Abstract
Little is known about how members of the indigenous microflora interact with their mammalian hosts to establish mutually beneficial relationships. We have used a gnotobiotic mouse model to show that Bacteroides thetaiotaomicron, a component of the intestinal microflora of mice and humans, uses a repressor, FucR, as a molecular sensor of l-fucose availability. FucR coordinates expression of an operon encoding enzymes in the l-fucose metabolic pathway with expression of another locus that regulates production of fucosylated glycans in intestinal enterocytes. Genetic and biochemical studies indicate that FucR does this by using fucose as an inducer at one locus and as a corepressor at the other locus. Coordinating this commensal’s immediate nutritional requirements with production of a host-derived energy source is consistent with its need to enter and persist within a competitive ecosystem.
Keywords: host–microbial cross-talk, germ-free mice, Bacteroides thetaiotaomicron, transcriptional repressors, epithelial fucosylated glycan synthesis
Humans must adapt to life in a microbial world. As adults, the number of microbes associated with our mucosal surfaces exceeds our total number of somatic and germ cells by more than an order of magnitude (1). The gastrointestinal tract is home to our most complex and populous society of microbes. The composition of the microflora varies along the length of the gut and during the life of the host. The microflora provides a functional barrier to colonization by pathogens (2, 3), plays an important role in normal nutrition and metabolism, and is thought to help shape development of the intestine’s mucosal immune system (4). Despite its importance, almost nothing is known about the molecular mechanisms that allow components of the microflora to interact with their hosts so as to establish relationships that are advantageous to both. Understanding such relationships is important in considering the origins of opportunistic infections, various immunopathologic states, and the propagation of antibiotic-resistant organisms (2–8).
Assembly of the gut microflora commences at birth. When space and nutrients are not limiting, commensals with high division rates predominate. As the population increases and nutrients are depleted, niches become occupied with more specialized species (4). One conceptualization of how this process may be orchestrated is that the distribution of early-colonizing gut commensals is defined by a preformed nutrient foundation that has been laid down by the host. The ability of other commensals to enter occupied habitats would depend on their ability to utilize these nutrient substrates more efficiently and/or to engineer alterations in the nutrient reservoir to better suit their own metabolic capacities. In a mutualistic relationship, coordination of microbial nutrient utilization and host nutrient production should be achieved at a minimal energetic cost to microbe and host and not be deleterious to either.
To test this notion at a cellular and molecular level, we created a simplified model intestinal ecosystem by using germ-free mice that were colonized with a genetically manipulable species that normally resides in the distal intestine of mice and humans. We reported previously (9) that the microflora orchestrates a developmentally regulated program of fucosylated glycan production in the distal small intestinal (ileal) epithelium of mice belonging to the NMRI strain. Conventionally raised mice initiate production of Fucα1,2Gal-containing glycans in the principal epithelial lineage (enterocytes) during the weaning period. Expression of these glycans begins in enterocytes overlying scattered ileal villi and can be detected with the Fucα1,2Galβ-specific lectin, Ulex europaeus type I agglutinin (UEA-I). Induction commences on postnatal day 17 (P17) and generalizes by P25 to involve all enterocytes overlying all ileal villi. This induction represents a replacement of UEA-I-negative enterocytes with new UEA-I-positive enterocytes and is accompanied by transcriptional activation of a host α1,2-fucosyltransferase gene. In the presence of a microflora, fucosylated glycoconjugate expression is sustained in rapidly renewing enterocytes throughout adulthood (9).
Germ-free mice are also able to initiate production of Fucα1,2Gal-glycans, beginning at the same time as conventionally raised mice. However, in the absence of a microflora, production ceases by the end of weaning (P28). Nonetheless, the capacity of ileal enterocytes to synthesize Fucα1,2Gal-glycans is maintained in adult germ-free animals because introduction of a conventional flora at any point after P28 reinitiates production (9).
Bacteroides thetaiotaomicron is an abundant member of the normal mouse and human distal small intestinal microflora (10, 11). It can scavenge a wide variety of host cell glycans in vitro and in vivo (12). Colonization of adult germ-free NMRI mice with B. thetaiotaomicron reproduces the cellular, spatial, and kinetic features of fucosylated glycan induction orchestrated by a complete microflora (9). Induction depends on the density of colonizing organisms and does not involve direct bacterial binding to the intestinal epithelium. Fu-4 is a B. thetaiotaomicron strain, generated by random transposon (Tn4351)-mediated mutagenesis, that is unable to grow in media containing l-fucose as the sole carbon source (12). The Fu-4 strain is also unable to signal host production of fucosylated glycans, even at high colonization densities (9). This report provides a molecular explanation for the connection between fucose utilization by B. thetaiotaomicron and its ability to instruct the host to produce hydrolyzable fucosylated glycans.
MATERIALS AND METHODS
Isolation of the B. thetaiotaomicron Fucose Utilization Gene Cluster.
Tn4351 contains a unique HindIII site 3.5 kilobases (kb) from its 3′ end, allowing the intact tetracycline-resistance gene (tetX) to be recovered within a single HindIII fragment that encompasses flanking chromosomal DNA (13). Southern blot analysis of Fu-4 DNA indicated that tetX was contained in a single 4.44-kb HindIII fragment. HindIII-digested Fu-4 DNA was size-fractionated, and the fragments were subcloned into pBluescript II KS (Stratagene). A plasmid (pBS-tetR), recovered from a tetracycline-resistant Escherichia coli transformant, contained a 4.44-kb insert that included 962 bp of genomic DNA flanking the 3′ end of the transposon.
To isolate the corresponding locus from the isogenic wild-type B. thetaiotaomicron strain VPI-5482 (Anaerobe Laboratory, Virginia Polytechnic Institute and State University, Blacksburg, VA), 325 bp of the cloned genomic sequence (probe A) was amplified from pBS-tetR by using PCR (primers: 5′-GATATTATGTAACCAGTCAGGT-3′ and 5′-TCATCAAAATCCTGACAGATAT-3′). The product hybridized to a 4-kb ClaI–SalI fragment generated from VPI-5482 DNA. Therefore, ClaI–SalIdigested genomic DNA was size-selected and subcloned into pBluescript II KS. Colony hybridization with probe A yielded pBS-bt1 with a 4-kb insert and pBS-bt4 with an overlapping 4.6-kb ScaI fragment containing the upstream portion of a fucose-utilization gene cluster. The downstream portion of the cluster was obtained by PCR amplification of a 402-bp fragment from the 3′ end of the pBS-bt1 insert (primers: 5′-TCGACAGGACTAATCCTGTTTC-3′ and 5′-TCTGCTATCGGGATGCTCGAAC-3′). This PCR product (probe B) was used to identify and subclone a 6.5-kb HindIII genomic fragment into pBluescript II KS (pBS-bt3).
Generation of Isogenic Mutant B. thetaiotaomicron Strains.
The suicide vector pGERM was produced by A. Salyers (Univ. of Illinois, Champaign-Urbana) by first blunt-ending a NcoI–PstI fragment carrying the erythromycin-resistance gene, ermG (14) and ligating it into the SspI site of pUC19 [pUC19(ermG)]. A HaeII fragment carrying oriT(RK2) (15) was then blunt-ended and ligated into the SapI site of pUC19(ermG) to yield pGERM.
An internal fragment from each ORF of the B. thetaiotaomicron fucose-utilization gene cluster was amplified by PCR using primers containing EcoRI sites at their 5′ ends and a single-bp mutation to produce an in-frame stop codon. The amplified fragments were cloned into the EcoRI site of pGERM. The resulting constructs were used to transform E. coli S17-1, which contains a chromosomal copy of the Tra+ plasmid RP4 (16). Transformants were used as donors in matings with B. thetaiotaomicron strain VPI-5482 (17). After plating on trypticase-yeast extract-glucose (TYG) agar containing erythromycin (10 μg/ml), transconjugants were assessed for their ability to grow on defined medium (18) containing 0.5% l-fucose or l-rhamnose as the sole carbon source. Genomic DNAs prepared from transconjugants were analyzed by Southern blotting for insertion of pGERM at the appropriate chromosomal locations.
Strains containing an E. coli β-glucuronidase (GUS) transcriptional reporter were constructed as follows. A 1.8-kb XbaI–HindIII fragment containing GUS was released by digestion from pGJL1 (kindly provided by A. Salyers). The fragment was then ligated to the XbaI–HindIII site of a suicide vector (pBT-1; ref. 19), yielding pBT1-GUS. The 422-bp sequence located directly upstream of the fucR start codon was inserted 5′ to GUS, yielding pBT1-FuP/GUS. Transconjugations were performed as above, and transconjugants were selected on TYG agar containing 3 μg/ml tetracycline (and 10 μg/ml erythromycin for the fucR∷pGERM recipient).
Enzymatic Assays.
Wild-type and mutant B. thetaiotaomicron strains were grown to mid-logarithmic phase in defined medium containing 0.5% l-rhamnose with or without 0.5% l-fucose. Cells were disrupted by sonication. l-fucose isomerase activity was measured in cleared bacterial lysates by assaying the rate of formation of l-fuculose with the cysteine-carbazole method (20). Permease activity was quantitated by monitoring the uptake of l-[6-3H]fucose by intact logarithmic-phase cells (12). α-l-Fucosidase activity in culture supernatants was assayed at pH 7 in 0.1 M sodium phosphate, by using 1 mM 4-methylumbelliferyl-α-l-fucopyranoside as the substrate (21).
ORFs from the B. thetaiotaomicron fucose-utilization operon were amplified by PCR, subcloned into pET3a (Stratagene), sequenced, and the plasmids were introduced into E. coli strain BL21 (DE3) (Stratagene). Isomerase, kinase, and aldolase activities were assayed in cleared lysates prepared from bacteria that had been grown to mid-logarithmic phase in defined medium (18) and treated with isopropyl-β-d-thiogalactoside (IPTG) for 3 h. l-Fuculose kinase activity was determined from the rate of ADP formation in a coupled assay by using β-NADH, pyruvate kinase, and lactate dehydrogenase (22). l-Fuculose-1-phosphate aldolase activity was assayed by measuring dihydroxyacetone phosphate formation in a coupled reaction with purified α-glycerophosphate dehydrogenase and β-NADH (23). All assays were done in the linear range of product formation.
Purification and Characterization of FucR.
E. coli strain BL21(DE3) containing pET3a-fucR was grown to mid-logarithmic phase, and FucR production was induced with IPTG. The protein was then purified to apparent homogeneity by using a protocol described for E. coli TreR (24). Antibodies to purified FucR were raised in rabbits by Cocalico Biologicals (Reamstown, PA).
Interactions between FucR and various monosaccharides were characterized by ligand-dependent quenching of the intrinsic fluorescence spectrum by using a PTI 01–610 fluorometer (Photon Technology International, Princeton). All assays were performed at 22°C in 2.5-ml reactions containing buffer A (100 mM KCl/10 mM MgCl2/50 mM Tris⋅HCl, pH 7.5), 1.1 μM purified FucR, and varying amounts of saccharide. Samples were excited at 280 nm, and emission was monitored at 340 nm.
The state of FucR oligomerization was determined by dynamic light scattering (DynaPro-801, Protein Solutions, Charlottesville, VA) and by sedimentation equilibrium analysis (Beckman XL-A Optima analytical ultracentrifuge). Data obtained from the sedimentation studies ([FucR] = 10 μM) were analyzed by using the program sednterp (25).
Interactions between FucR and its target DNA sequences were assessed initially by gel mobility-shift assays performed in 15-μl reactions containing buffer A, a 32P-labeled restriction fragment or double stranded oligonucleotide, purified FucR (0.18–180 nM), a potential monosaccharide ligand (0.1–1000 μM), 0.3 mg/ml BSA, 0.13 mg/ml poly(dI,dC), and 10% glycerol. After incubation at 30°C for 1 h, the labeled species were separated by electrophoresis through 4% polyacrylamide gels. Subsequent DNase I footprint analyses were carried out according to ref. 26.
Colonization of Germ-Free NMRI Mice.
Germ-free NMRI/KI mice (9) were housed in gnotobiotic isolators under a strict 12-h light cycle and fed an autoclaved chow diet (Lactamin, Vadstena, Sweden) ad libitum. Males (30–90 days old) were inoculated with wild-type or mutant B. thetaiotaomicron strains by spreading organisms from stationary-phase cultures on their fur. Animals were sacrificed 10 days later, and the number of colony-forming units (cfu) per ml of ileal contents was determined by serial dilution plating. All isogenic strains were tested in parallel with their wild-type parent (n = 4–5 animals per group per experiment; 2–3 experiments per mutant). Intestines from mice colonized with >107 cfu/ml ileal contents at the time of sacrifice were scored for Fucα1,2Gal-glycans by staining whole-mount preparations with peroxidase-conjugated UEA-I (9).
RESULTS
Tn4351 Disrupts the l-Fucose Isomerase Gene (fucI), Which Is Part of a Fucose-Utilization Gene Cluster.
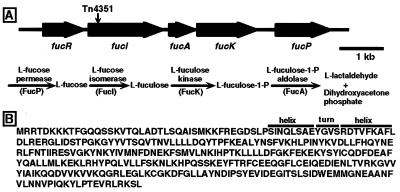
We began our analysis of the connection between fucose utilization by B. thetaiotaomicron and its ability to instruct the host to produce hydrolyzable fucosylated glycans by determining that Tn4351 had been inserted 445 bp downstream of the start codon of an ORF. This ORF encodes a 591-aa protein having 63% identity with E. coli l-fucose isomerase (27). The genomic sequence adjacent to the site of transposon insertion was used subsequently to isolate, from the wild-type strain VPI-5482 (9), a gene cluster containing five closely spaced ORFs. Three of the ORFs have homology to E. coli (and other bacterial) genes that encode the key enzymes in fucose metabolism: l-fucose isomerase, l-fuculose kinase, and l-fuculose-1-phosphate aldolase (Fig. 1A). The identities of the B. thetaiotaomicron genes were confirmed by overexpressing their protein products in E. coli. The genes were named fucI, fucA, and fucK based on the nomenclature used in E. coli (27). A fourth ORF situated immediately upstream of fucI encodes a 331-aa protein whose N-terminal sequence is similar to the helix–turn–helix motif of members of the gluconate repressor (GntR) family of bacterial regulatory proteins (ref. 28; Fig. 1B). This gene was named fucR. B. thetaiotaomicron FucR is not homologous to E. coli FucR, the transcriptional activator of E. coli’s fucose-utilization pathway genes (29). A fifth ORF begins 418 bp downstream from the stop codon of fucK and specifies a protein with homology to E. coli l-fucose permease (FucP) (Fig. 1A).
Figure 1.
B. thetaiotaomicron l-fucose-utilization gene cluster. (A) ORFs and the site of Tn4351 insertion. The bacterial l-fucose metabolic pathway is shown. The product of an ORF terminating 177 bp upstream of fucR shows homology to the 30S ribosomal protein S16 from several species. Another ORF beginning 187 bp downstream of fucP shares homology with several RNA polymerase σ factors. The GenBank accession number for this 10671-bp locus is AF137263. (B) Amino acid sequence of FucR. The underlined region encompasses a helix–turn–helix DNA binding region conserved in GntR family members.
Each of the five ORFs was disrupted by single-crossover homologous recombination of a suicide vector (pGERM) with strong transcription terminators. Integration of pGERM will have a polar effect on transcription of any downstream genes contained within an operon. Insertion of pGERM into fucR, fucI, fucA, fucK, or fucP abolished the ability of B. thetaiotaomicron to grow on defined medium containing l-fucose as the sole carbon source, but had no effect on the ability of the organism to grow on the related methyl pentose, l-rhamnose (data not shown).
FucR Functions as a Repressor of the fucRIAK Operon.
To examine how expression of this gene cluster is regulated, a DNA fragment encompassing the 422 bp 5′ to the fucR start codon was linked to E. coli GUS, and the construct was integrated, by single-crossover homologous recombination, just upstream of the gene cluster in isogenic wild-type and fucR∷pGERM strains. Integration resulted in duplication of the 422-bp sequence (Fig. 2A) and did not affect growth of the strain with a wild-type background on medium containing fucose as the sole carbon source.
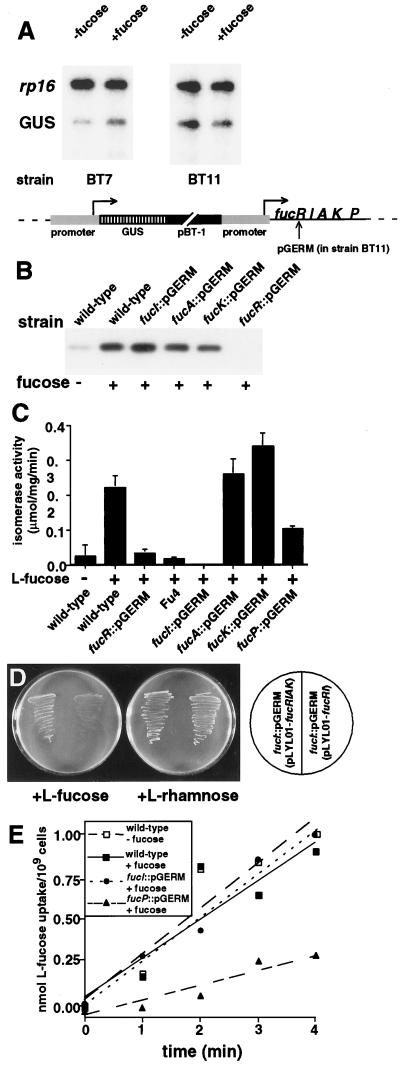
Figure 2.
FucR functions as a repressor of the fucRIAK operon. (A) Transcription reporter assay. Strains BT7 (wild-type background) and BT11(fucR∷pGERM background) contain an integrated chromosomal copy of GUS fused to 422 bp of sequence that is normally located directly upstream of the fucR start codon. Note that integration produces a duplication of the 422-bp element, with one copy linked to GUS and the other copy remaining at its endogenous location. GUS mRNA was detected by RNase protection, and signals were compared with those obtained by using a probe to 30S ribosomal protein S16 (rp16) mRNA. The results are representative of three independent experiments. (B) FucR expression. Strains were grown to mid-logarithmic phase on defined medium, with or without l-fucose. Soluble cellular proteins (10 μg per lane) were separated by SDS/PAGE. Western blots were prepared and probed with a rabbit antiserum generated against purified FucR. (C) Assays of cellular l-fucose isomerase activity from mid-logarithmic phase cultures of isogenic strains. (D) Evidence that fucA and fucK are expressed cotranscriptionally with fucRI. The shuttle vector pLYL01 containing fucRIAK rescues the ability of the fucI∷pGERM strain to grow on defined medium containing l-fucose, whereas fucRI does not. (E) l-Fucose permease activity in cells recovered from mid-logarithmic phase cultures of wild-type and mutant strains.
RNase protection assays indicated that GUS mRNA levels were higher when wild-type cells were grown in the presence of l-fucose than when grown in its absence. In contrast, GUS transcription in cells lacking FucR was equivalent in the presence or absence of fucose (Fig. 2A). The constitutive pattern of GUS expression in the strain lacking FucR demonstrates that FucR is a repressor that regulates transcription from a fucose-inducible promoter located upstream of its own ORF.
Western blot analysis confirmed that FucR expression in wild-type B. thetaiotaomicron is induced by l-fucose (Fig. 2B). Enzymatic assays demonstrated that FucI is also inducible by fucose and that disruption of fucI by pGERM reduces isomerase activity to below the limits of detection. A similar reduction was observed in the Fu-4 strain (Fig. 2C). Disruption of fucR by pGERM abolishes expression of FucR and markedly reduces isomerase activity (Fig. 2 B and C).
A genetic experiment established that fucRIAK forms an operon. The block in fucose utilization produced by disrupting fucR or fucI was not complemented by a shuttle vector containing a genomic fragment encompassing the fucose-responsive promoter plus fucRI but was overcome with a fragment encompassing the promoter plus fucRIAK (Fig. 2D).
Disruption of fucP results in a reduced rate of [3H]l-fucose uptake relative to the wild-type strain (Fig. 2E). [3H]l-Fucose uptake in the fucI∷pGERM strain is equivalent to that in the wild-type strain (Fig. 2E), indicating that fucP is regulated by a promoter that is distinct from PRIAK.
Fucose Binds to FucR and Blocks Its Ability to Bind Target DNA Sequences.
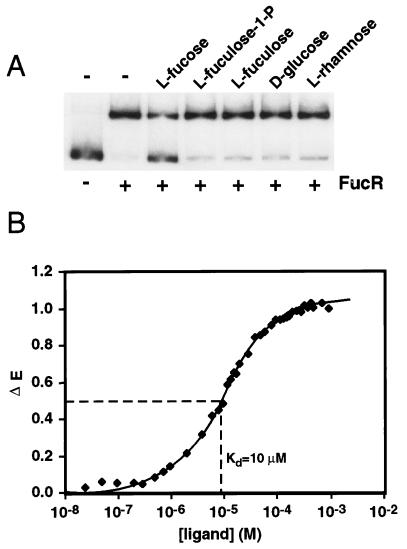
FucR was produced in E. coli and purified to apparent homogeneity. FucR binds to a DNA fragment encompassing the 283-bp sequence located upstream of the fucR initiator Met codon. Binding is inhibited by l-fucose in a dose-dependent fashion, but is not affected by l-fuculose, l-fuculose-1-phosphate, l-rhamnose, or d-glucose (Fig. 3A). DNA footprint analysis allowed us to identify the site of fucose-responsive FucR binding as 5′-TGCTGAGCTATGCTGTTGC-3′.
Figure 3.
l-Fucose binds to FucR and causes it to dissociate from the fucose pathway promoter. (A) Gel mobility-shift assay. A 32P-labeled DNA fragment, corresponding to the 283 bp of sequence 5′ to the initiator Met codon of fucR, was incubated with 30 ng of purified FucR and 1 mM of l-fucose, l-fuculose, l-fuculose-1-phosphate, d-glucose, or l-rhamnose. (B) Determination of l-fucose binding affinity by ligand-dependent quenching of FucR’s intrinsic fluorescence spectrum (see Materials and Methods).
Fluorescence spectroscopy revealed that FucR binds l-fucose with a Kd of 10 μM (Fig. 3B). Consistent with the results of the DNA gel-shift experiments, l-fuculose and l-fuculose-1-phosphate do not cause a significant change in the intrinsic fluorescence spectrum of FucR, even at 1 mM. Sedimentation equilibrium studies disclosed that the protein exists in solution as a dimer (Kd = 0.6 μM) with or without bound l-fucose (data not shown).
We concluded that in B. thetaiotaomicron, l-fucose induces expression of the fucose pathway enzymes (and FucR) by binding to the FucR repressor and reducing its interaction with a fucose-inducible promoter (PRIAK). Thus, expression of FucR is autoregulated. The organization and regulation of the B. thetaiotaomicron fucose-utilization gene cluster differs markedly from that of E. coli, where two divergently transcribed operons are regulated by an activator (29) and where l-fuculose-1-phosphate, rather than l-fucose, functions as the inducer (30).
FucR Regulates Another Genetic Locus Required for Microbial Signaling to the Host.
To understand how regulation of fucRIAK is related to generation of a microbial signal that induces host fucosylated glycan synthesis, we compared glycan production in germ-free NMRI mice that had been colonized with the various isogenic strains (n = 90 animals). Mice were sacrificed 10 days after strain inoculation, and Fucα1,2Gal-glycan production in the intestinal epithelium was defined by staining with UEA-I. Previous experiments (9) had demonstrated that at least 107 cfu of the wild-type strain per ml of ileal contents was necessary to induce full expression of host Fucα1,2Gal-glycans. Therefore, we only scored mice that had achieved that colonization density.
As expected, no UEA-I staining was seen in the small intestines of germ-free controls (n = 5; Fig. 4A). Six of seven mice with >107 cfu of wild-type B. thetaiotaomicron per ml of ileal contents exhibited uniform UEA-I staining of their ileal villus enterocytes. There was also uniform staining in four of seven mice colonized with fucA∷pGERM and in five of five mice harboring fucK∷pGERM. In contrast, all mice containing Fu-4 (n = 4) or fucI∷pGERM (n = 7) had no, or rare, UEA-I-positive villi (Fig. 4A). Retention of gene disruptions in bacteria recovered from ileal contents was confirmed by plating on defined medium containing fucose as the sole carbon source and by Southern blot analysis of chromosomal DNA.
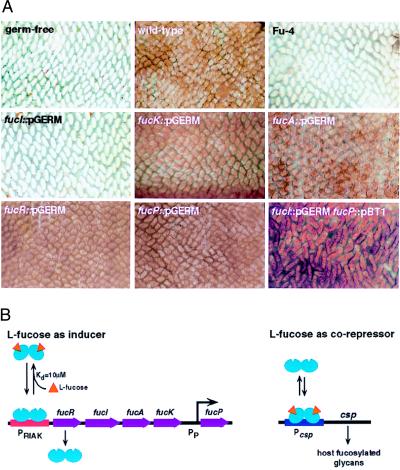
Figure 4.
Regulation of Fucα1,2Gal-containing glycan production in the ileum of germ-free NMRI mice. (A) Representative whole-mount preparations of ileum obtained 10 days after inoculation of the indicated strain and stained with peroxidase-conjugated UEA-I (brown). Villi appear as finger-like projections. (B). Model for the coordinate regulation of bacterial fucose metabolism with host fucosylated glycan production. l-fucose acts through FucR both as the inducer of transcription of fucRIAK and as a corepressor of transcription at csp, which is directly or indirectly responsible for production of a bacterial signal that results in increased production of host fucosylated glycoconjugates.
These results indicated that l-fucose isomerase plays a key role in determining whether this organism is able to signal its host to synthesize fucosylated glycans. The product of the isomerase, l-fuculose, could play a role in signal generation either by (i) interacting directly with the intestinal epithelium, (ii) becoming a substrate for another bacterial metabolic pathway that converts it to a signaling molecule, or (iii) acting as the inducer of a pathway involved in signal production through interactions with a transcriptional regulator encoded by another locus. To determine whether any of these possibilities applies, we tested the ability of the fucR∷pGERM strain to signal up-regulation of Fucα1,2Gal-glycans. As noted above, pGERM disruption of fucR eliminates production of FucR, FucI, FucA, and FucK, but not FucP. Remarkably, seven of seven germ-free mice colonized by fucR∷pGERM at >107 cfu/ml showed uniform UEA-I staining of the ileal epithelium (Fig. 4A). This observation establishes that l-fuculose is not directly involved in bacterial signal production and eliminates i–iii. It also indicates that FucR mediates repression of at least one other genetic locus, in addition to fucRIAK, that either regulates or is directly involved in production of the signal that leads to synthesis of Fucα1,2Gal-glycans. We designated this second FucR-responsive locus control of signal production (csp) and postulated that when FucR is absent, repression at csp is relieved and signaling to the epithelium ensues.
Genetic Evidence That l-Fucose Functions as a Corepressor with FucR at csp.
Fig. 4B presents a model consistent with the experimental results. A key feature of the model is that l-fucose functions as an inducer of the fucRIAK operon by reducing FucR binding to PRIAK and as a corepressor at csp by enhancing FucR binding to Pcsp. An autoregulatory bacterial repressor that functions to suppress metabolism of its inducer at one locus and uses this inducer as a corepressor at another locus would be novel. This regulatory strategy would result in maintenance of intracellular B. thetaiotaomicron fucose concentrations at a particular “set point”, determined in large part by the affinity of FucR for fucose (10 μM) and by the affinity of the fucose-bound form of FucR for the csp promoter. If intracellular fucose levels rise above this set point, the proportion of FucR in the fucose-bound form increases, resulting in decreased binding of the repressor to PRIAK and an increased rate of fucose degradation. The other consequence is increased binding of FucR at the csp promoter (i.e., there is no need to signal the host for more fucose). Conversely, if fucose levels fall below the setpoint, the proportion of unbound FucR will increase, resulting in increased binding to the fucose pathway promoter (reflecting a decreased need to metabolize fucose) and decreased binding at the csp promoter (a “request” to the host for more fucose). Because transcription of fucP is independent of fucRIAK transcription, disruption of fucI has no effect on the rate of l-fucose uptake (see Fig. 2E). According to the model, disruption of fucI results in a strain in which imported l-fucose cannot be metabolized, accumulates within the cell, and is available to interact with FucR to repress csp. The presence of the isomerase in strains that lack fucA and/or fucK would still allow conversion of l-fucose to l-fuculose, thereby preventing l-fucose from accumulating and decreasing repression of csp by FucR.
Because our model predicts that low intracellular fucose levels will stimulate signal production, we used the fucP∷pGERM strain to examine the effects of reducing fucose import on signaling. Three of four mice colonized with >107 cfu/ml of fucP∷pGERM exhibited Fucα1,2Gal-glycan production throughout their distal small intestine (Fig. 4A), supporting the model. A further prediction from the model is that abolishing fucose uptake in the fucI∷pGERM strain will convert it from a nonsignaling to a signaling strain. This prediction was confirmed when four of four mice colonized with >107 cfu/ml of a fucP∷pBT-1 fucI∷pGERM double mutant were found to have uniform UEA-1 staining of their ileal epithelium (Fig. 4A).
Evidence that l-Fucose Is an Important Carbon Source for B. thetaiotaomicron in the Distal Small Intestine Under Competitive, Nutrient-Limiting Conditions.
A final in vivo experiment indicated that the ability to utilize fucose confers a competitive advantage under nutrient-limiting conditions. Germ-free mice (n = 10; two independent experiments) were inoculated with fucI∷pGERM. This strain was selected because it lacks all three fucose-metabolizing enzymes. Ten days later, the animals were subjected to a 24-h fast to simulate a competitive ecosystem in which dietary carbohydrates are scarce. Control mice, colonized with the same strain, were given free access to chow during this 24-h period. Insertion of pGERM (containing an erythromycin-resistance gene) by single crossover into fucI not only results in loss of FucI, FucK, and FucA, but causes partial duplication of the targeted locus. In vitro cultivation of fucI∷pGERM cells for 40 generations in the absence of antibiotic disclosed a constant frequency of spontaneous vector removal (10−7) that did not confer a simple growth advantage. In vivo, we found that there was a statistically significant (P < 0.05) 3-fold increase in the number of fucose-utilizing/erythromycin-sensitive organisms after food deprivation. Southern blotting confirmed that the capacity to utilize fucose and erythromycin sensitivity were linked to loss of pGERM. These observations support the notion that fucose utilization is advantageous to this organism when it is subjected to nutrient deprivation in the gut. However, we cannot formally exclude the less likely possibility that fasting, per se, somehow leads to an enhanced frequency of reversion of a pGERM insertion.
Fucose is almost always a terminal, α-linked sugar in mammalian glycoconjugates. Therefore, a secreted bacterial α-fucosidase should be sufficient to obtain monosaccharidic fucose for import and metabolic processing (31, 32). Additional exo- or endoglycosidases are not needed, thereby minimizing the investment required for the microbe to obtain fucose as an energy source. Consistent with this notion, we found that wild-type, fucR∷pGERM, and fucI∷pGERM strains all secrete α-fucosidase activity when grown in defined medium, in either the presence or absence of fucose (data not shown).
DISCUSSION
By colonizing germ-free mice with B. thetaiotaomicron, we have been able to generate a simplified model ecosystem that allows molecular definition of the interactions between a resident bacterial species and the intestinal epithelium. In this partnership, the host appears to be an active participant in providing for the nutritional needs of at least one of its resident commensals. In addition, the bacterium is able to actively determine the degree to which its nutritional needs are met. A key feature of the signal-transduction pathway is that l-fucose acts through FucR both as an inducer of fucRIAK, and as a corepressor of another locus, control of signal production.
It is reasonable that B. thetaiotaomicron would turn to epithelial fucosylated glycans as a source of energy in the highly competitive intestinal ecosystem. Strict anaerobes, including Bacteroides species, typically colonize the mammalian intestine during weaning (1). The intestinal ecosystem is densely populated at this point with the preweaning microflora, likely limiting the availability of simple sugars needed for continued growth of the bacterial population. The ability to instruct the host to present a source of utilizable carbohydrate should confer a competitive advantage under these conditions. Fucose is a logical choice. It is a normal and abundant component of many host intestinal cell-surface glycoconjugates (31, 32). Moreover, many of the core oligosaccharide structures to which fucose is commonly linked are constitutively synthesized, so presentation of fucose may only require induction of one or more host fucosyltransferases.
Our findings are compatible with the notion that the intestine provides a nutrient foundation that helps program assembly and preservation of a microbial society during the weaning transition. At early stages of colonization, the B. thetaiotaomicron population density is presumably inadequate to generate sufficient amounts of a concentration-dependent microbial signal to increase the availability of hydrolyzable fucose in its habitat. However, the presence of a preformed pool of Fucα1,2Gal-containing glycans in scattered ileal villi may provide sufficient hydrolyzable fucose to promote initiation and early progression of B. thetaiotaomicron colonization. Once a critical density of organisms is attained, the population can signal sustained production of an adequate supply of host-derived fucose. Signaling to the host only when fucose availability declines below a certain set point ensures that B. thetaiotaomicron will avoid expending valuable energy on signal production when fucose is abundant. By tightly coordinating presentation of host-derived fucose with the rate of fucose utilization, an overabundance of hydrolyzable epithelial fucose is also avoided. This may minimize the risk of encroachment by pathogens that use fucosylated glycans as receptors for their adhesins (33, 34).
We do not know whether the relationship between B. thetaiotaomicron and its host is symbiotic (both partners benefit) or commensal (neither is harmed). The host may benefit in several ways from filling its intestinal habitat with B. thetaiotaomicron at the weaning transition. During this time of great compositional flux in the microflora, B. thetaiotaomicron could help direct assembly of a stabilized microflora that provides colonization resistance. Additional benefits may include provision of critical metabolic activities and shaping of the underlying mucosal immune system. The paradigm illustrated by this host–microbial relationship may apply to other nutrient sources, other components of our microflora, and other mucosal surfaces.
Acknowledgments
We are indebted to Abigail Salyers and Nadja Shoemaker (University of Illinois, Champaign-Urbana) for their gifts of pLYL01, pGERM, pBT1, and strain Fu-4, plus their many helpful suggestions throughout the course of our studies. A.-K. Persson assisted with colonization of germ-free mice. Chi-Huey Wong (Scripps Institute) generously supplied l-fuculose and l-fuculose-1-phosphate. This work was supported by grants from the National Institutes of Health (DK30292, DK37960). L.V.H. was supported by postdoctoral fellowships from the Lucille P. Markey Foundation and the National Institutes of Health (DK07120).
ABBREVIATIONS
- Fuc
fucose
- Gal
galactose
- UEA-1
Ulex europaeus type I agglutinin
- P
postnatal day
- kb
kilobases
- GUS
β-glucuronidase
- cfu
colony-forming units
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF137263).
Present address: Department of Gastrointestinal Pharmacology, AstraZeneca, S-431 83 Mölndal, Sweden.
References
- 1.Savage D C. Annu Rev Microbiol. 1977;31:107–133. doi: 10.1146/annurev.mi.31.100177.000543. [DOI] [PubMed] [Google Scholar]
- 2.Van der Waaij D. Annu Rev Microbiol. 1989;43:69–87. doi: 10.1146/annurev.mi.43.100189.000441. [DOI] [PubMed] [Google Scholar]
- 3.Araneo B A, Cebra J J, Beuth J, Fuller R, Heidt P J, Midvedt T, Nord C E, Nieuwenhuis P, Manson W L, Pulverer G, et al. Zentralbl Bakteriol. 1996;283:431–465. doi: 10.1016/s0934-8840(96)80122-8. [DOI] [PubMed] [Google Scholar]
- 4.Falk P G, Hooper L V, Midtvedt T, Gordon J I. Microbiol Mol Biol Rev. 1998;62:1157–1170. doi: 10.1128/mmbr.62.4.1157-1170.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kühn R, Löhler J, Rennick D, Rajewski K, Müller W. Cell. 1993;75:263–274. doi: 10.1016/0092-8674(93)80068-p. [DOI] [PubMed] [Google Scholar]
- 6.Sadlack B, Merz H, Schorle H, Schimpl A, Feller A C, Horak I. Cell. 1993;75:253–261. doi: 10.1016/0092-8674(93)80067-o. [DOI] [PubMed] [Google Scholar]
- 7.Taurog J D, Richardson J A, Croft J T, Simmons W A, Zhou M, Fernandez-Sueiro J L, Balish E, Hammer R E. J Exp Med. 1994;180:2359–2364. doi: 10.1084/jem.180.6.2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hunter A E, Haynes A P, Russell N H. J Antimicrob Chemother. 1995;36,Suppl. B:119–133. doi: 10.1093/jac/36.suppl_b.119. [DOI] [PubMed] [Google Scholar]
- 9.Bry L, Falk P G, Midtvedt T, Gordon J I. Science. 1996;273:1380–1383. doi: 10.1126/science.273.5280.1380. [DOI] [PubMed] [Google Scholar]
- 10.Moore W E C, Holdeman L V. Appl Microbiol. 1974;27:961–979. doi: 10.1128/am.27.5.961-979.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ushijima T, Takahashi M, Tatewaki K, Ozaki Y. Microbiol Immunol. 1983;27:985–993. doi: 10.1111/j.1348-0421.1983.tb02929.x. [DOI] [PubMed] [Google Scholar]
- 12.Salyers A A, Pajeau M. Appl Environ Microbiol. 1989;55:2572–2578. doi: 10.1128/aem.55.10.2572-2578.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cheng Q, Hwa V, Salyers A A. J Bacteriol. 1992;174:7185–7193. doi: 10.1128/jb.174.22.7185-7193.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cooper A J, Shoemaker N B, Salyers A A. Antimicrob Agents Chemother. 1996;40:506–508. doi: 10.1128/aac.40.2.506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guiney D G, Yakobson E. Proc Natl Acad Sci USA. 1983;80:3595–3598. doi: 10.1073/pnas.80.12.3595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Simon R, Priefer U, Puhler A. Bio/Technology. 1983;1:784–791. [Google Scholar]
- 17.Shoemaker N B, Getty C, Gardner J F, Salyers A A. J Bacteriol. 1986;166:929–936. doi: 10.1128/jb.165.3.929-936.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McCarthy R E, Salyers A A. Appl Environ Microbiol. 1986;52:9–16. doi: 10.1128/aem.52.1.9-16.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tancula E, Feldhaus M J, Bedzyk L A, Salyers A A. J Bacteriol. 1992;174:5609–5616. doi: 10.1128/jb.174.17.5609-5616.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dische Z, Borenfreund E. J Biol Chem. 1951;192:583–587. [PubMed] [Google Scholar]
- 21.Aviles M, Abascal I, Martinez-Menarguez J A, Castells M T, Skalaban S R, Ballesta J, Alhadeff J A. Biochem J. 1996;318:821–831. doi: 10.1042/bj3180821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Anderson R L, Wood W A. J Biol Chem. 1962;237:1029–1033. [PubMed] [Google Scholar]
- 23.Ghalambor M A, Heath E C. J Biol Chem. 1962;237:2427–2433. [PubMed] [Google Scholar]
- 24.Horlacher R, Boos W. J Biol Chem. 1997;272:13026–13032. doi: 10.1074/jbc.272.20.13026. [DOI] [PubMed] [Google Scholar]
- 25.Herr, A. B. & Waksman, G. (1999) Methods Mol. Med., in press.
- 26.Brenowitz M, Senear D F, Shea M A, Ackers G K. Methods Enzymol. 1986;130:132–181. doi: 10.1016/0076-6879(86)30011-9. [DOI] [PubMed] [Google Scholar]
- 27.Lu Z, Lin E C C. Nucleic Acids Res. 1989;17:4883–4884. doi: 10.1093/nar/17.12.4883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Haydon D J, Guest J R. FEMS Microbiol Lett. 1991;63:291–295. doi: 10.1016/0378-1097(91)90101-f. [DOI] [PubMed] [Google Scholar]
- 29.Chen Y-M, Zhu Y, Lin E C C. Mol Gen Genet. 1987;210:331–337. doi: 10.1007/BF00325702. [DOI] [PubMed] [Google Scholar]
- 30.Bartkus J M, Mortlock R P. J Bacteriol. 1986;165:710–714. doi: 10.1128/jb.165.3.710-714.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Björk S, Breimer M E, Hansson G C, Karlsson K-A, Leffler H. J Biol Chem. 1987;262:6758–6765. [PubMed] [Google Scholar]
- 32.Finne J, Breimer M E, Hansson G C, Karlsson K-A, Leffler H, Vliegenthart J F, van Halbeek H. J Biol Chem. 1989;264:5720–5735. [PubMed] [Google Scholar]
- 33.Benitez J A, Spelbrink R G, Silva A, Phillips T E, Stanley C M, Boesman-Finkelstein M, Finkelstein R A. Infect Immun. 1997;65:3474–3477. doi: 10.1128/iai.65.8.3474-3477.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brassart D, Woltz A, Golliard M, Nesser J R. Infect Immun. 1991;59:1605–1613. doi: 10.1128/iai.59.5.1605-1613.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]