas proteolysis by endoproteinase Glu-C is shown on lanes 5 and 6.
Secondary structure analysis
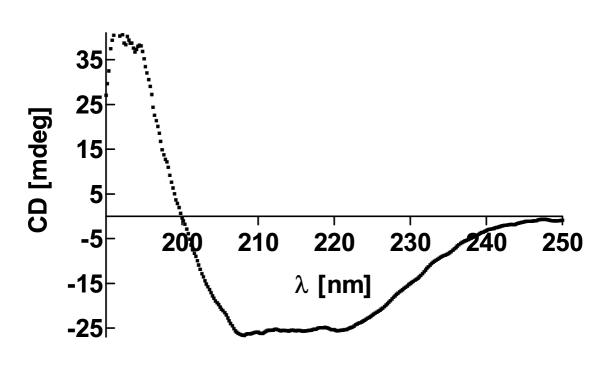
To determine the overall content of secondary structure elements of purified aaTHEP1, far UV circular dichroism spectroscopy was performed. A typical spectrum is given in figure 11. Analysis of the CD spectrum using the Jasco Spectrum Analyzer software revealed an alpha helix content of 34.3 % and 33.6 % beta-sheets.
Figure 11.
aaTHEP1 consists of 33.6% beta-sheets and 34.3% alpha-helix. Secondary structure analysis of purified aaThep1 by far UV circular dichroism spectroscopy.
Discussion
Our approach is based on the assumption that proteins abundant in thermophilic and rare in mesophilic genomes are the most attractive targets for further biochemical investigations to understand the physiology specific for thermophiles in more detail. Consequently, our first goal was to identify such candidates via bioinformatic methods and as the most suitable protein THEP1 was selected for this study. The high ranking thermophile-specificity of THEP1 among procaryotes may be explained by an essential physiological role in thermophiles that is of no functional relevance for almost all mesophilic microorganisms. As an alternative explanation, a function also present in mesophilic organisms could be carried out by a protein that was not able to adapt to higher temperatures and in the course of convergent evolution, THEP1 could have taken over this particular function. Methanosarcina acetivorans str. C2A is the only mesophilic organism containing THEP1 (MA3402). Since the genome of M. acetivorans reveals extensive metabolic and physiological diversity and there are thermophilic Methanosarcinae [12], one may take into consideration the possibility that M. acetivorans facultatively could be thermophilic. In addition to phylogenetic patterns search we also performed BLAST with the aaTHEP1 sequence. However, although no significant homologies to further sequences from mesophilic unicellular organisms could be detected, we discovered homologies to 6 multicellular eucaryotes. Consequently, THEP1s could belong to a class of proteins that are conserved in Archeae and Eukarya (with losses) and have been passed to thermophilic bacteria by lateral gene transfer.
The present data show that aaTHEP1 catalyzes ATP and GTP hydrolysis in vitro as predicted by Cort et al. [6]. In contrast, the annotated nucleotide kinase activity could not be confirmed experimentally.
As expected, the observed turnover rates are too low to represent a physiological in vivo situation where the free energy of ATP hydrolysis is coupled to energy consuming tasks. In vitro, similar turnover rates of purified NTPase are published in the literature: 1.1 × 10-2 sec-1 for PilT from A. aeolicus [13], 1.2 × 10-2 sec-1 for TadA for Actinobacillus actinomycetemcomitans [14], 3 × 10-3 sec-1 for TrwD from Escherichia coli [15], and 6.9 × 10-4 sec-1 for PilQ from Escherichia coli [16]. In vivo, additional proteins might be needed to activate aaTHEP1 by protein-protein interactions and take up the free energy released by NTP-hydrolysis e. g. for motion, active transport or another energy consuming cellular function. Alternatively, aaTHEP1 could catalyze an NTP driven thermodynamically unfavourable anabolic reaction of a yet undiscovered cosubstrate or play a role in cellular regulation.
ATP-hydrolysis could be inhibited by all other 7 nucleosidetriphosphates under investigation in a competitive manner which may be interpreted that in addition to GTP the other nucleotides are also substrates for isolated aaTHEP1. Possibly the enzyme lost a certain in vivo substrate specificity upon isolation. Since GDP inhibits ATP-hydrolysis whereas GMP, AMP and UMP do not, we propose that aaTHEP1 recognizes the β- and γ-phosphates rather than the nucleoside moieties.
A low activity of a recombinant protein may also be explained by trace amounts of E. coli enzymes still present after purification. For the measured NTPase activity of aaTHEP1 we exclude this possibility because we do not expect an E. coli enzyme exhibiting the same temperature dependence as shown in figure 6. There are even more indications that aaTHEP1 is a thermostable protein. The elution during gel filtration at a position corresponding to a lower relative molecular weight than the calculated 20,555 Da for a monomer and the resistance to limited proteolysis suggest that aaTHEP1 is a highly compact folded one-domain protein, a well known feature for thermophilic proteins. No cooperative behaviour in the kinetic experiments is also expected for a monomeric enzyme.
Although this study clearly defines a biochemical in vitro activity of aaTHEP1, there are many possible in vivo functions and bioinformatic analysis allows to make some predictions. Based on the genomic context, gene functions can be predicted by searching for the conservation of operons and gene orders because genes found in gene strings, particularly in multiple genomes, can be legitimately assumed to be functionally linked [17]. For THEP1, we indeed detected 4 genomes where the THEP1-gene immediately is followed on the same strand by a COG1867 protein (N2, N2-dimethylguanosine tRNA methyltransferase). Furthermore, COG1867 also belongs to the group of THEPs indicating a functional link (table 1).
Conclusions
This study experimentally confirms the hypothesis of Cort et al. who suggested THEP1s being a novel family of ATPases in both sequence and structural terms [6]. On the other hand, we refute the theoretical prediction that THEP1s are nucleotide kinases. In addition to the experimental work, a list of further THEPs as potential targets to study microbial thermophily is provided (tables 1 and 2).
Table 2.
Functional predictions of high-scoring COGs.
| COG name | Predicted function |
| COG1618 | Predicted nucleotide kinase |
| COG3635 | Predicted phosphoglycerate mutase, AP superfamily |
| COG1355 | Predicted dioxygenase |
| COG1371 | Uncharacterized conserved protein |
| COG2078 | Uncharacterized conserved protein |
| COG1980 | Archaeal fructose 1,6-bisphosphatase |
| COG1144 | Pyruvate:ferredoxin oxidoreductase and related 2-oxoacid:ferredoxin oxidoreductases, delta subunit |
| COG1820 | N-acetylglucosamine-6-phosphate deacetylase |
| COG1730 | Predicted prefoldin, molecular chaperone implicated in de novo protein folding |
| COG1503 | Peptide chain release factor 1 (eRF1) |
| COG1867 | N2, N2-dimethylguanosine tRNA methyltransferase |
| COG1581 | Archaeal DNA-binding protein |
| COG1888 | Uncharacterized protein conserved in archaea |
| COG1350 | Predicted alternative tryptophan synthase beta-subunit (paralog of TrpB) |
| COG1909 | Uncharacterized protein conserved in archaea |
| COG1110 | Reverse gyrase |
| COG1318 | Predicted transcriptional regulators |
| COG1630 | Uncharacterized protein conserved in archaea |
| COG2250 | Uncharacterized conserved protein related to C-terminal domain of eukaryotic chaperone, SACSIN |
Descriptions are taken from the current COG-database.
Methods
Cloning, expression and purification of aaTHEP1 from A. aeolicus
Genomic DNA from A. aeolicus was kindly provided by Dr. R. Huber, Regensburg, Germany. aq_1292 was amplified by PCR using the primers 5'-CACCATGAAAATCATCATAACCGGTGA-3' and 5'-TTACCGCTCAAGAAGTGAGAGAAT-3'. The PCR fragment was inserted into the pre-linearized plasmid pET101/D-TOPO (Invitrogen). For propagation and maintenance, the E. coli strain TOP10 (Invitrogen) was used. The correct sequence of the insert as well as its orientation were verified by sequence analysis using the reverse T7-primer 5'-TAGTTATTGCTCAGCGGTGG-3'.
To express aq_1292, E. coli BL21 Star™ (DE3) was used. According to the instructions of the manufacturer, freshly transformed cells were grown by transferring the entire transformation mixture to 10 ml Luria-Bertani broth (LB) medium containing 50 μg/ml carbenicillin and 1% glucose. After growing for 4 h at 37°C, the preculture was added to 40 ml of fresh medium, grown for additional 16 h and then 30 ml were used to inoculate a 2 l main culture. At A600 = 0.7, protein expression was induced by 1 mM isopropyl β-D-thiogalactopyranoside (IPTG, Roth). Cells were harvested at A600 = 1.3, yielding approximately 4 g after centrifugation at 6,000 × g for 10 min. The pellet was stored at -20°C before use.
Buffer A for protein purification was 50 mM Tris/HCl, 25 mM MgCl2, 5 mM KCl, 1 mM DL-dithiothreitol (DTT), 0.1 mM ethylenediaminetetraacetic acid (EDTA), pH 7.0. To disrupt the cell walls, 20 ml of buffer A containing 10 instead of 50 mM Tris, 5 mM sodium deoxycholate, 40,000 U/ml lysozyme, and 50 μg/ml DNaseI were added to 1 g of thawed cells. Lysis was performed by stirring the suspension at 20°C for 1 h and the cell debris were removed by centrifugation at 20,000 × g for 30 min. To precipitate the bulk of E. coli proteins, the supernatant was heated to 75°C for 10 min and immediately chilled on ice. Denatured proteins were removed by centrifugation at 20,000 × g and 4°C for 30 min. To remove nucleic acids and further purify recombinant aaTHEP1, the supernatant was loaded at 0.5 ml/min onto a 1 ml HiTrap™ SP HP cation exchange column (Amersham Biosciences AB) equilibrated with buffer A. The column was washed with buffer A containing 150 mM KCl and aaTHEP1 was eluted at 500 mM KCl. To the collected peak a 4-fold volume of buffer A containing 2.438 M (NH4)2SO4 (62.5 % saturation) instead of KCl was added. After centrifugation at 20,000 × g for 30 min, the resulting protein solution was applied to a 1 ml HiTrap™ (low sub) Phenyl Sepharose™ FF (Amersham Biosciences AB) equilibrated in buffer A containing 1.85 M (NH4)2SO4 (50 % saturation) at 0.5 ml/min. The column was washed at 40 % and eluted at 20 % (NH4)2SO4 saturation. Finally, (NH4)2SO4 was removed from aaTHEP1 by using a NAP™ 5 column (Amersham Biosciences AB) equilibrated in buffer A. The purified protein could be stored in buffer A for several weeks at -20°C without significant loss in activity. Protein concentrations were determined by densitometry [18] of SDS-Gels [19] using the Sigma bovine serum albumin protein micro standard. Gels were analyzed via an imaging densitometer (Bio-Rad GS-700) and the Molecular Analyst software.
Steady state kinetics
To measure ATP or GTP hydrolysis, the release of [γ-32P] from [γ-32P]ATP or [γ-32P]GTP was determined. Assays were performed using purified aaTHEP1 in 25 μl buffer A. Aliquots of the reaction mixture were stopped after different times of incubation by adding 25 μl of 40 % formic acid and the mixture was separated by thin layer chromatography on PEI-cellulose in 0.5 M potassium phosphate pH 3.9. Quantification was performed by scanning exposed and developed Hyperfilm™ – βmax films (Amersham Biosciences AB) using an imaging densitometer (Bio-Rad GS-700) and the Molecular Analyst software. Each catalytic activity was determined by the average of end point measurements divided by time of at least three different times of incubation under steady-state conditions. Blank values determined in the absence of aaTHEP1 were subtracted from each data point. Unless stated otherwise, the experiments were performed at 70°C in a Biometra T3 thermocycler. Competition experiments were measured at a constant concentration of 5 μM ATP. Nonlinear regressions to determine kcat- and Km-values were performed with the GraphPad Prism™ software using the Michaelis-Menten equation. Ki-values were determined by fitting the data points to
V = (Vmax × [ATP])/([ATP] + Km × (1 + [Inhibitor]/Ki)).
Nucleotide kinase activities were assayed at 5 and 1000 μM ATP by adding 5 as well as 1000 μM of GDP, UMP, AMP, and GMP respectively.
Gel filtration
To determine if purified aaTHEP1 is in mono- or oligomeric form, a 1 × 50 cm column filled with superose 6 prep grade (Amersham Biosciences AB) was calibrated with cytochrom C, carbonic anhydrase monomer and dimer [20] and bovine serum albumin monomer and dimer [21] as molecular weight standards. Proteins were run at 0.2 ml/min in buffer A containing 150 mM KCl. Approximately 80 μg of aaTHEP1 or marker protein in 0.5 ml were applied to the column. To dissociate possible cold-induced unspecific aggregates, aaTHEP1 was heated for 10 min at 75°C prior to gel filtration.
Limited proteolysis
In order to probe the domain structure of aaTHEP1, limited proteolysis by trypsin and endoproteinase Glu-C was performed. The reactions were carried out in buffer A containing 4 μg/ml protease and 40 μg/ml protein substrate. After incubation at 20°C for 1 h, samples were analyzed by SDS-PAGE. The β-subunit of tryptophan synthase from E. coli was prepared as described earlier [22].
Circular dichroism
Far UV circular dichroism spectra were recorded using a J-810 CD spectralpolarimeter (Jasco) at 30°C. 2.5 μg of purified aaTHEP1 was measured in 200 μl of buffer A in an 1 mm quartz cuvette. Helix- and beta-sheet contents were calculated using the Spectrum Analyzer software (Jasco).
Bioinformatics
COGs were analyzed by extended phylogenetic patterns search (EPPS) [23,24] using the march 5, 2003 release of the COG database. Multiple sequence alignments were performed at EMBL Outstation [25] using CLUSTALW 1.81 [26] and the data was visualized using BOXSHADE v3.21 [27]. BLAST searches were performed at NCBI [28].
Authors' contributions
CK carried out the molecular biology experiments, the purification of aaTHEP1, and the gel filtrations. MR performed the CD-measurements, RH performed the activity measurements and limited proteolysis experiments. MK carried out the bioinformatics, conceived of the study, and participated in its design and coordination. All authors read and approved the final manuscript.
Acknowledgments
Acknowledgements
We thank Andreas Savelsberg for his help in performing the kinetic measurements and Reiner Würdinger for assistance during the CD-measurements at Jasco GmbH Germany. We greatly appreciate discussions with Marina V. Rodnina on all aspects of the manuscript.
Contributor Information
Claudia Klinger, Email: cklinger@uni-wh.de.
Michael Roßbach, Email: miro@uni-wh.de.
Rebecca Howe, Email: rebeccah@uni-wh.de.
Michael Kaufmann, Email: mika@uni-wh.de.
References
- Tatusov RL, Koonin EV, Lipman DJ. A genomic perspective on protein families. Science. 1997;278:631–637. doi: 10.1126/science.278.5338.631. [DOI] [PubMed] [Google Scholar]
- Tatusov RL, Natale DA, Garkavtsev IV, Tatusova TA, Shankavaram UT, Rao BS, Kiryutin B, Galperin MY, Fedorova ND, Koonin EV. The COG database: new developments in phylogenetic classification of proteins from complete genomes. Nucleic Acids Res. 2001;29:22–28. doi: 10.1093/nar/29.1.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phylogenetic patterns search http://www.ncbi.nlm.nih.gov/COG/phylox.html
- Forterre P. A hot story from comparative genomics: reverse gyrase is the only hyperthermophile-specific protein. Trends Genet. 2002;18:236–237. doi: 10.1016/S0168-9525(02)02650-1. [DOI] [PubMed] [Google Scholar]
- Makarova KS, Wolf YI, Koonin EV. Potential genomic determinants of hyperthermophily. Trends Genet. 2003;19:172–176. doi: 10.1016/S0168-9525(03)00047-7. [DOI] [PubMed] [Google Scholar]
- Cort JR, Yee A, Edwards AM, Arrowsmith CH, Kennedy MA. Structure-based functional classification of hypothetical protein MTH538 from Methanobacterium thermoautotrophicum. J Mol Biol. 2000;302:189–203. doi: 10.1006/jmbi.2000.4052. [DOI] [PubMed] [Google Scholar]
- Deckert G, Warren PV, Gaasterland T, Young WG, Lenox AL, Graham DE, Overbeek R, Snead MA, Keller M, Aujay M, Huber R, Feldman RA, Short JM, Olsen GJ, Swanson RV. The complete genome of the hyperthermophilic bacterium Aquifex aeolicus. Nature. 1998;392:353–358. doi: 10.1038/32831. [DOI] [PubMed] [Google Scholar]
- Leipe DD, Wolf YI, Koonin EV, Aravind L. Classification and evolution of P-loop GTPases and related ATPases. J Mol Biol. 2002;317:41–72. doi: 10.1006/jmbi.2001.5378. [DOI] [PubMed] [Google Scholar]
- Ridder IS, Dijkstra BW. Identification of the Mg2+-binding site in the P-type ATPase and phosphatase members of the HAD (haloacid dehalogenase) superfamily by structural similarity to the response regulator protein CheY. Biochem J. 1999;339 ( Pt 2):223–226. doi: 10.1042/0264-6021:3390223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann M, Schwarz T, Jaenicke R, Schnackerz KD, Meyer HE, Bartholmes P. Limited proteolysis of the beta 2-dimer of tryptophan synthase yields an enzymatically active derivative that binds alpha-subunits. Biochemistry. 1991;30:4173–4179. doi: 10.1021/bi00231a010. [DOI] [PubMed] [Google Scholar]
- Hogberg-Raibaud A, Goldberg ME. Preparation and characterization of a modified form of beta2 subunit of Escherichia coli tryptophan synthetase suitable for investigating protein folding. Proc Natl Acad Sci U S A. 1977;74:442–446. doi: 10.1073/pnas.74.2.442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galagan JE, Nusbaum C, Roy A, Endrizzi MG, Macdonald P, FitzHugh W, Calvo S, Engels R, Smirnov S, Atnoor D, Brown A, Allen N, Naylor J, Stange-Thomann N, DeArellano K, Johnson R, Linton L, McEwan P, McKernan K, Talamas J, Tirrell A, Ye W, Zimmer A, Barber RD, Cann I, Graham DE, Grahame DA, Guss AM, Hedderich R, Ingram-Smith C, Kuettner HC, Krzycki JA, Leigh JA, Li W, Liu J, Mukhopadhyay B, Reeve JN, Smith K, Springer TA, Umayam LA, White O, White RH, Conway de Macario E, Ferry JG, Jarrell KF, Jing H, Macario AJ, Paulsen I, Pritchett M, Sowers KR, Swanson RV, Zinder SH, Lander E, Metcalf WW, Birren B. The genome of M. acetivorans reveals extensive metabolic and physiological diversity. Genome Res. 2002;12:532–542. doi: 10.1101/gr.223902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herdendorf TJ, McCaslin DR, Forest KT. Aquifex aeolicus PilT, homologue of a surface motility protein, is a thermostable oligomeric NTPase. J Bacteriol. 2002;184:6465–6471. doi: 10.1128/JB.184.23.6465-6471.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharjee MK, Kachlany SC, Fine DH, Figurski DH. Nonspecific adherence and fibril biogenesis by Actinobacillus actinomycetemcomitans: TadA protein is an ATPase. J Bacteriol. 2001;183:5927–5936. doi: 10.1128/JB.183.20.5927-5936.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivas S, Bolland S, Cabezon E, Goni FM, de la Cruz F. TrwD, a protein encoded by the IncW plasmid R388, displays an ATP hydrolase activity essential for bacterial conjugation. J Biol Chem. 1997;272:25583–25590. doi: 10.1074/jbc.272.41.25583. [DOI] [PubMed] [Google Scholar]
- Sakai D, Horiuchi T, Komano T. Atpase activity and multimer formation of Pilq protein are required for thin pilus biogenesis in plasmid R64. J Biol Chem. 2001;276:17968–17975. doi: 10.1074/jbc.M010652200. [DOI] [PubMed] [Google Scholar]
- Wolf YI, Rogozin IB, Kondrashov AS, Koonin EV. Genome alignment, evolution of prokaryotic genome organization, and prediction of gene function using genomic context. Genome Res. 2001;11:356–372. doi: 10.1101/gr.GR-1619R. [DOI] [PubMed] [Google Scholar]
- Wadsworth C. A simple stained protein assay (SPA) for rapid total protein determination applied to spinal fluids. Scand J Immunol. 1976;5:1209–1213. doi: 10.1111/j.1365-3083.1976.tb00264.x. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Cleland JL, Wang DI. Transient association of the first intermediate during the refolding of bovine carbonic anhydrase B. Biotechnol Prog. 1992;8:97–103. doi: 10.1021/bp00014a002. [DOI] [PubMed] [Google Scholar]
- Levi V, Gonzalez Flecha FL. Reversible fast-dimerization of bovine serum albumin detected by fluorescence resonance energy transfer. Biochim Biophys Acta. 2002;1599:141–148. doi: 10.1016/S1570-9639(02)00414-4. [DOI] [PubMed] [Google Scholar]
- Bartholmes P, Kirschner K, Gschwind HP. Cooperative and noncooperative binding of pyridoxal 5'-phosphate to tryptophan synthase from Escherichia coli. Biochemistry. 1976;15:4712–4717. doi: 10.1021/bi00666a027. [DOI] [PubMed] [Google Scholar]
- Reichard K, Kaufmann M. EPPS: mining the COG database by an extended phylogenetic patterns search. Bioinformatics. 2003;19:784–785. doi: 10.1093/bioinformatics/btg089. [DOI] [PubMed] [Google Scholar]
- Universität Witten/Herdecke: EPPS http://www.uni-wh.de/epps
- ClustalW http://www.ebi.ac.uk/clustalw/
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BoxShade Server http://www.ch.embnet.org/software/BOX_form.html
- NCBI BLAST Home Page http://www.ncbi.nlm.nih.gov/BLAST/